Abstract
Host–parasite evolutionary interactions are typically considered in a pairwise species framework. However, natural infections frequently involve multiple parasites. Altering parasite diversity alters ecological and evolutionary dynamics as parasites compete and hosts resist multiple infection. We investigated the effects of parasite diversity on host–parasite population dynamics and evolution using the pathogen Pseudomonas aeruginosa and five lytic bacteriophage parasites. To manipulate parasite diversity, bacterial populations were exposed for 24 hours to either phage monocultures or diverse communities containing up to five phages. Phage communities suppressed host populations more rapidly but also showed reduced phage density, likely due to interphage competition. The evolution of resistance allowed rapid bacterial recovery that was greater in magnitude with increases in phage diversity. We observed no difference in the extent of resistance with increased parasite diversity, but there was a profound impact on the specificity of resistance; specialized resistance evolved to monocultures through mutations in a diverse set of genes. In summary, we demonstrate that parasite diversity has rapid effects on host–parasite population dynamics and evolution by selecting for different resistance mutations and affecting the magnitude of bacterial suppression and recovery. Finally, we discuss the implications of phage diversity for their use as biological control agents.
Keywords: Bacteria, bacteriophage, community, experimental evolution, host/parasite
Evolution between hosts and their parasites is hypothesized to play a central role in several biological phenomena, including the maintenance of biodiversity (Haldane 1949) and virulence (Best et al. 2009). Host–parasite evolutionary interactions are largely studied in a pairwise species framework, both empirically across a wide range of experimental systems (e.g. King et al. 2009; Schulte et al. 2010; Thrall et al. 2012; Decaestecker et al. 2013; Duncan et al. 2013) and theoretically (Bell and Smith 1987; Kouyos et al. 2009). However, in nature, most host–parasite interactions will be driven within a network of ecological interactions. For example, coinfection of a host individual by a diversity of parasite species (Telfer et al. 2010) and competition between these parasites for host resources (Ulrich and Schmid‐Hempel 2012) are common. Evolutionary interactions between multiple parasites can also have important consequences for parasite transmission and virulence (Ben‐Ami et al. 2008; Balmer et al. 2009; Griffiths et al. 2011; Kwan and Ernst 2011). Introducing multiple parasite species into the standard host–parasite evolutionary framework may thus provide useful insights as to how evolution operates in natural host–parasite communities (Mitchell‐Olds and Bergelson 2000; Mostowy et al. 2010).
Here, we use experimental evolution to test for the effects of parasite diversity on host–parasite population dynamics and evolution using the bacterium Pseudomonas aeruginosa and its bacteriophages. P. aeruginosa is an opportunistic human pathogen. The strain used in these experiments (PAO1) was originally isolated from a burn wound infection from a patient in Melbourne (Holloway 1955). Contemporary strains of P. aeruginosa cause pneumonia in hospitals (Jones 2010) and in particular cause mortality in patients with cystic fibrosis (Behrends et al. 2013). Bacteriophages, the viral parasites of bacteria, are ideally suited to laboratory studies of evolution (Buckling et al. 2009). Bacteriophages are the most numerous form of life on the planet (Chibani‐Chennoufi et al. 2004). They are ecologically important as they are responsible for killing an estimated 50% of the total bacterial population on earth every 48 hours (Fischetti et al. 2006). Their ability to depress bacterial populations was recognized early in the 20th century (D'Herelle 1922) when they were used to treat many diseases including staphylococcal skin infections, dysentery, and the bubonic plague (Sulakvelidze et al. 2001).
Host resistance to bacteriophages can arise through mutation or reduced expression of the receptor molecule (Samson et al. 2013). Changes in gene expression can be coordinated by quorum sensing molecules that reduce the expression of a bacteriophage receptor in the whole bacterial population (Høyland‐Kroghsbo et al. 2013). Lysogenic bacteriophage are known to provide resistance against superinfection (Berngruber et al. 2010) and lytic phage may also be capable of excluding superinfection by their competitors. For example the pilus‐binding phage LUZ19 down regulates LPS genes in PAO1 upon infection (Lavigne et al. 2013), potentially reducing the ability of any LPS‐binding competitor phage from entering the same cell. However, resistance is not a binary trait: phages can infect and replicate within partially resistant cells without having a large impact on bacterial growth (Levin and Bull 2004).
Early investigations of phage‐bacteria evolution focused on single bacterial strains evolving with a single phage (Buckling and Rainey 2002; Mizoguchi et al. 2003; Brockhurst et al. 2007; Forde et al. 2008; Poullain et al. 2008). However, diverse phage communities exist in the human microbiome (Minot and Bryson 2013; Naidu et al. 2014; Liu et al. 2015), in the environment (Paul et al. 2002; Williamson et al. 2005), and are artificially assembled for phage therapy (Merabishvili et al. 2009; Chan et al. 2013). While one study showed that multiple phages can engage in within‐host competition giving an advantage to fast‐replicating phage (Refardt 2011), the presence of additional enemies (predators, parasites, or competitors) in the bacteria‐phage pairwise framework has been shown to affect host populations and their evolution. Adding a resistant bacterial competitor to a pairwise phage‐bacteria interaction resulted in greater suppression of bacterial growth, occasionally leading to host extinction, an observation not commonly made in bacterial monocultures (Harcombe and Bull 2005). Similarly, combining bacteriophage into three‐phage communities suppresses bacterial host growth more than the constituent phages can achieve alone (Turki et al. 2012). Multiple enemies may also shape the degree to which resistance is enemy specific in evolved host populations. For example combining the well‐studied P. fluorescens SBW25–Φ2 bacteriophage system with the predator, Tetrahymena thermophila, can select for divergent outcomes: either specialists, whereby hosts are good at resisting either the phage or predator (Friman and Buckling 2013)—or for a population of generalists that can resist both (Örmälä‐Odegrip et al. 2015). Phage communities have also been shown to select for the same resistance specialist strategies as the single phage that make up the community rather than a generalist phenotype (Koskella et al. 2012).
In our study, we manipulated phage diversity by mixing all combinations of a panel of five naturally isolated lytic phages with their bacterial host, P. aeruginosa PAO1, and allowed them to evolve in response to those phage combinations. We predicted that, relative to phage monocultures, increasingly diverse phage communities would (i) reduce phage density due to more intense phage competition in communities, (ii) be more effective at suppressing host growth, (iii) favor general, and perhaps costly, resistance mechanisms, and thereby (iv) reduce selection for resistance to phages. We found that diverse phage communities initially acted synergistically and reduced bacterial host population growth within the first three hours of interaction. This synergy between phages eventually transitioned into antagonism as phage community size reduced and host populations recovered most efficiently in diverse phage communities. Host populations rapidly evolved resistance across all phage community treatments, a finding confirmed by whole‐genome sequencing, with more generalized resistance mechanisms selected for by the five‐phage community and specific resistance to phage receptors evolving against phage monocultures. Finally, we found that resistance evolved as rapidly against a diversity of parasites as against a single parasite species, all within 24 hours. Our results indicate that diverse parasite communities shape host and parasite population dynamics and evolutionary responses. We further discuss the implications of our results for applying a diversity of phages as biological control agents.
Materials and Methods
STRAINS, STORAGE, AND CULTURE CONDITIONS
P. aeruginosa PAO1 was used with a panel of five, field‐isolated, lytic bacteriophage; PEV2, LMA2, 14/1, LUZ7, and LUZ19. These phages have previously been used separately in evolution experiments with P. aeruginosa, making them ideally suited for combination into experimental communities. LUZ19 is believed to rely on type IV pili for attachment (Chibeu et al. 2009; Lammens et al. 2009). The receptors for PEV2 and LUZ7 (classified as N4‐like phages) are unknown, however in order to attach to their hosts, these phages may rely on either LPS (Ceyssens et al. 2009b) or NrfA (Kiino and Rothman‐denes 1989), a membrane protein with homology to TadD (McPartland 2008) that is involved type IV pili formation (Bernard et al. 2009). LMA2 and 14/1 are related to PB1 (Ceyssens et al. 2009a) that relies on LPS for attachment (Kropinski et al. 1977). Isogenic bacterial stocks were prepared by inoculating 15 mL tubes containing 3 mL King's B (KB) medium (King et al. 1954) with twice streaked bacterial colonies from a frozen (–80°C) stock. The tubes were incubated in a MaxQ 8000 (Thermo Scientific, Waltham, MA, USA) at 225RPM for 24 hours at 37°C. In the absence of phage but otherwise the condition of this experiment, this would allow for 4–5 bacterial doublings before stationary phase but the lysis caused by phage makes accurate estimation of doubling time almost impossible. Previous work with these phages has shown that 24 hours is sufficient for considerable phenotypic change (Betts et al. 2013, 2014).
Bacteriophages were purified from bacteria by using 0.1 vol. chloroform and centrifugation at 13,000 g to kill and pellet the bacteria. Working bacteriophage stocks were prepared from frozen isogenic aliquots; a 1 μL loop was used to inoculate a 3 mL bacterial suspension (OD600 = 0.3). This was incubated for 24 hours at 225 RPM and 37°C before purification and filtration using a 0.22 μm syringe filter. The filtrates were diluted in M9 buffer to 1 × 108 plaque forming units (PFU) mL−1 and stored at +4°C for experimental use.
EXPERIMENTAL DESIGN
All combinations of the five phages (one, two, three, four, and five‐phage communities) were prepared such that the phages were at 5 × 105 PFU mL−1. Mixed phage communities were prepared to a final titre of 5 × 105 PFU mL−1 with each phage present at equal densities. These phages were then mixed with fresh, diluted overnight culture in KB. Each overnight culture was prepared from a separate colony and used to inoculate each replicate to a starting concentration of 5 × 107 colony forming units per mL (CFU mL−1). These mixtures were added to 96‐well plates, 200 μL per well, until there were six replicates of every possible phage combination and a phage‐free control (where the phage stock was substituted for M9 buffer). The six replicates were split evenly across two plates and incubated at 37°C for 24 hours.
MEASURING PHAGE GROWTH
Phage population size was measured at the end of the experiment for every treatment by counting plaque forming units (PFU), which provides a measure of viron density, using a soft agar overlay method (e.g., Buckling and Rainey 2002).
MEASURING BACTERIAL GROWTH
The experiment was conducted in a Synergy 2 plate reader (BioTek Instruments, Winooski, VT, USA). OD600 was measured every 2.5 minutes for the duration of the 24‐hour experiment. All populations were then frozen at –80°C. Endpoint CFU counts were also performed.
MEASURING RESISTANCE
Evolved populations were streaked out on KB agar and incubated overnight at 37°C in a static incubator to present single colonies. These colonies were picked, streaked out for a second time, and similarly incubated to present single colonies. This process ensured that each colony was both isogenic and free of phage. Populations evolved with a single phage type were sampled by taking eight colonies each from the 12 populations (two populations per phage type, one from each plate). Six colonies were sampled from each of the 20 populations that evolved with three‐phage types (two populations per phage combination, one from each plate). The populations evolved with five‐phage types and the phage‐free controls were sampled by taking six from all six replicates of each.
These bacterial colonies were separately incubated for 24 hours in 5 mL tubes containing 3 mL KB. Then 100 μL from each tube was mixed separately with 2.9 mL of soft (0.75%) KB agar and poured over the surface of a (1.5%) KB agar plate. Separate 4 μL droplets of purified phage were then applied to this soft agar lawn, allowed to dry, and incubated for 24 hours, subsequently assessed for zones of inhibition to indicate susceptibility or resistance. Zones with no visible inhibition were scored as resistant ( = 1), with some visible inhibition were scored as partially resistant ( = 0.5), and with complete inhibition were scored as susceptible ( = 0). These data were then analyzed as the proportion of clones that were resistant or susceptible to a given phage isolate.
WHOLE GENOME SEQUENCING OF EVOLVED BACTERIA
A subsample of the twice‐streaked bacterial colonies scored for resistance were grown in 3 ml KB in a 5‐ml tube overnight at 37°C to provide the starting material for the extraction process. DNA was extracted from 1 ml of culture using a Dneasy Blood and Tissue kit (Qiagen, Inc., Chatworth, California, USA), quantified using the QuantiFluor dsDNA system (Promega, Madison, WI, USA) and the salt ratio verified by Nano Drop (Thermo Scientific, Waltham, MA, USA). Library preparation and sequencing was performed at the Wellcome Trust Centre for Human Genetics (Oxford, UK) on an Illumina MiSeq in a 2 × 300 paired end reaction using v3 reagents.
In total, an ancestral clone and 17 evolved clones were sequenced: six phage‐free evolved clones (one from each replicate) from 24 hours to control for possible laboratory adaptation, one from each replicate of the five‐phage treatment and one from the first replicate of each one‐phage population. The sequenced clones were randomly selected from those also scored for resistance. Full details of the bioinformatics are provided in the supplementary materials.
STATISTICAL ANALYSIS
Analysis and graphing was performed using JMP 10.0 (SAS Institute Inc. 2012). All data were analyzed using ANOVA without transformation. All post‐hoc contrasts were t‐tests with a significance threshold of 0.05. Between‐treatment final phage population size significance was analyzed by ANOVA. The OD600 data were analyzed by calculating the area under the curve using the trapz function within pracma in R 3.1.1 (R Core Team 2013), and then ANOVA where the same model was used on two subsets of the data; 0–3 hours and 21–24 hours. These time windows captured the initial suppression of host growth and the recovery of host populations where growth of resistant mutants had exceeded the maximum reached before the populations were suppressed. For the PFU data and the OD600 data, the number of phage types in the community was treated as a categorical explanatory variable. The resistance of bacteria evolved with single, three, and the five‐phage communities to the five ancestral phages were compared in two separate ANOVA tests. The resistance of evolved bacteria was compared to their sympatric ancestral phage, then to all five phage types, both allopatric and sympatric.
Results
EFFECTS OF PHAGE DIVERSITY ON BACTERIAL GROWTH
To measure the impact of phage diversity on bacterial population dynamics, we measured the optical density of bacterial populations that were exposed to phage relative to phage‐free control populations (0–3 h: F1,191 = 18.4 P < 0.01; 21–24 h: F1,191 = 32.2 P < 0.01). There was a significant effect of the number of phages both in early and late growth (0–3 h: F4,191 = 6.6 P < 0.01; 21–24 h: F4,191 = 8.3 P < 0.01) however the relationship between the number of phage and bacterial growth differed between the two time points (Fig. 1). In early growth (0–3 h), higher phage diversity was associated with a greater depression of bacterial population growth. Post‐hoc contrasts revealed that bacterial growth was suppressed significantly more by the four‐ and five‐phage treatments, which were not significantly different from one another. The three‐phage treatment was the next lowest, but the one and two‐phage treatments did not significantly differ. Conversely, in late growth (21–24 h), lower phage diversity was associated with a greater depression of bacterial population growth. The one‐phage treatment reduced bacterial growth more significantly than all other treatments, followed by the two‐phage treatment. The three‐, four‐, and five‐phage treatments did not significantly differ from one another. To confirm that optical density provides an appropriate measure of population density, we performed endpoint CFU counts, which corroborated the OD data (Fig. S1).
Figure 1.
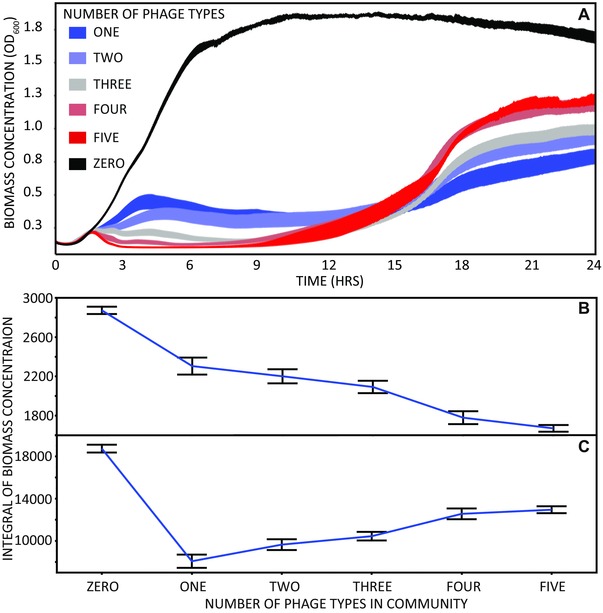
(A) Shows the bacterial biomass concentration (OD600) over time for the different phage containing treatments and the phage‐free control. The areas under the curves for OD600 during the first 3 (B) and last 3 (C) hours of growth split by the number of phage types in the community. The area under the curve reflects how much bacterial biomass was present within the time window. Error bars show 1 standard error.
EFFECTS OF PHAGE DIVERSITY ON PHAGE GROWTH
Although increasing phage diversity has the potential to increase bacterial mortality rates, increasing phage diversity also introduces the possibility for competition between phage types. To test for this effect, we measured the final phage titres (PFU ml−1) after 24 hours incubation (Fig. 2). There was a significant effect of phage community diversity on phage titer (F4,180 = 4.17, P < 0.01). Post‐hoc contrasts revealed that the single‐phage treatment had the highest mean titer, while all other phage communities were lower and not significantly different from each other.
Figure 2.
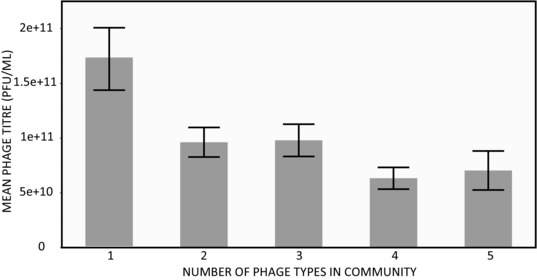
Shows the mean phage titers measured after 24 hours where five bacteriophages were cultured in every possible combination up to a five‐phage community. There were six replicates of each possible combination split across two 96‐well plates. Error bars show one standard error.
EFFECTS ON BACTERIAL RESISTANCE EVOLUTION
Predation by phage generates selection for phage resistance in bacterial populations; increasing phage diversity is predicted to alter both the rate and mechanisms of resistance evolution. To test for resistance evolution, we measured the ability of the five ancestral phage to infect bacterial clones that were sampled from the end of our experiment. We found phenotypic resistance across all treatments and not the phage‐free control.
Firstly, we hypothesized that increasing phage diversity should decrease the rate of resistance evolution to sympatric phage (Fig. 3). There was a significant effect of phage community diversity (single‐, three‐, or five‐phage types) on mean resistance to sympatric phage (F2,605 = 80.2.58 P < 0.001). However, host resistance to the five ancestral phage types differed significantly (F4,605 = 73.8 P < 0.001) and resistance to the ancestral phages differed depending on treatment(F8,605 = 12.65 P < 0.001).
Figure 3.
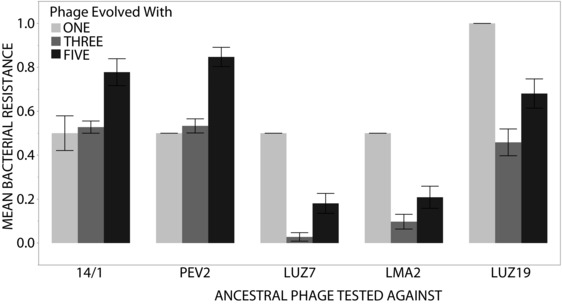
Shows the mean bacterial resistance to the five ancestral phage types for those that had evolved with one, three and five phage types, Only sympatric data are included, that is only bacteria evolved with PEV2 were scored against PEV2. Error bars show 1 standard error.
The post‐hoc comparisons (t‐tests with a significance threshold of 0.05) revealed groupings among the five phage types. The degree of resistance in bacteria evolved against 14/1 and PEV2 differed from all others, but were not significantly different from each other. A similar pattern was revealed for bacterial populations evolved against LUZ7 and LMA2. In contrast, the degree of resistance in bacterial populations evolved against LUZ19 was significantly different from populations evolved against other phages. Within each phage type, the post‐hoc contrasts revealed a significant difference between evolved bacterial resistance among the phage communities, except for those evolved against PEV2 and 14/1 where the one and three phage populations did not differ.
Secondly, we hypothesized that parasite diversity would select for more generalized resistance mechanisms. We compared the resistance of the evolved bacteria from the single‐phage treatments to all five ancestral phages (Fig. 4). Here evolved bacterial isolates from the single, three, and five‐phage populations were exposed to all five phage types. The number of phage types with which bacteria had evolved significantly affected the mean resistance against the five phage types (F2,1164 = 52.6 P < 0.001). Nested within the single‐phage populations, the different phage types evolved with selected for different levels of resistance to allopatric phage (F13,1164 = 4.48 P < 0.001). Post‐hoc contrasts revealed similar groupings among the single‐phage populations. Populations evolved with 14/1 and PEV2 had selected for the highest resistance against the five ancestral phage, and were significantly different from all others, but not from each other. Similarly, LUZ19, LMA2, and LUZ7 had selected for the lowest resistance to the five ancestral phage, but did not differ from each other
Figure 4.
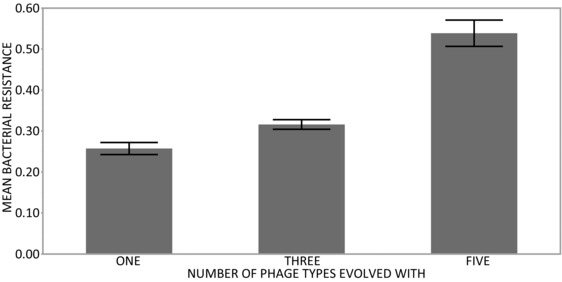
Shows the resistance of evolved bacteria averaged across the five different ancestral phage types against which they were challenged. The data are shown separately for the bacterial populations evolved with one, three and five phages in a single community. Error bars shows 1 standard error.
To confirm that the observed changes in resistance were due to evolution and not phenotypic plasticity, we sequenced bacterial clones to look for evidence of genomic changes. All of the evolved clones possessed a single chromosomal mutation relative to the ancestor revealing that genomic evolution had occurred within 24 hours. No mutations were observed in the clones sequenced from the phage‐free control populations. We observed parallel evolution in genes known to play roles in phage/bacteria interactions (e.g. LPS production). The details are summarized in Table 1.
Table 1.
Shows the mutations detected in the evolved bacteria
| MUTATION INFORMATION | PHAGE RESISTANCE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EVOLUTION TREATMENT | Gene ID | Gene | Function | Mutation | PEV2 | LUZ7 | LUZ19 | LMA2 | 14/1 | |
| ONE‐PHAGE | PEV2 | PA1959‐PA2216 | – | 257 genes | mgd | 0.5 | 0.5 | 0 | 0.5 | 0.5 |
| 14/1 | PA3154 | wzy | LPS | G94* | 0.5 | 0 | 0 | 0.5 | 0.5 | |
| LUZ7 | PA2911 | – | TonB receptor | L20P | 0 | 0.5 | 0 | 0.5 | 0 | |
| LMA2 | PA2911 | – | TonB receptor | A26fs | 0 | 0.5 | 0 | 0.5 | 0 | |
| LUZ19 | PA3805 | pilF | Type IV pili | R169_L170insRL | 0 | 0 | 1 | 0 | 0 | |
| FIVE‐PHAGE | PA3154 | wzy | LPS | W207fs | 1 | 0 | 1 | 0 | 1 | |
| PA3154 | wzy | LPS | A43Lfs | 1 | 0 | 1 | 0 | 1 | ||
| PA3154 | wzy | LPS | A43Lfs | 1 | 0 | 1 | 0 | 1 | ||
| PA0938 | wzz2 | LPS | T263M | 1 | 0 | 1 | 0 | 1 | ||
| PA1893‐PA2206 | – | 314 genes | mgd | 0.5 | 0.5 | 0 | 0.5 | 0.5 | ||
| PA0763 | mucA | Alginate | Q192fs | 1 | 1 | 0.5 | 1 | 0.5 | ||
Each row shows the result for a single evolved clone. No mutations were detected in the phage‐free lines but a single mutation was detected in every phage‐evolved clones, one clone for each single‐phage treatment and 6 clones from the five‐phage treatment (fs–frameshift, mgd–multi‐gene deletion, *–stop codon). The resistance of sequenced clones to the five ancestral phage types are also given. Clones were scored as either completely resistant ( = 1), partially resistant ( = 0.5), or completely susceptible ( = 0).
Discussion
Here, we show that phage diversity drives significant changes in bacterial host ecology and evolution. While bacterial populations are initially depressed in size when exposed to a diversity of phages, they recover rapidly thereafter within a few hours. Recovery is likely due to a combination of resistance evolution and competition‐induced lower phage population sizes. The greater the phage diversity in the community, the more rapidly the hosts were initially suppressed suggesting an additive effect of phages on the host population. For example, mixed phage infections can result in increased infection success if one phage affects the host in such a way that it is then more susceptible to a second phage (Schmerer et al. 2014).
Conversely, after less than 24 hours this synergy gave way to antagonism between phages as phage diversity resulted in a lower phage population size, enhancing bacterial recovery. Competition between phages could take place both between and within hosts. Phages of the same type could compete over attachment sites on the host cell surface (Lenski 1988) or modify host gene expression to prevent other phages from attaching to the same cell (Pedruzzi et al. 1998). Additionally, when there is multiple‐infection, host death can occur prematurely, without phage production, as a result of damage caused by the sheer number of penetrations (Abedon 2011) thereby reducing both bacterial and phage population sizes. On the other hand, competition within the host could arise when phages differ in lysis time (Refardt 2011), and the slower phage fails to complete its life cycle. Such competition is important for bacteriophage ecology and can drive inter‐phage coevolutionary arms races (Wichman et al. 2005).
The increased recovery of bacterial populations evolving with the five‐phage communities could also be due to early bottlenecking. Strong initial selection for resistant clones left them to grow with little competition, consequently by 24 hours they had reached a higher biomass concentration than the other treatments. Interestingly, the greater bacterial recovery observed in bacterial populations evolving with the five‐phage communities relative to the single‐phage populations suggests that despite the diversity of parasites, the evolution of resistance was not delayed. A diversity of phages instead applied a stronger selection pressure and allowed for greater bacterial recovery. Parasite competition affects the dynamics of infection in other systems (Hughes et al. 2004; Ulrich and Schmid‐Hempel 2012) but interactions between parasites can also lead to more virulent infections, for example in the case of HIV enhancing the TB epidemic (Kwan and Ernst 2011).
Our results are consistent with the hypothesis that hosts evolving with a diverse parasite assemblage are more likely to evolve generalized resistance mechanisms, while pairwise evolution selects for specific resistance (Thrall et al. 2007). For example, a single mutation in pilF (involved in type IV pili formation) occurred in the bacterial population that evolved with LUZ19 (which is type IV pili‐dependent phage). This mutation conferred complete resistance to LUZ19 and did not occur in any of the sampled bacterial populations subjected to attack by five‐phage communities (Table 1). However, a single mutation in mucA was observed that seemed to confer partial resistance to multiple phage types. MucA represses alginate production and its inactivation is associated with conversion to mucoidy (Boucher et al. 1997). Mucoid phenotypes have been shown to evolve in response to bacteriophage selection pressure in other systems (Vogwill et al. 2011; Tazzyman and Hall 2014), but such a mutation was not present here in the single‐phage treatments. These data will enable the formation of more developed hypotheses in our future work.
When evolved bacterial populations from all treatments were tested against all five ancestral phage, we observed the greatest breadth of resistance in those populations that had evolved with all five‐phage types. This result suggests that the greater the phage diversity, the more bacterial populations are likely to evolve a broad resistance phenotype. When we focus on sympatric bacteria‐phage comparisons only, we observe a strong phage‐dependent effect such that for PEV2 and 14/1 combination into five‐phage communities increases the amount of resistance evolved to these phage whereas the amount of resistance to LUZ7, LMA2, and LUZ19 decreases when they are combined with other phage. Interestingly, for all phage the level of evolved resistance was always lower for the three‐phage communities than the five‐phage communities, suggesting that there are important interactions between phages when in communities, perhaps in the form of competition for hosts (as suggested by the observed reduction in phage population size in communities) and evolution of cross‐resistance (Fig. S2).
Future work will need to establish the effects of combining phage types on longer term host–parasite coevolutionary dynamics. Here, we show widespread evolution of resistance, but our data also indicate that a degree of partial susceptibility remained. Thus, it is possible that some individuals at lower frequency remained completely susceptible (Schrag and Mittler 1996; Bohannan and Lenski 2000), providing the host reservoirs necessary for coevolution with phages. All the phages herein have been shown to exhibit the ability to coevolve with P. aeruginosa (Betts et al. 2014), and the nature of host mutations observed has interesting implications for potential long‐term coevolutionary dynamics with bacteriophage communities. When coevolution was restricted to pairwise interactions in this study, mutations occurred at different loci depending on the phage type. For instance, two out of five clones showed LPS mutations with the remaining three in either type IV pili‐ or TonB‐dependent receptors. In contrast, five out of six clones from bacteria populations evolved with the five‐phage communities had mutations in LPS, and one additional mucoidy‐associated mutation. When different LPS phages were combined, they can coselect for the same resistance mechanism. The LPS phages PEV2 and 14/1 both selected for genes that effect LPS production and the five‐phage cocktail selected for similar mutations. Alternatively, there may be no genetic correlation in resistance to LPS phage and type IV pili; specific resistance to LUZ19 (type IV pili dependent) via a single mutation in a pili gene confers no protection to any of the other phage and so combining them might allow for diffuse coevolution. A mixture of diffuse and pairwise coevolution may take place when host populations coevolve with a diverse parasite community (Brockhurst et al. 2014).
There are implications in our results for the application of a “cocktail” of phage types to treat bacterial infections. In phage therapy, phages may be administered as monocultures (Skurnik and Strauch 2006) or in multiphage communities (Chan et al. 2013). The single phage has the advantage of simpler kinetics and less regulatory burden (Lu and Koeris 2011), but it is limited by its host range (Hyman and Abedon 2010) and may only be effective against a narrow range of pathogen genotypes (Scanlan et al. 2013). However, the phage cocktail community can have a broader host range (Gu et al. 2012) and may delay the evolution of host resistance (Barbosa et al. 2013; Fischer et al. 2013). In addition to phage diversity, the phage titers are also important (Nilsson 2014) as phage therapy is more effective with greater multiplicity of infection (Beeton et al. 2015). Although our experiments were conducted in vitro and with a strain that is genetically divergent from those of current clinical relevance, they are novel in exposing bacteria to all phage types separately and simultaneously, for a clinically relevant period of time and for considering the effects on phage productivity. We observed that diverse phage communities did not delay the evolution of host resistance and eventually resulted in reduced phage titers. Our data thus suggest that boosting phage diversity in a cocktail might not automatically increase the efficacy of phage therapy.
In summary, we demonstrate that diverse parasite communities comprised of multiple phage types more rapidly depress host growth than the constituent phages alone and that increasing phage diversity did not delay the evolution of resistance. Our results therefore support the hypothesis that diverse parasite communities can impose stronger selection on host populations than single parasite species. Host resistance evolution also depends on the biology of individual parasite species in the community. Overall, our results demonstrate the potential for interesting ecological and evolutionary dynamics in bacteria‐phage communities beyond lab investigations of pairwise phage–host interactions. Exploring the effect of parasite community diversity will therefore be a significant step toward creating host–parasite coevolutionary mosaics (King et al. 2009; Thrall et al. 2012) more representative of nature and testing hypotheses on host–parasite coevolution in a community context.
Associate Editor: A.L. Laine
Handling Editor: J. Conner
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Figure S1: This figure shows the final bacterial population density measured by counting CFU per ML.
Figure S2: Demonstrates the evolution of cross resistance to multiple phage following exposure to a single phage.
ACKNOWLEDGMENTS
A.B. was supported by funding from the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/J014427/1]. R.C.M. and D.R.G. were supported by funding from the Royal Society. K.C.K. was supported by a Leverhulme Research Project Grant. We thank the High‐Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics funded by Wellcome Trust grant reference 090532/Z/09/Z and Medical Research Council Hub grant G0900747 91070 for generation of the high‐throughput sequencing data.
LITERATURE CITED
- Abedon, S. T. 2011. Lysis from without. Bacteriophage 1:46–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer, O. , Stearns S. C., Schötzau A., and Brun R.. 2009. Intraspecific competition between co‐infecting parasite strains enhances host survival in African trypanosomes. Ecology 90:3367–3378. [DOI] [PubMed] [Google Scholar]
- Barbosa, C. , Venail P., Holguin A. V., and Vives M. J.. 2013. Co‐evolutionary dynamics of the bacteria Vibrio sp. CV1 and phages V1G, V1P1, and V1P2: implications for phage therapy. Microb. Ecol. 66:897–905. [DOI] [PubMed] [Google Scholar]
- Beeton, M. L. , Alves D. R., Enright M. C., and Jenkins A. T. A.. 2015. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 46:196–200. [DOI] [PubMed] [Google Scholar]
- Behrends, V. , Ryall B., Zlosnik J. E. A., Speert D. P., Bundy J. G., and Williams H. D.. 2013. Metabolic adaptations of Pseudomonas aeruginosa during cystic fibrosis chronic lung infections. Environ. Microbiol. 15:398–408. [DOI] [PubMed] [Google Scholar]
- Bell, G. , and Smith J. M.. 1987. Short‐term selection for recombination among mutually antagonistic species. Nature 328:66–68. [Google Scholar]
- Ben‐Ami, F. , Mouton L., and Ebert D.. 2008. The effects of multiple infections on the expression and evolution of virulence in a Daphnia‐endoparasite system. Evolution 62:1700–1711. [DOI] [PubMed] [Google Scholar]
- Bernard, C. S. , Bordi C., Termine E., Filloux A., and de Bentzmann S.. 2009. Organization and PprB‐dependent control of the Pseudomonas aeruginosa tad Locus, involved in Flp pilus biology. J. Bacteriol. 191:1961–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berngruber, T. W. , Weissing F. J., and Gandon S.. 2010. Inhibition of superinfection and the evolution of viral latency. J. Virol. 84:10200–10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, A. , White A., and Boots M.. 2009. The implications of coevolutionary dynamics to host‐parasite interactions. Am. Nat. 173:779–791. [DOI] [PubMed] [Google Scholar]
- Betts, A. , Kaltz O., and Hochberg M. E.. 2014. Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proc. Natl. Acad. Sci. USA 111:11109–11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts, A. , Vasse M., Kaltz O., and Hochberg M. E.. 2013. Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol. Appl. 6:1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannan, B. J. M. , and Lenski R. E.. 2000. The relative importance of competition and predation varies with productivity in a model community. Am. Nat. 156:329–340. [DOI] [PubMed] [Google Scholar]
- Boucher, J. C. , Yu H., Mudd M. H., and Deretic V.. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst, M. A. , Chapman T., King K. C., Mank J. E., Paterson S., and Hurst G. D. D.. 2014. Running with the Red Queen: the role of biotic conflicts in evolution Darwin review. Proc. R. Soc. B Biol. Sci 281:20141382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst, M. A. , Morgan A. D., Fenton A., and Buckling A.. 2007. Experimental coevolution with bacteria and phage The Pseudomonas fluorescens—Phi 2 model system. Infect. Genet. Evol. 7:547–552. [DOI] [PubMed] [Google Scholar]
- Buckling, A. , Craig Maclean R., Brockhurst M. A., and Colegrave N.. 2009. The Beagle in a bottle. Nature 457:824–829. [DOI] [PubMed] [Google Scholar]
- Buckling, A. , and Rainey P.. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. B Biol. Sci. 269:931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyssens, P.‐J. , Miroshnikov K., Mattheus W., Krylov V., Robben J., Noben J.‐P., Vanderschraeghe S., Sykilinda N., Kropinski A. M., Volckaert G., et al. 2009a. Comparative analysis of the widespread and conserved PB1‐like viruses infecting Pseudomonas aeruginosa . Environ. Microbiol. 11:2874–2883. [DOI] [PubMed] [Google Scholar]
- Ceyssens, P.‐J. , Noben J.‐P., Ackermann H.‐W., Verhaegen J., De Vos D., Pirnay J.‐P., Merabishvili M., Vaneechoutte M., Chibeu A., Volckaert G., et al. 2009b. Survey of Pseudomonas aeruginosa and its phages: de novo peptide sequencing as a novel tool to assess the diversity of worldwide collected viruses. Environ. Microbiol. 11:1303–1313. [DOI] [PubMed] [Google Scholar]
- Chan, B. K. , Abedon S. T., and Loc‐Carrillo C.. 2013. Phage cocktails and the future of phage therapy. Future Microbiol. 8:769–783. [DOI] [PubMed] [Google Scholar]
- Chibani‐Chennoufi, S. , Bruttin A., Dillmann M. L., and Brussow H.. 2004. Phage‐host interaction: an ecological perspective. J. Bacteriol. 186:3677–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibeu, A. , Ceyssens P.‐J., Hertveldt K., Volckaert G., Cornelis P., Matthijs S., and Lavigne R.. 2009. The adsorption of Pseudomonas aeruginosa bacteriophage phiKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol. Lett. 296:210–218. [DOI] [PubMed] [Google Scholar]
- D'Herelle, F. 1922. The bacteriophage its role in immunity. Williams & Wilkins, Baltimore, USA. [Google Scholar]
- Decaestecker, E. , De Gersem H., Michalakis Y., and Raeymaekers J.. 2013. Damped long‐term host‐parasite Red Queen coevolutionary dynamics: a reflection of dilution effects? Ecol. Lett. 16:1455–1462. [DOI] [PubMed] [Google Scholar]
- Duncan, A. B. , Gonzalez A., and Kaltz O.. 2013. Stochastic environmental fluctuations drive epidemiology in experimental host–parasite metapopulations. Proc. R. Soc. B Biol. Sci. 280:20131747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, S. , Kittler S., Klein G., and Glünder G.. 2013. Impact of a single phage and a phage cocktail application in broilers on reduction of Campylobacter jejuni and development of resistance. PLoS One 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti, V. A , Nelson D., and Schuch R.. 2006. Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24:1508–1511. [DOI] [PubMed] [Google Scholar]
- Forde, S. E. , Thompson J. N., Holt R. D., and Bohannan B. J. M.. 2008. Coevolution drives temporal changes in fitness and diversity across environments in a bacteria‐bacteriophage interaction. Evolution 62:1830–1839. [DOI] [PubMed] [Google Scholar]
- Friman, V.‐P. , and Buckling A.. 2013. Effects of predation on real‐time host‐parasite coevolutionary dynamics. Ecol. Lett. 16:39–46. [DOI] [PubMed] [Google Scholar]
- Griffiths, E. C. , Pedersen A. B., Fenton A., and Petchey O. L.. 2011. The nature and consequences of coinfection in humans. J. Infect. 63:200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J. , Liu X., Li Y., Han W., Lei L., Yang Y., Zhao H., Gao Y., Song J., Lu R., Sun C., and Feng X.. 2012. A method for generation phage cocktail with great therapeutic potential. PLoS One 7:e31698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S. 1949. Disease and evolution. Ric Sci Suppl. A 19:68–76. [Google Scholar]
- Harcombe, W. , and Bull J.. 2005. Impact of phages on two‐species bacterial communities. Appl. Environ. Microbiol. 71:5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa . J. Gen. Microbiol. 13:572–581. [DOI] [PubMed] [Google Scholar]
- Høyland‐Kroghsbo, N. , Mærkedahl R., and Svenningsen S.. 2013. A quorum‐sensing‐induced bacteriophage defense mechanism. MBio 4(1):e00362–e00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, W. O. H. , Petersen K. S., Ugelvig L. V, Pedersen D., Thomsen L., Poulsen M., and Boomsma J. J.. 2004. Density‐dependence and within‐host competition in a semelparous parasite of leaf‐cutting ants. BMC Evol. Biol. 4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman P., and Abedon S. T.. 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70:217–248 [DOI] [PubMed] [Google Scholar]
- Jones, R. 2010. Microbial etiologies of hospital‐acquired bacterial pneumonia and ventilator‐associated bacterial pneumonia. Clin. Infect. Dis. 51(Suppl 1):S81–S87. [DOI] [PubMed] [Google Scholar]
- Kiino, D. R. , and Rothman‐denes L. B.. 1989. Genetic analysis of bacteriophage N4 adsorption. J. Bacteriol. 171:4595–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E. , Ward M., and Raney D.. 1954. 2 simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307. [PubMed] [Google Scholar]
- King, K. C. , Delph L. F., Jokela J., and Lively C. M.. 2009. The geographic mosaic of sex and the Red Queen. Curr. Biol. 19:1438–1441. [DOI] [PubMed] [Google Scholar]
- Koskella, B. , Lin D. M., Buckling A., and Thompson J. N.. 2012. The costs of evolving resistance in heterogeneous parasite environments. Proc. R. Soc. B Biol. Sci. 279:1896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos, R. D. , Salathé M., Otto S. P., and Bonhoeffer S.. 2009. The role of epistasis on the evolution of recombination in host‐parasite coevolution. Theor. Popul. Biol. 75:1–13. [DOI] [PubMed] [Google Scholar]
- Kropinski, A. M. , Chan L., Jarrell K., and Milazzo F. H.. 1977. The nature of Pseudomonas aeruginosa strain PAO bacteriophage receptors. Can. J. Microbiol. 23:653–658. [DOI] [PubMed] [Google Scholar]
- Kwan, C. , and Ernst J. D.. 2011. HIV and tuberculosis: a deadly human syndemic. Clin. Microbiol. Rev. 24:351–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens, E. , Ceyssens P.‐J., Voet M., Hertveldt K., Lavigne R., and Volckaert G.. 2009. Representational difference analysis (RDA) of bacteriophage genomes. J. Microbiol. Methods 77:207–213. [DOI] [PubMed] [Google Scholar]
- Lavigne, R. , Lecoutere E., Wagemans J., Cenens W., Aertsen A., Schoofs L., Landuyt B., Paeshuyse J., Scheer M., Schobert M., et al. 2013. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. MBio 4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R. E. 1988. Dynamics of interactions between bacteria and virulent bacteriophage. Adv. Microb. Ecol. 10:1–44. [Google Scholar]
- Levin, B. R. , and Bull J. J.. 2004. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2:166–173. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Yan R., Zhong Q., Ngo S., Bangayan N. J., Nguyen L., Lui T., Liu M., Erfe M. C., Craft N., et al. 2015. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 9:2078–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, T. K. , and Koeris M. S.. 2011. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 14:524–531. [DOI] [PubMed] [Google Scholar]
- McPartland, J. A. 2008. The tail sheath of bacteriophage N4 is required for adsorption to the host. 1st ed. Proquest, Michigan, USA. [Google Scholar]
- Merabishvili, M. , Pirnay J.‐P., Verbeken G., Chanishvili N., Tediashvili M., Lashkhi N., Glonti T., Krylov V., Mast J., Van Parys L., et al. 2009. Quality‐controlled small‐scale production of a well‐defined bacteriophage cocktail for use in human clinical trials. PLoS One 4:e4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot, S. , and Bryson A.. 2013. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA 110:12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell‐Olds, T. , and Bergelson J.. 2000. Biotic interactions: genomics and coevolution. Curr. Opin. Plant Biol. 3:273–277. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, K. , Morita M., Fischer C. R., Yoichi M., Tanji Y., and Unno H.. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157: H7 in continuous culture. Appl. Environ. Microbiol. 69:170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy, R. , Salathe M., Kouyos R. D., and Bonhoeffer S.. 2010. On the evolution of sexual reproduction in hosts coevolving with mutliple parasites. Evolution 64:1644–1656. [DOI] [PubMed] [Google Scholar]
- Naidu, M. , Robles‐Sikisaka R., Abeles S. R., Boehm T. K., and Pride D. T.. 2014. Characterization of bacteriophage communities and CRISPR profiles from dental plaque. BMC Microbiol. 14:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, A. S. 2014. Phage therapy—constraints and possibilities. Ups. J. Med. Sci. 119:192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örmälä‐Odegrip, A.‐M. , Ojala V., Hiltunen T., Zhang J., Bamford J. K., and Laakso J.. 2015. Protist predation can select for bacteria with lowered susceptibility to infection by lytic phages. BMC Evol. Biol. 15:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, J. H. , Sullivan M. B., Segall A. M., and Rohwer F.. 2002. Marine phage genomics. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133:463–476. [DOI] [PubMed] [Google Scholar]
- Pedruzzi, I. , Rosenbusch J. P., and Locher K. P.. 1998. Inactivation in vitro of the Escherichia coli outer membrane protein FhuA by a phage T5‐encoded lipoprotein. FEMS Microbiol. Lett. 168:119–125. [DOI] [PubMed] [Google Scholar]
- Poullain, V. , Gandon S., Brockhurst M. A., Buckling A., and Hochberg M. E.. 2008. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 62:1–11. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3‐900051‐07‐0, URL http://www.R‐project.org/
- Refardt, D. 2011. Within‐host competition determines reproductive success of temperate bacteriophages. ISME J. 5:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, J. E. , Magadán A. H., Sabri M., and Moineau S.. 2013. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11:675–687. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc . 2012. New Features in JMP 10. Cary, NC.
- Scanlan, P. D. , Hall A. R., Burlinson P., Preston G., and Buckling A.. 2013. No effect of host‐parasite co‐evolution on host range expansion. J. Evol. Biol. 26:205–209. [DOI] [PubMed] [Google Scholar]
- Schmerer, M. , Molineux I. J., and Bull J. J.. 2014. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2:e590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag, S. J. , and Mittler J. E.. 1996. Host‐parasite coexistence: the role of spatial refuges in stabilizing bacteria‐phage interactions. Am. Nat. 148:348–377. [Google Scholar]
- Schulte, R. D. , Makus C., Hasert B., Michiels N. K., and Schulenburg H.. 2010. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl. Acad. Sci. USA 107:7359–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik, M. , and Strauch E.. 2006. Phage therapy: facts and fiction. Int. J. Med. Microbiol. 296:5–14. [DOI] [PubMed] [Google Scholar]
- Sulakvelidze, A. , Alavidze Z., and Morris J.. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazzyman, S. J. , and Hall A. R.. 2014. Lytic phages obscure the cost of antibiotic resistance in Escherichia coli . ISME J. 9:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, S. , Lambin X., Birtles R., Beldomenico P., Burthe S., Paterson S., and Begon M.. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330:243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall, P. H. , Hochberg M. E., Burdon J. J., and Bever J. D.. 2007. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol. Evol. 22:120–126. [DOI] [PubMed] [Google Scholar]
- Thrall, P. H. , Laine A.‐L., Ravensdale M., Nemri A., Dodds P. N., Barrett L. G., and Burdon J. J.. 2012. Rapid genetic change underpins antagonistic coevolution in a natural host‐pathogen metapopulation. Ecol. Lett. 15:425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turki, Y. , Ouzari H., Mehri I., Ben Ammar A., and Hassen A.. 2012. Evaluation of a cocktail of three bacteriophages for the biocontrol of Salmonella of wastewater. Food Res. Int. 45:1099–1105. [Google Scholar]
- Ulrich, Y. , and Schmid‐Hempel P.. 2012. Host modulation of parasite competition in multiple infections. Proc. R. Soc. B Biol. Sci. 279:2982–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill, T. , Fenton A., and Brockhurst M. A.. 2011. Coevolving parasites enhance the diversity‐decreasing effect of dispersal. Biol. Lett. 7:578–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman, H. A. , Millstein J., and Bull J. J.. 2005. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics 170:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, K. E. , Radosevich M., and Wommack K. E.. 2005. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71:3119–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Figure S1: This figure shows the final bacterial population density measured by counting CFU per ML.
Figure S2: Demonstrates the evolution of cross resistance to multiple phage following exposure to a single phage.


