Abstract
Background
Phytophthora root and stem rot is one of the most yield-limiting diseases of soybean [Glycine max (L.) Merr], caused by the oomycete Phytophthora sojae. Partial resistance is controlled by several genes and, compared to single gene (Rps gene) resistance to P. sojae, places less selection pressure on P. sojae populations. Thus, partial resistance provides a more durable resistance against the pathogen. In previous work, plant introductions (PIs) originating from the Republic of Korea (S. Korea) have shown to be excellent sources for high levels of partial resistance against P. sojae.
Results
Resistance to two highly virulent P. sojae isolates was assessed in 1395 PIs from S. Korea via a greenhouse layer test. Lines exhibiting possible Rps gene immunity or rot due to other pathogens were removed and the remaining 800 lines were used to identify regions of quantitative resistance using genome-wide association mapping. Sixteen SNP markers on chromosomes 3, 13 and 19 were significantly associated with partial resistance to P. sojae and were grouped into seven quantitative trait loci (QTL) by linkage disequilibrium blocks. Two QTL on chromosome 3 and three QTL on chromosome 19 represent possible novel loci for partial resistance to P. sojae. While candidate genes at QTL varied in their predicted functions, the coincidence of QTLs 3-2 and 13-1 on chromosomes 3 and 13, respectively, with Rps genes and resistance gene analogs provided support for the hypothesized mechanism of partial resistance involving weak R-genes.
Conclusions
QTL contributing to partial resistance towards P. sojae in soybean germplasm originating from S. Korea were identified. The QTL identified in this study coincide with previously reported QTL, Rps genes, as well as novel loci for partial resistance. Molecular markers associated with these QTL can be used in the marker-assisted introgression of these alleles into elite cultivars. Annotations of genes within QTL allow hypotheses on the possible mechanisms of partial resistance to P. sojae.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2918-5) contains supplementary material, which is available to authorized users.
Keywords: Glycine max, GWAS, Haplotype, Linkage disequilibrium, Partial resistance, Phytophthora sojae, Single nucleotide polymorphism
Background
Phytophthora root and stem rot was the second most yield-limiting disease of soybean [Glycine max (L.) Merr] between 1996 and 2009 [1, 2]. This disease, caused by the soil-borne oomycete pathogen Phytophthora sojae [3], is prevalent when soil conditions become saturated [4], allowing the asexual, motile zoospores to chemotactically travel to soybean roots [5, 6]. Upon infection, P. sojae will produce haustoria and acquire nutrients in a hemi-biotrophic manner [7]. Susceptible plants will develop lesions, experience wilting and chlorosis, and, in severe cases, plant death [4].
Genetic resistance is an effective strategy to manage P. sojae in regions with high levels of inoculum and favorable environments [8]. Soybean breeding programs have primarily utilized single, dominant Rps-mediated resistance in which recognition of P. sojae effector proteins initiate effector-triggered immunity, resulting in complete resistance. To date, 21 Rps genes or alleles have been identified and mapped to five chromosomes [9–18]. Though potentially highly effective, Rps-mediated resistance is race-specific and effectiveness of a given Rps gene is dependent on the population of P. sojae present. Additionally, deployment of Rps genes places high selection pressures on the P. sojae populations causing the population to adapt and potentially gain virulence such that the Rps gene is no longer effective. Widespread deployment of Rps genes in soybean cultivars has resulted in the evolution of highly diverse P. sojae populations [19] with more than 200 physiological races (55 described) identified in the US [20–24]. Pathogen diversity and adaptation limits the efficacy of an Rps gene to eight to twenty years [8, 25] thus, breeders cannot rely solely on Rps genes.
In contrast to Rps-mediated resistance, partial resistance is a quantitative trait, controlled by multiple genes at numerous loci, each contributing a small effect or a few loci contributing a moderate effect [26–28]. Partial resistance to P. sojae has been shown to be effective against numerous pathotypes of P. sojae [29, 30]. Unlike Rps-mediated resistance, partial resistance to P. sojae is incomplete, and allows some pathogen growth and reproduction [30]. This is believed to place minimal selection pressure on the P. sojae populations exposed to cultivars possessing partial resistance. For this reason partial resistance is predicted to be more durable with examples such as powdery mildew management in winter wheat indicating effectiveness on the scale of decades [31, 32]. Multiple mechanisms for partial resistance have been broadly hypothesized [26–28]. Mechanistic studies specific to this pathosystem have provided evidence for the involvement of R-genes [33], components of defense signal transduction pathways [33–35] and genes regulating plant physiology or morphology [33, 35], including suberin deposition in the root [36, 37] in partial resistance to P. sojae.
Improved levels of partial resistance against P. sojae in US soybean cultivars can be achieved through the introgression of novel alleles. Over 1000 soybean plant introductions (PIs) were previously evaluated as potential novel sources of resistance to P. sojae and those originating from The Republic of Korea (S. Korea) were associated with high levels of partial resistance [38]. Therefore, it was proposed that high genetic diversity for P. sojae resistance may exist in PIs from S. Korea as a result of the potential co-evolution between soybean and P. sojae that may have occurred in this region [38].
Identification of QTL for partial resistance against P. sojae in soybean has been limited to the cultivar Conrad [39–44], southern germplasm V71-370 [45], and eight accessions originating from Asia [46–50]; including four PIs originating from S. Korea [47–50]. Among the PIs from S. Korea, QTL for partial resistance to P. sojae were identified on all chromosomes (Chrs), except 5, 6, 11, 17 and 19, with between two and eight QTL identified in each population and most QTL contributing less than 10 % of the phenotypic variation (PVE) [47–50]. Interestingly, in a recombinant inbred line (RIL) population derived from a cross between PI 427105B and the susceptible breeding line OX20-8, a QTL with the largest PVE (up to 45 %) was identified on Chr 18 [47].
Given the success in identifying and diversity of QTL contributing to partial resistance from PIs, it is pertinent to further evaluate a broader array of PIs for partial resistance and to identify the common alleles that may be contributing to resistance within this germplasm source. In this study 1395 PIs originating from S. Korea were evaluated for partial resistance to P. sojae. A genome-wide association (GWA) analysis was performed using high-density genetic markers from the Soy50KSNPChip [51]. In this study, the extent of root rot, root weight, shoot weight and plant height from inoculated plants were combined with genotypic data to identify 16 markers significantly associated with these traits across three chromosomes. Associated markers were grouped into seven QTL according to linkage disequilibrium and candidate genes were identified within these regions.
Results and discussion
Phenotypic data
Over 1300 PIs originating from S. Korea were evaluated for their response to two virulent isolates of P. sojae, C2S1 (vir 1a, 1b, 1c, 1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, 7 and 8) and OH12108_6.3 (OH121; vir 1a, 1b, 1c, 1d, 1k, 2, 3a, 3c, 4, 5, 6, 7 and 8), using the greenhouse-based layer test assay. Phenotypic data were collected for inoculated root rot score (IRRS; 1, no rot, to 9, completely rotted) as well as for root weight (RW), shoot weight (SW) and plant height (PH) from inoculated (I) and non-inoculated (N) assays. A total of 266 PIs were excluded from the final data set due to poor germination or missing data. There was no observable root rot in 306 PIs (IRRS ≤ 1.5) following inoculation with one or both isolates of P. sojae and these were excluded from analysis in order to eliminate any possible Rps-mediated resistance response. In addition, 23 PIs possessed a mean non-inoculated root rot score > to 1.5 and were removed from the analysis to exclude any effects from potential seed-borne pathogens. The remaining 800 PIs had a mean IRRS of 3.5 which ranged from 1.6 to 8.0. Best linear unbiased predictor (BLUP) values were calculated for each PI, representing the PI’s genotypic value for each trait. Similarly, best linear unbiased estimator (BLUE) values were calculated for each check class, which was treated as a fixed effect. In comparison to check cultivars, the PIs generally exhibited high levels of partial resistance. Twenty-seven PIs had lower IRRS BLUP values than the mean IRRS BLUE values for highly resistant checks (Conrad, PI407861A and PI398841); whereas no PIs had IRRS BLUP values that exceeded the mean BLUE values for either the moderately susceptible check (Sloan) or the highly susceptible check (OX20-8). IRW, ISW, IPH showed normal distributions and IRRS had a slightly positive skew (Fig. 1).
Fig. 1.
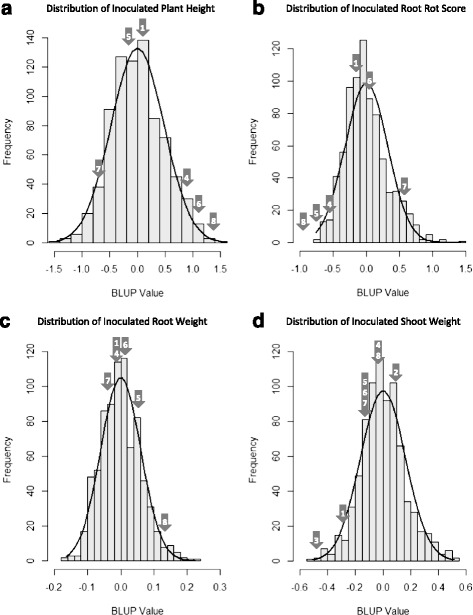
Distribution of BLUP values for inoculated traits. Histograms are depicted for inoculated plant height (a), inoculated root rot score (b), inoculated root weight (c) and inoculated shoot weight (D). Numbered arrows in histograms indicate BLUE values of checks (1 = Conrad, 2 = L83-570, 3 = OX-20, 4 = PI398841, 5 = PI407861A, 6 = Resnik, 7 = Sloan, and 8 = Williams 79)
All four inoculated traits were significantly correlated with each other, where IRW, ISW and IPH were positively correlated, and IRRS, in which a lower value indicates greater resistance, was negatively correlated to IRW, ISW and IPH (Table 1). There was significant genetic variance for the four traits in both inoculated and non-inoculated treatments (Table 2). The non-inoculated traits, NRW, NSW and NPH were all highly heritable, greater than 0.720. The heritability for the four inoculated traits ranged from moderately low at 0.334 for IRW to moderately high at 0.605 for ISW (Table 2). In an analysis of assays with each isolate of P. sojae separately, resistance towards C2S1 had lower heritability and lower disease incidence compared to OH121, where the mean of the raw IRRS for C2S1 and OH121 was 2.85 and 4.13, respectively. While the disparity in heritability between isolates are similar to observations in previous reports [48, 52] and can likely be attributed to the reduced disease development from the C2S1 isolate, it emphasizes the need to carry out assays for partial resistance with multiple isolates. Although the C2S1 isolate was virulent in the hypocotyl test and moderately aggressive in a tray test (Additional file 1: Table S1), the aggressiveness of P. sojae isolates can vary depending upon which component of partial resistance is measured in the phenotypic disease assay [30, 52].
Table 1.
Pearson’s correlation coefficients (top right) and p-values (bottom left) for inoculated root rot score (IRRS), inoculated root weight (IRW), inoculated shoot weigh (ISW) and inoculated plant height (IPH)
| IRRS | IRW | ISW | IPH | |
|---|---|---|---|---|
| IRRS | −0.67 | −0.62 | −0.5 | |
| IRW | <0.0001 | 0.78 | 0.51 | |
| ISW | <0.0001 | <0.0001 | 0.58 | |
| IPH | <0.0001 | <0.0001 | <0.0001 |
Table 2.
Genetic variance (σg 2), variance of isolate (σi 2), genotype by isolate variance (σgi 2), variance of error (σe 2) and heritability (H 2) of each trait
| Isolate | Traita | σg 2 | σi 2 | σgi 2 | σε 2 | H 2 |
|---|---|---|---|---|---|---|
| C2S1 and OH121 b | IRRS | 0.29*** | 0.83 | 0.22*** | 1.52*** | 0.23 |
| IRW | 0.01*** | 0.00 | 0.01** | 0.08*** | 0.20 | |
| ISW | 0.05*** | 0.00 | 0.01** | 0.10*** | 0.78 | |
| IPH | 0.65*** | 0.00 | 0.82*** | 2.75*** | 0.71 | |
| OH121 | IRRS | 0.75*** | NAc | NA | 1.51*** | 0.50 |
| IRW | 0.03*** | NA | NA | 0.06*** | 0.44 | |
| ISW | 0.07*** | NA | NA | 0.07*** | 0.67 | |
| IPH | 1.43*** | NA | NA | 3.75*** | 0.43 | |
| C2S1 | IRRS | 0.24** | NA | NA | 1.55*** | 0.24 |
| IRW | 0.01** | NA | NA | 0.09*** | 0.23 | |
| ISW | 0.05*** | NA | NA | 0.12*** | 0.43 | |
| IPH | 1.45*** | NA | NA | 1.88*** | 0.61 | |
| NA | NRW | 0.04*** | NA | NA | 0.06*** | 0.72 |
| NSW | 0.09*** | NA | NA | 0.10*** | 0.78 | |
| NPH | 1.15*** | NA | NA | 1.90*** | 0.71 |
Asterisks indicate the level of statistical significance: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001
a IRRS inoculated root rot score, IRW inoculated root weight, ISW inoculated shoot weight, IPH inoculated plant height, NRW non-inoculated root weight, NSW non-inoculated shoot weight, NPH non-inoculated plant height
b OH121: OH121086.3
cNot applicable
Population structure and linkage disequilibrium
Analysis of population structure among the 800 PIs that were included in the GWA analysis using ADMIXTURE [53] indicated a continued decline in cross-validation (CV) error as values of K were tested from 1 to 22 (Additional file 1: Figure S1A). However, subpopulations defined at a local minimum of K = 3 (Additional file 1: Figure S1B) corresponded to differentiation of PIs by principle component analysis and was determined to be the most likely number of subpopulations (Additional file 1: Figure S1C). A total of 19,303 polymorphic markers were used to carry out GWA mapping in this population. Markers were at an average genome-wide density of one marker every 50.5 kb or, more specifically, at a density of one marker every 29.9 and 124.4 kb in euchromatic and heterochromatic regions, respectively. LD decayed to half of its initial value at approximately 7.32 and 17.7 kb in the euchromatin and heterochromatic regions, respectively. Due to LD among markers, the 19,303 markers could be condensed into 12,313 effective markers, or an average of one effective marker approximately every 80 kb. Linkage disequilibrium (LD) decayed to an r2 of 0.2 at approximately 172.3 and 556.6 kb in euchromatin and heterochromatic regions, respectively (Fig. 2), indicating that, on average, marker density is sufficient to capture the majority of the genome. This effective number of markers (Meff) was considered the number of independent tests and employed in a correction for multiple testing in which marker trait associations were considered significant when the P-value was less than α/Meff, 4.06 × 10−6 [54].
Fig. 2.
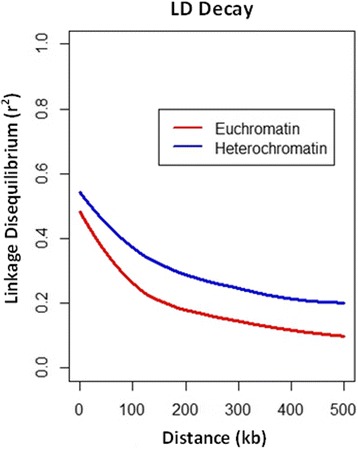
Linkage disequilibrium (r 2) as a function of physical distance (kb)
GWA analysis
GWA analysis of partial resistance to P. sojae has been limited to two studies, one which used 174 soybean accessions from the mini core collection of cultivated soybean from China with 495 SSR loci [55] and recently a second study used 472 accessions from a Chinese breeding program with nearly 60,000 SNPs [56]. As a result, little information is available regarding the genetic distribution of alleles for partial resistance in a breadth of germplasm. The present study utilized nearly 800 PIs and employed the largest population of any previously GWA analyses performed for disease resistance in soybean [55–60]. A total of 16 significant marker-trait associations were identified for IRRS and IRW (Table 2). These mapped to genomic regions on Chrs 3, 13 and 19 (Figs. 3 and 4). While the large number of accessions assayed and limited availability of seed prohibited an evaluation of a potential Rps gene mediated response for each isolate and accession combination, the methods used in this study applied several approaches to avoid Rps-mediated responses. These approaches included the selection of complex isolates, the removal of accessions exhibiting limited root rot in inoculations with either C2S1 or OH121, as well as the inclusion of a genotype-by-isolate interaction term in the model applied. In addition, hypocotyl assays carried out on 94 randomly selected accessions included in the GWA analysis showed a virulent reaction of C2S1, indicating a lack of Rps-mediated resistance within these accessions (Additional file 1: Table S2). Within this group of accessions there is one or more individuals possessing the resistance allele for 11 out of the 16 significant markers. Due to these approaches implemented in this study, it is expected that the significant marker-trait associations represent partial resistance loci. However, it cannot be ruled out that Rps-mediated resistance has been observed in a quantitaive manner due to incomplete resistance as observed in the root gene Rps2 [30] or errors associated with scoring of root rot.
Fig. 3.
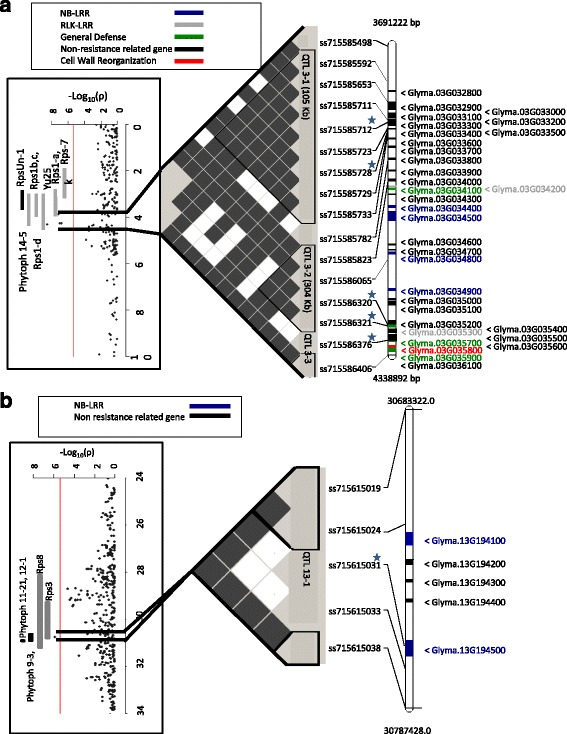
QTLs identified for root rot score on chromosome 3 (a) and for inoculated root weight on chromosome 13 (b). The far left image is a Manhattan plot indicating the level of marker association with either root rot score or inoculated root weight. The central image contains a visualization of linkage disequilibrium (white is an r2 value of zero, black is an r2 value of 1). Significant haplotype blocks are outlined in black. The far right image is a physical map of markers (stars indicate markers significantly associated with inoculated root rot score) and candidate genes with text color indicate the general annotation categories related to resistance
Fig. 4.
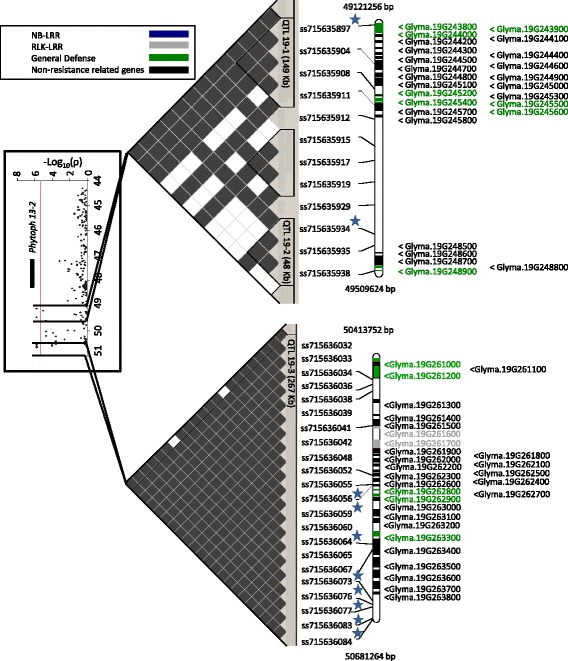
QTLs identified on chromosome 19 for root rot score. The far left image is a Manhattan plot indicating the level of marker association with either root rot score or inoculated root weight. The central image contains a visualization of linkage disequilibrium (white is an r2 value of zero, black is an r2 value of 1). Significant haplotype blocks are outlined in black. The far right image is a physical map of markers (stars indicate markers significantly associated with inoculated root rot score) and candidate genes with text color indicate the general annotation categories related to resistance
Genomic regions on Chrs 3 and 19 were identified with significant marker-trait associations for IRRS. Five SNPs on Chr 3 (ss715585728, ss715585712, ss715586321, ss715586320, ss715586376) were located within a 0.4 Mb region (3.9–4.3 Mb) (Fig. 3a). ss715586376 was the most significantly associated SNP with a P-value of 3.27 × 10−9 (Table 3). Ten SNPs on Chr 19 (ss715635897, ss715635934, ss715636056, ss715636059, ss715636064, ss715636073, ss715636076, ss715636077, ss715636083, ss715636084) were significantly associated with IRRS and were located within a 1.6 Mb region (49.4–50.7 Mb) (Fig. 4), with all ten SNPs possessing a P-value of 1.82 × 10−6 (Table 3).
Table 3.
Significant marker-trait associations with identified genomic regions and chromosome location for each inoculated trait and model
| Trait | Marker | Chr.a | Chr. position (bpb) | QTL | Minor allele frequency | Additive effect | PVEc | p-value |
|---|---|---|---|---|---|---|---|---|
| Inoculated Root Rot Score | ss715585712 | 3 | 3852888 | 3-1 | 0.18 | 0.096 | 3.162 | 9.14E-08 |
| ss715585728 | 3 | 3865730 | 3-1 | 0.18 | 0.0967 | 3.212 | 7.26E-08 | |
| ss715586320 | 3 | 4276534 | 3-2 | 0.21 | 0.0922 | 3.018 | 1.77E-07 | |
| ss715586321 | 3 | 4277380 | 3-2 | 0.21 | 0.0936 | 3.133 | 1.04E-07 | |
| ss715586376 | 3 | 4315512 | 3-3 | 0.2 | 0.1039 | 3.895 | 3.27E-09 | |
| Inoculated Root weight | ss715615031 | 13 | 30766058 | 13-1 | 0.42 | −0.0695 | 2.468 | 1.44E-06 |
| Inoculated Root Rot Score | ss715635897 | 19 | 49121258 | 19-1 | 0.01 | −0.4622 | 2.513 | 1.82E-06 |
| ss715635934 | 19 | 49461582 | 19-2 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636056 | 19 | 50544363 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636059 | 19 | 50555433 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636064 | 19 | 50604933 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636073 | 19 | 50663466 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636076 | 19 | 50666563 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636077 | 19 | 50668662 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636083 | 19 | 50679714 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 | |
| ss715636084 | 19 | 50681263 | 19-3 | 0.01 | −0.4622 | 2.513 | 1.82E-06 |
a Chr chromosome
b bp basepair position in the Glyma.Wm82.a2 assembly
c PVE percent variance explained
A single SNP (ss715615031) at 30.7 Mb on Chr 13 was significantly associated with IRW (P-value = 1.44 × 10−6) (Table 3; Fig. 3b). While no significant marker-trait associations were identified for ISW or IPH, for IPH several near significant markers at ~3.9 Mb on Chr 3 and ~30 Mb on Chr 13 were noted (Additional file 1: Figure S2). Near significance of ss715615031, the significant marker for IRW, on Chr 13 (P-value = 4.83 × 10−6) and near significance of ss715585728, one of the significant markers for IRRS, on Chr 3 (P-value = 1.11 × 10−5) were observed for IPH.
No significant marker-trait associations were found in the NRW or NPH (Additional file 1: Figure S3). Five genomic regions were identified with significant marker associations for NSW on Chr 2 (3.4–4.6 Mb), 3 (5.2–5.6 Mb), 4 (6.1–6.6 Mb), 17 (8.5 Mb), and 18 (51.7–53.0 Mb). However, none of the significant markers for NSW were coincident with the markers identified for the inoculated traits, IRRS and IRW.
Grouping of significantly associated SNPs into QTL
A QTL was defined as a haplotype block possessing marker(s) identified as significantly associated with a trait. Based on this criterion, the 16 markers significantly associated with IRRS or IRW were grouped into seven QTL ranging in size from 176 to 48 kb. The extensive historical recombination present in a population of PIs can lead to the identification of relatively narrow QTL in GWA analysis in comparison to mapping conducted in bi-parental populations. In previous mapping studies conducted with RIL and NAM populations, identified QTL spanned an average of 6132.6 kb, with the largest QTL encompassing 940 genes [39–50, 52]. In contrast, the largest QTL in this study is 304 kb in length and contains 13 genes.
Candidate genes and coincident traits for QTL 3-1, 3-2 and 3-3
The five significant marker-trait associations on Chr 3 were grouped into two haplotype blocks, each possessing two significant marker-IRRS associations. A fifth marker which was significantly associated with IRRS, was not in significant LD with neighboring markers. These regions are referred to as QTL 3-1, 3-2 and 3-3, respectively (Fig. 3a). QTL 3-1 is coincident with a previously identified QTL (Phytoph 14-5; www.soybase.org [61]) for lesion length in a tray test of partial resistance to P. sojae where the resistance allele was from a PI originating from S. Korea [52]. A QTL associated with resistance to the necrotrophic pathogen, Sclerotinia sclerotiorum, was also in close proximity to QTL 3-1 [59, 62]. Additionally, QTL 3-1 partially overlaps with Rps1a, b, c, d and k; RpsYu25; and RpsUn1 and is also nearby the putative position of Rps7 [9, 10, 12, 15, 63] (Fig. 3a). QTL 3-1 contains eight predicted genes in a 105.5 kb region (Glyma.Wm82.a2.v1, accessed Phytozome v10; Additional file 2: Table S3). Interestingly, while QTL 3-1 partially overlaps with a number of Rps genes, none of the eight predicted genes from the Williams 82 reference sequence within this QTL were conventionally considered to be related to defense or resistance. Therefore, QTL 3-1 may be conferred by non-canonical R-genes or, more likely, is located upstream of the Rps genes and associated with a novel mechanism for partial resistance.
The second QTL (3-2) is 238.1 kb downstream of QTL 3-1 and is coincident with the mapped positions of Rps1d and RpsUn1 [10, 15]. This region does not coincide with any QTL for partial resistance to P. sojae (Fig. 3a). Thus, QTL 3-2 may represent a novel QTL for partial resistance to P. sojae. QTL 3-2 encompasses 304 kb and 13 predicted genes (Additional file 2: Table S3), seven of which have putative functions relating to disease resistance or defense. Four of these genes (Glyma.03G034400, Glyma.03G034500, Glyma.03G034800 and Glyma.03G034900) putatively encode a nucleotide binding (NB) domain characteristic of R-genes [64]. In addition, there are two leucine-rich repeat receptor-like kinase (LRR-RLK)encoding genes in this region (Glyma.03G034200 and Glyma.03G035300), a class of genes involved in basal defense and plant developmental responses [65–68]. Finally, Glyma.03G034100 is homologous to a Sec61 protein transporter-encoding gene, which is involved in activation of systemic-acquired resistance in Arabidopsis [69].
Marker ss715586376 is located 238.1 kb downstream of QTL 3-2 and was significantly associated with IRRS. Marker ss715586376 was not in LD with nearby markers, thus, the QTL 3-3 region was defined by the two flanking markers, ss715586321 and ss715586406 (Fig. 3a). There are a total of seven genes between these markers (Additional file 2: Table S3), of which three have possible roles in defense. Glyma.03G035700 putatively encodes an abscisic acid responsive stress related protein [70]. Glyma.03G035800 encodes a putative Alpha-expansin involved in cell wall extension and growth [71, 72]. Glyma.03G035900 is a CAD1 encoding gene with Mac/perforin domains. The CAD1 encoding genes have been shown to be involved in plant defense in Arabidopsis by activating the salicylic acid pathway causing a hypersensitive response [73].
Candidate genes and coincident traits for QTL 13-1
Located on Chr 13, ss71565031 was significantly associated with IRW (Fig. 3b). Similar to QTL 3-3, ss71565031 was not located in a haplotype block. Therefore, QTL 13-1 region was defined by the two flanking markers, ss715615924 and ss715615033 and spans 49.2 kb. QTL 13-1 is coincident with Rps8 and Rps3 [9, 17], three previously identified QTL for partial resistance against P. sojae (Phytoph 11-21, 9-3, 12-1 [61]) [43, 48, 52], a QTL for flood tolerance [48], and QTL for resistance against S. sclerotiorum [62]. The region contains a total of five predicted genes (Additional file 2: Table S3), of which two (Glyma.13G194100 and Glyma.13G194500) are NB-LRR encoding genes.
Candidate genes and coincident traits for QTL 19-1, 19-2 and 19-3
A total of ten significant markers for the IRRS were located on Chr 19. The significant markers were grouped into three QTL based on haplotype blocks, QTL 19-1, QTL 19-2 (Fig. 4) and QTL 19-3 (Fig. 4). No previously identified QTL associated with resistance to P. sojae or other pests or pathogens were coincident with the three QTL on Chr 19. However, marker ss715635897 from QTL 19-1 is located approximately 1.1 Mb from a previously identified QTL (Phytoph13-2 [61]) for partial resistance to P. sojae through a tray test disease assay [52].
QTL 19-1 spans 149 kb and contains a total of 21 predicted genes (Fig. 4; Additional file 2: Table S3), of which seven are putative defense related genes. The defense genes in the region include Glyma.19G245400, Glyma.19G245500 and Glyma.19G245600, which encode putative PR4-related chitin-binding proteins [74]. Glyma.19G243800 encodes a putative ribose 5-phosphate isomerase. Homologs in Arabidopsis have been shown to function in cellulose synthase [75], which has been shown to be involved in the regulation of jasmonic acid and ethylene [76] with mutants displaying enhanced resistance to bacterial and fungal pathogens [77]. Other genes in the region include Glyma.19G244000 that encodes a putative MATE efflux protein potentially involved in defense signaling pathways or in transporting toxic compounds from infected cells [78, 79] as well as Glyma.19G245200 that encodes a putative auxin responsive gene [70]. Finally, Glyma.19G244400 encodes a putative ammonium transporter, homologs of which have been implicated in interactions with root endophytes [80–82] and can act as a negative regulator of basal defense responses in Arabidopsis [83].
QTL 19-2 is 48 kb in length and located 218 kb downstream of QTL 19-1 (Fig. 4). This QTL encompassed five predicted genes (Additional file 2: Table S3) including Glyma.19G248900 that encodes a putative ethylene/JA responsive transcription factor [70].
The third QTL identified on Chr 19 (QTL 19-3) is 267 kb in length and is located 904 kb downstream of QTL 19-2 (Fig. 4). It encompasses a total of 29 predicted genes (Additional file 2: Table S3), six of which have annotations representative of possible defense function. These genes include Glyma.19G261000 and Glyma.19G261700 that putatively encode LRR-RLKs, Glyma.19G261200 that is a dicer family protein putatively involved in the control of gene silencing [84], Glyma.19G262800 and Glyma.19G262900 that putatively encode GDSL esterase/lipases which can be associated with the ethylene/JA responsive defense pathways [85–87], and Glyma.19G263300 that encodes a putative ethylene/JA responsive lipoxygenase [88].
Inferences about mechanisms of partial resistance
It has been hypothesized that partial disease resistance can be controlled through developmental or morphological mechanisms, basal defense genes, production of antimicrobials or detoxification of phytotoxins (chemical warfare), components of defense signal transduction pathways, weak R-gene responses or other unknown mechanisms [27]. While functional gene analysis is required to identify mechanisms, the co-localization of annotated genes and QTLs can theoretically provide evidence in support of particular mechanisms. In this study, co-localization of annotated genes and QTLs provides varying levels of support for each of the aforementioned hypotheses.
Mechanisms of partial resistance associated with developmental or morphological mechanisms are difficult to assess through annotations of co-localized genes because there are a limited number of clearly defined pathways for these complex traits. However, QTLs 3-3 and 19-1 contain candidate genes putatively involved in morphology and development, including cell growth and cellulose production. Evidence for basal defense was found through co-localization of QTLs 3-2 and 19-3 with LRR-RLK-encoding genes. However, while LRR-RLKs are known to be involved in recognition of microbe associated molecular patterns leading to basal defense responses [65–67], LRR-RLK-encoding genes have also been implicated in a range of functions, including plant development [68]. Transport of toxic compounds is a potential function of a MATE efflux-encoding gene within QTL 19-1, providing a candidate for the chemical warfare hypothesis. QTLs 3-2, 3-3, 19-1 and 19-3 encompass genes involved in defense signal transduction, with putative functions ranging from negative regulators of basal defense to the control of gene silencing. Among the largest class of genes within these QTL are NB-LRR-encoding genes [64]. In support of the weak R-gene hypothesis, QTLs 3-2 and 13-1 are coincident with six NB-LRR-encoding genes and overlap with estimated positions of Rps genes [9, 10, 12, 15, 63]. Finally, QTL 3-1 does not contain any genes normally associated with defense or development pertinent to P. sojae resistance and therefore supports the idea that quantitative defense can be conferred by a yet unknown class of genes [27].
Consistent with previous studies [33], the QTL encompass genes functioning in a wide range of processes, potentially indicating that a number of different mechanisms contribute to quantitative defense, making this germplasm a welcome resource to diversify resistance genes. The current study along with previous QTL mapping studies have identified 33 genetic regions on 18 chromosomes with QTL for partial resistance to P. sojae [33, 39–50, 52, 56, 61]. Of the 33 regions, four are coincident with Rps-genes [9, 10, 12, 15, 63] five are coincident with QTL for resistance to Sudden Death Syndrome [89–93], root disease caused by members of the fungus Fusarium viguliforme, 11 are coincident with QTL/genes for resistance to Soybean Cyst Nematode [94–103], and 11 are coincident with QTL for resistance to the necrotrophic fungal pathogen S. sclerotiorum causing Sclerotinia stem rot [59, 62, 104, 105]. The coincidence of QTL for partial resistance to P. sojae with QTL and R-genes for resistance to pathogens with varied lifestyles provides evidence that partial resistance is likely conferred through a variety of different mechanisms.
Conclusions
In the present study, a GWA analysis was performed to detect genomic regions contributing to quantitative resistance to P. sojae using PIs from the Republic of Korea. In addition to identifying five novel QTL, QTLs that coincide with previously reported QTLs for resistance to P. sojae were identified. Candidate genes and coincident QTL were identified to explore mechanistic hypotheses of partial resistance, providing evidence towards a number of different hypotheses including a weakened R-gene response and genes involved in morphology and development, basal defense and signal transduction. To fully characterize the genes conferring resistance in these regions, functional analyses of candidate genes is necessary or in process.
Methods
Seed material
A collection of 1345 PIs from S. Korea were obtained from the National Plant Germplasm System, consisting of 50 seeds per inbred line ranging from maturity groups I to IV. In addition to these lines, checks (‘Conrad’, ‘Sloan’, ‘OX20-8’, ‘Williams 79’, ‘Resnik’, ‘L83-570’, PI 398841 and PI 407861A) with known and varied levels of resistance to P. sojae were included in the experimental design. All seeds were vapor phase sterilized following a chlorine gas protocol adapted from Olhoft et al. [106] prior to disease assays.
Disease assays
Pathogenicity tests were conducted prior to the phenotypic disease assay for selection of two P. sojae isolates based on virulence and aggressiveness. Twenty-seven isolates were tested on 15 soybean differentials in a hypocotyl assay for Rps-mediated resistance [38] (Additional file 1: Table S1). Virulence was measured by the percentage of susceptible (>90 % compatible reaction), and resistant (<10 % incompatible reaction) responses in 10 soybean seedlings of each differential. Using a growth chamber based tray test [39], the aggressiveness of each isolate was measured according to the mean lesion length of soybean taproots in ten inoculated seedlings from each cultivar, ‘Sloan’ and ‘Conrad’, possessing moderate susceptibility and a high level of partial resistance, respectively [33]. Isolates OH121 (vir 1a, 1b, 1c, 1d, 1k, 2, 3a, 3c, 4, 5, 6, 7 and 8) and C2S1 (vir 1a, 1b, 1c, 1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, 7 and 8) were both recovered from soybean in Ohio and were identified as moderately aggressive such that adequate separation of accession with low and high levels of resistance would be expected. OH121 and C2S1 also possesses complex pathotypes (Additional file 1: Table S1) for eliminating Rps-mediated responses enabling partial resistance in the greenhouse based layer test [107] could be observed. In order to further assess the potential of Rps-mediated response, hypocotyl assays were conducted. Due to the limited seed quantities available for each accession, hypocotyl assays were conducted for only a single isolate (C2S1) and for 94 randomly selected lines which were included in the GWA analysis (Additional file 1: Table S2).
Phenotypic disease assays were conducted on 1398 PIs originating from S. Korea following a layer test protocol adapted from Dorrance et al. [108] to evaluate partial resistance to P. sojae. IPH and NPH were averaged from three seedlings per cup. IRW, NRW, ISW, NSW, IRRS and NRRS were measured two weeks after planting. IRRS and NRRS were rated on the 1–9 scale according to Dorrance et al. [107]. Fresh weights of the roots or shoots were adjusted by dividing by the total number of plants per cup to calculate IRW, NRW, ISW and NSW. The full experiment was repeated twice for each isolate. The 1395 PIs were first evaluated with C2S1. Lack of root rot is a characteristic of immunity imparted by Rps-mediated resistance. Isolates OH121 possesses a pathotype capable of detecting (incompatible, avirulent reaction) Rps3b and both isolates may detect novel Rps genes. Therefore, in order to limit genetic associations to those involved in partial resistance, PIs that exhibited little to no root rot (mean IRRS ≤ 1.5) were removed from the subsequent disease assays with OH121. PIs exhibiting limited root rot (mean IRRS ≤ 1.5) with OH121 were also removed from the dataset. In addition, PIs with NRRS ≥ 1.5 were removed from the dataset, eliminating possible disease due to contamination or seed-borne pathogens. A total of 800 PIs with a mean IRRS > 1.5 for both isolates and a mean NRRS ≤ 1.5 were further analyzed for heritability and GWA analysis.
Phenotypic data analysis
BLUP values were generated for each PI for IRRS, IRW, ISW, IPH, NRW, NSW and NPH using PROC MIXED procedure in the software SAS (SAS 9.3, SAS Institute 163 Inc., Cary, NC, USA). Data were analyzed with two models. Using observations from both isolates, the model included interactions with isolate: Yhjklm = μ + Ih + R(I)hj + K(IR)hjk + Cl + G(C)lm + IG(C)hlm + IChl + εhjklm, where μ is the overall mean, Ih is effect of the hth isolate, R(I)hj is the effect of the jth experimental replicate for the hth isolate, K(IR)hjk is the effect of the kth bench in the jth experimental replicate for the hth isolate, Cl is the effect of the lth class of entry [1–9 for Conrad (rps, high levels of partial resistance), L83-570 (Rps3a, moderate levels of partial resistance), OX20-8 (Rps1a, highly susceptible), PI 398841 (high levels of partial resistance), PI 407861A (high levels of partial resistance), Resnik (Rps1k, moderate levels of partial resistance), Sloan (rps, moderately susceptible), Williams 79 (Rps1c, moderate levels of partial resistance), and experimental lines, respectively], G(C)lm is the effect of the mth genotype within class for experimental lines (genotypic variance, σg2), IG(C)hlm is the effect of the hth isolate with the mth genotype within the lth class for the experimental lines (genotypic x isolate variance, σgi2), IChl is the effect of the hth isolate with the Ith class entry, εhjklm is the experimental error (σε2). Broad-sense heritability (H2) was calculated for each trait as follows: H2 = σg2/(σg2 + σgi2/i + σε2/ir), where σg2 represents the genetic variance, σgi2 represents the genotype × isolate variance, σε2 represents the error variance, r is the number of experimental replicates, and i is the number of isolates.
Genotypic data analysis
Genotypic data [109] collected using the Illumina Infinium SoySNP50K iSelect BeadChip developed by the Beltsville, MD, USDA Soybean Genomics and Improvement Lab were downloaded from Soybase [61]. The genotypic data consisted of 42,509 SNPs [51]. Monomorphic markers and markers with > 5 % missing data or >10 % missing plus heterozygous allele calls were removed. A total of 19,303 polymorphic markers, including 11,126 with minor allele frequencies (MAF) ≥ 5 %, were included in the association analyses. Missing genotypes were imputed with fastPHASE [108]. A genome-wide estimation of LD decay in euchromatic and heterochromatic regions was plotted as physical distance (kbp) vs r2. Population structure was examined using the software ADMIXTURE with five-fold cross-validation [53].
GWA analyses
GWA analyses of the phenotypic and genotypic data sets for the 797 non-immune cultivars were conducted using the GAPIT (Genome Association Prediction Tool) package [110] in R software [111] with a compressed mixed linear model and population parameters previously determined [112]. The optimal number of principle components for inclusion in the model was determined with GAPIT by Bayesian Information Criterion. The significance threshold for marker-trait associations was determined by a modified Bonferroni adjustment in which Meff was calculated using simpleM [113] Genome-wide threshold levels for each of the two datasets were determined by α/Meff, where α = 0.05.
Haplotype blocks were constructed based on the following criteria: 1) markers in significant LD (four-gamete method) [114]; were grouped into haplotype blocks, 2) adjacent blocks separated by < 10 kb were combined. However, no haplotype blocks met criteria 2, thus, no haplotype blocks were combined. For this study, a significant QTL was defined as a haplotype block possessing marker(s) identified through GWA analysis to be significantly associated with a trait. The haplotype block determination and visualization were carried out with Haploview Version 4.2 [115].
Abbreviations
BLUP, best-linear unbiased predictor; CMLM, compressed mixed linear model; GAPIT, Genome Association Prediction Tool; GWAS, genome-wide association study; IPH, inoculated plant height; IRRS, inoculated root rot score; IRW, inoculated root weight; ISW, inoculated shoot weight; JA, Jasmonic acid; kb, kilobase; LD, linkage disequilibrium; LRR-RLK, leucine rich repeat-receptor like kinase; MAPK mitogen activated protein kinase; Meff, effective number of markers; NB-LRR, nucleotide binding-leucine rich repeat; NPH, non-inoculated plant height; NRW, non-inoculated root weight; NSW, non-inoculated shoot weight; PCA, principle component analysis; PI, plant introduction; QTL, quantitative trait loci; RILs, recombinant inbred line; SNPs, single nucleotide polymorphisms
Acknowledgements
The authors are grateful to Tom Fitz Gibbon, Andika Gunadi, Allen Honerlaw, Anna Stasko, Christine Balk, Dr. Sungwoo Lee, Sarah Lewis, Scott McIntyre, Jaqueline Huzar Novakowiski, Charlotte Smith, Lisa Sutton and Deloris Veney for assistance with phenotyping and to G. John Lazur for provision of custom scripts for data handling.
Funding
Salaries and research support for this project were provided by the United Soybean Board, the Ohio Soybean Council, The Ohio State University Center for Applied Plant Sciences, and the USDA National Institute of Food and Agriculture, Hatch projects OHO01279 and OHO01303.
Availability of data and materials
The data sets supporting the results of this article are available with the article and additional files or on request from the corresponding author.
Authors’ contributions
LKM, AED and RS conceived and designed experiments. RS executed experiments. RS, WR and LKM analyzed data. QS and PC designed SNP markers and performed genotyping. RS, WR, AED and LKM wrote the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Summary of mean lesion lengths from tray tests and virulence profiles from hypocotyl inoculations utilizing 22 different isolates of P. sojae. Table S2a Hypocotyl assay with isolate C2S1 and genotypic data of markers within QTL for 94 randomly selected accessions included in the GWA analysis. Table S2b Hypocotyl assay of differentials for isolate C2S1. Figure S1 Examination of population structure of 800 Plant Introductions. Figure S2 Manhattan plot of the soybean genome depicting the extent of associations of 19,138 SNPs with inoculated root rot score, inoculated root weight, inoculated shoot weight and inoculated plant height. Figure S3 Manhattan plot of the soybean genome depicting the extent of associations of 19,138 SNPs with non-inoculated root weight, non-inoculated shoot weight and non-inoculated plant height. (PDF 962 kb)
Annotations for candidate genes within QTLs. (XLSX 44 kb)
References
- 1.Koenning SR, Wrather JA. Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Prog. 2010 [Google Scholar]
- 2.Wrather JA, Koenning SR. Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Prog. 2009 [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufmann MJ, Gerdemann J. Root and stem rot of soybean caused by Phytophthora sojae n. sp. Phytopathology. 1958;48:201–208. [Google Scholar]
- 4.Dorrance A, Mills D, Robertson A, Draper M, Giesler L, Tenuta A. Phytophthora root and stem rot of soybean. Plant Health Instr. 2007 [Google Scholar]
- 5.Morris PF, Ward E. Chemoattraction of zoospores of the soybean pathogen, Phytophthora sojae, by isoflavones. Physiol Mol Plant Pathol. 1992;40:17–22. doi: 10.1016/0885-5765(92)90067-6. [DOI] [Google Scholar]
- 6.Morris PF, Bone E, Tyler BM. Chemotropic and contact responses of Phytophthora sojae hyphae to soybean isoflavonoids and artificial substrates. Plant Physiol. 1998;117:1171–1178. doi: 10.1104/pp.117.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moy P, Qutob D, Chapman BP, Atkinson I, Gijzen M. Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant-Microbe Interact. 2004;17:1051–1062. doi: 10.1094/MPMI.2004.17.10.1051. [DOI] [PubMed] [Google Scholar]
- 8.Schmitthenner A. Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 1985;69:362–368. doi: 10.1094/PD-69-362. [DOI] [Google Scholar]
- 9.Demirbas A, Rector B, Lohnes D, Fioritto R, Graef G, Cregan P, Shoemaker R, Specht J. Simple sequence repeat markers linked to the soybean genes for Phytophthora resistance. Crop Sci. 2001;41:1220–1227. doi: 10.2135/cropsci2001.4141220x. [DOI] [Google Scholar]
- 10.Weng C, Yu K, Anderson TR, Poysa V. Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J Hered. 2001;92:442–446. doi: 10.1093/jhered/92.5.442. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Zhang B, Shi S, Zhao J, Feng Y, Na G, Gai J, Han X. Identification, genetic analysis and mapping of resistance to Phytophthora sojae of Pm28 in soybean. Agric Sci China. 2011;10:1506–1511. doi: 10.1016/S1671-2927(11)60145-4. [DOI] [Google Scholar]
- 12.Sun S, Wu X, Zhao J, Wang Y, Tang Q, Yu D, Gai J, Xing H. Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed. 2011;130:139–143. doi: 10.1111/j.1439-0523.2010.01794.x. [DOI] [Google Scholar]
- 13.Zhang J, Xia C, Wang X, Duan C, Sun S, Wu X, Zhu Z. Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor Appl Genet. 2013;126:1555–1561. doi: 10.1007/s00122-013-2073-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Xia C, Duan C, Sun S, Wang X, Wu X, Zhu Z. Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS One. 2013;8:e69799. doi: 10.1371/journal.pone.0069799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin F, Zhao M, Ping J, Johnson A, Zhang B, Abney TS, Hughes TJ, Ma J. Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor Appl Genet. 2013;126:2177–2185. doi: 10.1007/s00122-013-2127-4. [DOI] [PubMed] [Google Scholar]
- 16.Sugimoto T, Yoshida S, Kaga A, Hajika M, Watanabe K, Aino M, Tatsuda K, Yamamoto R, Matoh T, Walker DR, Biggs AR, Ishimoto M. Genetic analysis and identification of DNA markers linked to a novel Phytophthora sojae resistance gene in the Japanese soybean cultivar Waseshiroge. Euphytica. 2011;182:133–145. doi: 10.1007/s10681-011-0525-8. [DOI] [Google Scholar]
- 17.Gordon SG, St Martin SK, Dorrance AE. 8 Maps to a Resistance Gene Rich Region on Soybean Molecular Linkage Group F. Crop Sci. 2006;46:168–173. doi: 10.2135/cropsci2004.04-0024. [DOI] [Google Scholar]
- 18.Sandhu D, Gao H, Cianzio S, Bhattacharyya MK. Deletion of a disease resistance nucleotide-binding-site leucine-rich- repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics. 2004;168:2157–2167. doi: 10.1534/genetics.104.032037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorrance A, McClure S, DeSilva A. Pathogenic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis. 2003;87:139–146. doi: 10.1094/PDIS.2003.87.2.139. [DOI] [PubMed] [Google Scholar]
- 20.Schmitthenner A, Hobe M, Bhat R. Phytophthora sojae races in Ohio over a 10-year interval. Plant Dis. 1994;78:269–276. doi: 10.1094/PD-78-0269. [DOI] [Google Scholar]
- 21.Yang X, Ruff R, Meng X, Workneh F. Races of Phytophthora sojae in Iowa soybean fields. Plant Dis. 1996;80:1418–1420. doi: 10.1094/PD-80-1418. [DOI] [Google Scholar]
- 22.Abney T, Melgar J, Richards T, Scott D, Grogan J, Young J. New races of Phytophthora sojae with Rps 1-d virulence. Plant Dis. 1997;81:653–655. doi: 10.1094/PDIS.1997.81.6.653. [DOI] [PubMed] [Google Scholar]
- 23.Leitz R, Hartman G, Pedersen W, Nickell C. Races of Phytophthora sojae on soybean in Illinois. Plant Dis. 2000;84:487. doi: 10.1094/PDIS.2000.84.4.487D. [DOI] [PubMed] [Google Scholar]
- 24.Kaitany R, Hart L, Safir G. Virulence composition of Phytophthora sojae in Michigan. Plant Dis. 2001;85:1103–1106. doi: 10.1094/PDIS.2001.85.10.1103. [DOI] [PubMed] [Google Scholar]
- 25.Grau CR, Dorrance AE, Bond J, Russin J. Fungal Diseases. In: Boerma R, Specht JE, editors. Soybeans: Improvement, Production and Uses. Madison: Agronomy Monograph No. 16, American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 2004. pp. 679–763. [Google Scholar]
- 26.Kou Y, Wang S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol. 2010;13:181–185. doi: 10.1016/j.pbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 2009;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 28.St. Clair DA. Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopathol. 2010;48:247–268. doi: 10.1146/annurev-phyto-080508-081904. [DOI] [PubMed] [Google Scholar]
- 29.Tooley P, Grau C. Field characterization of rate-reducing resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology. 1984;74:1201–1208. doi: 10.1094/Phyto-74-1201. [DOI] [Google Scholar]
- 30.Mideros S, Nita M, Dorrance AE. Characterization of components of partial resistance, Rps2, and root resistance to Phytophthora sojae in soybean. Phytopathology. 2007;97:655–662. doi: 10.1094/PHYTO-97-5-0655. [DOI] [PubMed] [Google Scholar]
- 31.Shaner G. Evaluation of slow-mildewing resistance of Knox wheat in the field. Phytopathology. 1973;63:1307–1311. doi: 10.1094/Phyto-63-1307. [DOI] [Google Scholar]
- 32.Tucker DM, Griffey CA, Liu S, Brown-Guedira G, Marshall DS, Saghai Maroof MA. Confirmation of three quantitative trait loci conferring adult plant resistance to powdery mildew in two winter wheat populations. Euphytica. 2007;155:1–13. doi: 10.1007/s10681-006-9295-0. [DOI] [Google Scholar]
- 33.Wang H, Wijeratne A, Wijeratne S, Lee S, Taylor CG, St Martin SK, McHale L, Dorrance AE. Dissection of two soybean QTL conferring partial resistance to Phytophthora sojae through sequence and gene expression analysis. BMC Genomics. 2012;13:428. doi: 10.1186/1471-2164-13-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vega-Sánchez ME, Redinbaugh MG, Costanzo S, Dorrance AE. Spatial and temporal expression analysis of defense-related genes in soybean cultivars with different levels of partial resistance to Phytophthora sojae. Plant Pathol. 2005;66:175–182. [Google Scholar]
- 35.Zhou L, Mideros SX, Bao L, Hanlon R, Arredondo FD, Tripathy S, Krampis K, Jerauld A, Evans C. St. Martin SK, Maroof SMA, Hoeschele I, Dorrance AE, Tyler BM. Infection and genotype remodel the entire soybean transcriptome. BMC Genomics. 2009;10:49. doi: 10.1186/1471-2164-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas R, Fang X, Ranathunge K, Anderson TR, Peterson CA, Bernards MA. Soybean root suberin: Anatomical distribution, chemical composition, and relationship to partial resistance to Phytophthora sojae. Plant Physiol. 2007;144:299–311. doi: 10.1104/pp.106.091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranathunge K, Thomas R, Fang X, Peterson CA, Gijzen M, Bernards MA. Soybean root suberin and partial resistance to root rot caused by Phytophthora sojae. Phytopathology. 2008;98:1179–1189. doi: 10.1094/PHYTO-98-11-1179. [DOI] [PubMed] [Google Scholar]
- 38.Dorrance A, Schmitthenner A. New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 2000;84:1303–1308. doi: 10.1094/PDIS.2000.84.12.1303. [DOI] [PubMed] [Google Scholar]
- 39.Burnham K, Dorrance A, VanToai T, St Martin S. Quantitative trait loci for partial resistance to Phytophthora sojae in Soybean. Crop Sci. 2003;43:1610–1617. doi: 10.2135/cropsci2003.1610. [DOI] [Google Scholar]
- 40.Weng C, Yu K, Anderson TR, Poysa V. A quantitative trait locus influencing tolerance to Phytophthora root rot in the soybean cultivar ‘Conrad’. Euphytica. 2007;158:81–86. doi: 10.1007/s10681-007-9428-0. [DOI] [Google Scholar]
- 41.Han Y, Teng W, Yu K, Poysa V, Anderson T, Qiu L, Lightfoot DA, Li W. Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica. 2008;162:231–239. doi: 10.1007/s10681-007-9558-4. [DOI] [Google Scholar]
- 42.Li X, Han Y, Teng W, Zhang S, Yu K, Poysa V, Anderson T, Ding J, Li W. Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’ and ‘Hefeng 25’. Theor Appl Genet. 2010;121:651–658. doi: 10.1007/s00122-010-1337-2. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Waller L, Tripathy S, St Martin SK, Zhou L, Krampis K, Tucker DM, Mao Y, Hoeschele I, Maroof S, Tyler BM, Dorrance AE. Analysis of genes underlying soybean quantitative trait loci conferring partial resistance to Phytophthora sojae. Plant Genome. 2010;3:23–40. doi: 10.3835/plantgenome2009.12.0029. [DOI] [Google Scholar]
- 44.Stasko A, Wickramasinghe D, Nauth B, Acharya B, Ellis M, Taylor C, McHale L, Dorrance A. High density mapping of resistance QTL towards Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Sci. 2016 [Google Scholar]
- 45.Tucker D, Maroof S, Mideros S, Skoneczka J, Nabati D, Buss G, Hoeschele I, Tyler B, St. Martin S, Dorrance A. Mapping quantitative trait loci for partial resistance to Phytophthora sojae in a soybean Interspecific Cross. Crop Sci. 2010;50:628–635. doi: 10.2135/cropsci2009.03.0161. [DOI] [Google Scholar]
- 46.Wu X, Zhou B, Zhao J, Guo N, Zhang B, Yang F, Chen S, Gai J, Xing H. Identification of quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Plant Breed. 2011;130:144–149. doi: 10.1111/j.1439-0523.2010.01799.x. [DOI] [Google Scholar]
- 47.Lee S, Mian MR, Sneller CH, Wang H, Dorrance AE, McHale LK. Joint linkage QTL analyses for partial resistance to Phytophthora sojae in soybean using six nested inbred populations with heterogeneous conditions. Theor Appl Genet. 2014;127:429–444. doi: 10.1007/s00122-013-2229-z. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen V, Vuong T, VanToai T, Lee J, Wu X, Mian M, Dorrance A, Shannon J, Nguyen H. Mapping of quantitative trait loci associated with resistance to Phytophthora sojae and flooding tolerance in soybean. Crop Sci. 2012;52:2481–2493. doi: 10.2135/cropsci2011.09.0466. [DOI] [Google Scholar]
- 49.Lee S, Mian MR, McHale LK, Wang H, Wijeratne AJ, Sneller CH, Dorrance AE. Novel quantitative trait loci for partial resistance to Phytophthora sojae in soybean PI 398841. Theor Appl Genet. 2013;126:1121–1132. doi: 10.1007/s00122-013-2040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Mian R, McHale LK, Sneller CH, Dorrance AE. Identification of quantitative trait loci conditioning partial resistance to Phytophthora sojae in soybean PI 407861A. Crop Sci. 2013;53:1022–1031. doi: 10.2135/cropsci2012.10.0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One. 2013;8:e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, St Martin SK, Dorrance AE. Comparison of phenotypic methods and yield contributions of quantitative trait loci for partial resistance to in soybean. Crop Sci. 2012;52:609–622. doi: 10.2135/cropsci2011.06.0323. [DOI] [Google Scholar]
- 53.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 55.Sun J, Guo N, Lei J, Li L, Hu G, Xing H. Association mapping for partial resistance to Phytophthora sojae in soybean (Glycine max (L.) Merr.) J Genet. 2014;93:355–363. doi: 10.1007/s12041-014-0383-y. [DOI] [PubMed] [Google Scholar]
- 56.Li L Gua N, Niu J, Wang Z, Cui X, Sun J, Zhao T, Xing H. Loci and candidate gene identification for resistance to Phytophthora sojae via association analysis in soybean [Glycine max (L.) Merr.]. Mol Genet Genomics. 2016. doi: 10.1007/s00438-015-1164-x. [DOI] [PubMed]
- 57.Bastien M, Sonah H, Belzile F. Genome wide association mapping of Sclerotinia sclerotiorum resistance in soybean with a genotyping-by-sequencing approach. Plant Genome. 2014;7:1–13. doi: 10.3835/plantgenome2013.10.0030. [DOI] [Google Scholar]
- 58.Wen Z, Tan R, Yuan J, Bales C, Du W, Zhang S, Chilvers MI, Schmidt C, Song Q, Cregan PB. Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genomics. 2014;15:809. doi: 10.1186/1471-2164-15-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iquira E, Humira S, Francois B. Association mapping of QTLs for sclerotinia stem rot resistance in a collection of soybean plant introductions using a genotyping by sequencing (GBS) approach. BMC Plant Biol. 2015;15:5. doi: 10.1186/s12870-014-0408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han Y, Zhao X, Cao G, Wang Y, Li Y, Liu D, Teng W, Zhang Z, Li D, Qiu L. Genetic characteristics of soybean resistance to HG type 0 and HG type 1.2. 3.5. 7 of the cyst nematode analyzed by genome-wide association mapping. BMC Genomics. 2015;16(16):1. doi: 10.1186/s12864-015-1800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant D, Nelson RT, Cannon SB, Shoemaker RC. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38:D843–D846. doi: 10.1093/nar/gkp798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arahana VS, Graef GL, Specht JE, Steadman JR, Eskridge KM. Identification of QTLs for Resistance to Sclerotinia sclerotiorum in Soybean. Crop Sci. 2001;41:180–188. doi: 10.2135/cropsci2001.411180x. [DOI] [Google Scholar]
- 63.Sugimoto T, Yoshida S, Watanabe K, Aino M, Kanto T, Maekawa K, Irie K. Identification of SSR markers linked to the Phytophthora resistance gene Rps1‐d in soybean. Plant Breed. 2008;127:154–159. doi: 10.1111/j.1439-0523.2007.01440.x. [DOI] [Google Scholar]
- 64.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gómez-Gómez L, Boller T. FLS2 an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/S1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 66.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 67.Kemmerling B, Halter T, Mazzotta S, Mosher S, Nürnberger T. A genome-wide survey for Arabidopsis leucine-rich repeat receptor kinases implicated in plant immunity. Front Plant Sci. 2011;2:88. doi: 10.3389/fpls.2011.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becraft PW. Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol. 2002;18:163–192. doi: 10.1146/annurev.cellbio.18.012502.083431. [DOI] [PubMed] [Google Scholar]
- 69.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 70.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 71.Matern U, Grimmig B, Kneusel RE. Plant cell wall reinforcement in the disease-resistance response: molecular composition and regulation. Can J Bot. 1995;73:511–517. doi: 10.1139/b95-290. [DOI] [Google Scholar]
- 72.Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015;25:162–172. doi: 10.1016/j.pbi.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morita-Yamamuro C, Tsutsui T, Sato M, Yoshioka H, Tamaoki M, Ogawa D, Matsuura H, Yoshihara T, Ikeda A, Uyeda I, Yamaguchi J. The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol. 2005;46:902–912. doi: 10.1093/pcp/pci095. [DOI] [PubMed] [Google Scholar]
- 74.Ferreira RB, Monteiro S, Freitas R, Santos CN, Chen Z, Batista LM, Duate J, Borges A, Teixeira AR. The role of plant defence proteins in fungal pathogenesis. Mol Plant Pathol. 2007;8:677–700. doi: 10.1111/j.1364-3703.2007.00419.x. [DOI] [PubMed] [Google Scholar]
- 75.Howles PA, Birch RJ, Collings DA, Gebbie LK, Hurley UA, Hocart CH, Arioli T, Williamson RE. A mutation in an Arabidopsis ribose 5‐phosphate isomerase reduces cellulose synthesis and is rescued by exogenous uridine. Plant J. 2006;48:606–618. doi: 10.1111/j.1365-313X.2006.02902.x. [DOI] [PubMed] [Google Scholar]
- 76.Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hernandez-Blanco C, Feng DX, Hu J, Sanchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sanchez-Rodriguez C, Anderson LK, Somerville S, Marco Y, Molina A. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell. 2007;19:890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci. 2006;27:587–593. doi: 10.1016/j.tips.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Nawrath C, Heck S, Parinthawong N, Metraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 2011;23:3812–3823. doi: 10.1105/tpc.111.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lahrmann U, Ding Y, Banhara A, Rath M, Hajirezaei MR, Dohlemann S, von Wiren N, Parniske M, Zuccaro A. Host-related metabolic cues affect colonization strategies of a root endophyte. Proc Natl Acad Sci U S A. 2013;110:13965–13970. doi: 10.1073/pnas.1301653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Favre P, Bapaume L, Bossolini E, Delorenzi M, Falquet L, Reinhardt D. A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol. 2014;14:333. doi: 10.1186/s12870-014-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pastor V, Gamir J, Camañes G, Cerezo M, Sánchez-Bel P, Flors V. Disruption of the ammonium transporter AMT1.1 alters basal defenses generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina. Front Plant Sci. 2014;5:231. doi: 10.3389/fpls.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Waterhouse PM, Wang M, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 85.Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi DS, Kim YJ, Hwang BK. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta. 2008;227:539–558. doi: 10.1007/s00425-007-0637-5. [DOI] [PubMed] [Google Scholar]
- 86.Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell. 2005;17:2832–2847. doi: 10.1105/tpc.105.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee DS, Kim BK, Kwon SJ, Jin HC, Park OK. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem Biophys Res Commun. 2009;379:1038–1042. doi: 10.1016/j.bbrc.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 88.Blée E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002;7:315–322. doi: 10.1016/S1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- 89.Njiti V, Lightfoot D. Genetic analysis infers Dt loci underlie resistance to Fusarium solani f. sp. glycines in indeterminate soybeans. Can J Plant Sci. 2006;86:83–90. doi: 10.4141/P05-046. [DOI] [Google Scholar]
- 90.Iqbal M, Meksem K, Njiti V, Kassem MA, Lightfoot D. Microsatellite markers identify three additional quantitative trait loci for resistance to soybean sudden-death syndrome (SDS) in Essex × Forrest RILs. Theor Appl Genet. 2001;102:187–192. doi: 10.1007/s001220051634. [DOI] [Google Scholar]
- 91.Njiti V, Meksem K, Iqbal M, Johnson J, Kassem MA, Zobrist K, Kilo V, Lightfoot D. Common loci underlie field resistance to soybean sudden death syndrome in Forrest, Pyramid, Essex, and Douglas. Theor Appl Genet. 2002;104:294–300. doi: 10.1007/s001220100682. [DOI] [PubMed] [Google Scholar]
- 92.Kazi S, Shultz J, Afzal J, Johnson J, Njiti V, Lightfoot DA. Separate loci underlie resistance to root infection and leaf scorch during soybean sudden death syndrome. Theor Appl Genet. 2008;116:967–977. doi: 10.1007/s00122-008-0728-0. [DOI] [PubMed] [Google Scholar]
- 93.Abdelmajid KM, Ramos L, Leandro L, Mbofung G, Hyten DL, Kantartzi SK, Njiti VN, Cianzio S, Meksem K. The ‘PI 438489B’ by ‘Hamilton’ SNP-based genetic linkage map of soybean [Glycine max (L.) Merr.] identified quantitative trait loci that underlie seedling SDS resistance. J Plant Genome Sci. 2012;1:18–30. doi: 10.5147/jpgs.2012.0053. [DOI] [Google Scholar]
- 94.Wu X, Blake S, Sleper DA, Shannon JG, Cregan P, Nguyen HT. QTL, additive and epistatic effects for SCN resistance in PI 437654. Theor Appl Genet. 2009;118:1093–1105. doi: 10.1007/s00122-009-0965-x. [DOI] [PubMed] [Google Scholar]
- 95.Concibido VC, Lange DA, Denny RL, Orf JH, Young ND. Genome mapping of soybean cyst nematode resistance genes in ‘Peking’, PI 90763, and PI 88788 using DNA markers. Crop Sci. 1997;37:258–264. doi: 10.2135/cropsci1997.0011183X003700010046x. [DOI] [Google Scholar]
- 96.Vuong T, Sleper D, Shannon J, Wu X, Nguyen H. Confirmation of quantitative trait loci for resistance to multiple-HG types of soybean cyst nematode (Heterodera glycines Ichinohe) Euphytica. 2011;181:101–113. doi: 10.1007/s10681-011-0430-1. [DOI] [Google Scholar]
- 97.Yue P, Arelli P, Sleper D. Molecular characterization of resistance to Heterodera glycines in soybean PI 438489B. Theor Appl Genet. 2001;102:921–928. doi: 10.1007/s001220000453. [DOI] [Google Scholar]
- 98.Ferreira MFDS, Cervigni GDL, Ferreira A, Schuster I, Santana FA, Pereira WD, Barros EG, Moreira MA. QTLs for resistance to soybean cyst nematode, races 3, 9, and 14 in cultivar Hartwig. Pesq Agrop Brasileira. 2011;46:420–428. doi: 10.1590/S0100-204X2011000400012. [DOI] [Google Scholar]
- 99.Winter SM, Shelp BJ, Anderson TR, Welacky TW, Rajcan I. QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor Appl Genet. 2007;114:461–472. doi: 10.1007/s00122-006-0446-4. [DOI] [PubMed] [Google Scholar]
- 100.Chang W, Dong L, Wang Z, Hu H, Han Y, Teng W, Zhang H, Guo M, Li W. QTL underlying resistance to two HG types of Heterodera glycines found in soybean cultivar ‘L-10’. BMC Genomics. 2011;12:233. doi: 10.1186/1471-2164-12-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang D, Diers B, Arelli P, Shoemaker R. Loci underlying resistance to race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor Appl Genet. 2001;103:561–566. doi: 10.1007/PL00002910. [DOI] [Google Scholar]
- 102.Qiu B, Arelli P, Sleper D. RFLP markers associated with soybean cyst nematode resistance and seed composition in a ‘Peking’ × ‘Essex’ population. Theor Appl Genet. 1999;98:356–364. doi: 10.1007/s001220051080. [DOI] [Google Scholar]
- 103.Schuster I, Abdelnoor R, Marin S, Carvalho V, Kiihl R, Silva J, Sediyama C, Barros E, Moreira M. Identification of a new major QTL associated with resistance to soybean cyst nematode (Heterodera glycines) Theor Appl Genet. 2001;102:91–96. doi: 10.1007/s001220051622. [DOI] [Google Scholar]
- 104.Guo X, Wang D, Gordon SG, Helliwell E, Smith T, Berry SA, St Martin SK, Dorrance AE. Genetic mapping of QTLs underlying partial resistance to Sclerotinia sclerotiorum in soybean PI 391589A and PI 391589B. Crop Sci. 2008;48:1129–1139. doi: 10.2135/cropsci2007.04.0198. [DOI] [Google Scholar]
- 105.Huynh T, Bastien M, Iquira E, Turcotte P, Belzile F. Identification of QTLs associated with partial resistance to white mold in soybean using field-based inoculation. Crop Sci. 2010;50:969–979. doi: 10.2135/cropsci2009.06.0311. [DOI] [Google Scholar]
- 106.Olhoft PM, Donovan CM, Somers DA. Soybean (Glycine max) transformation using mature cotyledonary node explants. Agrobacter Protoc. 2006;343:385–396. doi: 10.1385/1-59745-130-4:385. [DOI] [PubMed] [Google Scholar]
- 107.Dorrance AE, Berry SA, Anderson TR, Meharg C. Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant Health Prog. 2008 [Google Scholar]
- 108.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song QJ, Hyten DL, Jia GF, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Fingerprinting soybean germplasm and its utility in genomic research. G3-Genes Genom Genet. 2015;5:1999–2006. doi: 10.1534/g3.115.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 111.R Core Team. R . A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 112.Zhang Z, Ersoz E, Lai C, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guo X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:362. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 114.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are available with the article and additional files or on request from the corresponding author.


