Abstract
Course-based undergraduate research experiences (CUREs) provide an opportunity for students to engage in experiments with outcomes that are unknown to both the instructor and students. These experiences allow students and instructors to collaboratively bridge the research laboratory and classroom, and provide research experiences for a large number of students relative to traditional individual mentored research. Here, we describe a molecular biology CURE investigating the impact of clinically relevant mutations found in the bromodomain of the p300 transcriptional regulator on acetylated histone interaction. In the CURE, students identified missense mutations in the p300 bromo-domain using the Catalogue of Somatic Mutations in Cancer (COSMIC) database and hypothesized the effects of the mutation on the acetyl-binding function of the domain. They cloned and purified the mutated bromodomain and performed peptide pulldown assays to define its potential to bind to acetylated histones. Upon completion of the course, students showed increased confidence performing molecular techniques and reported positively on doing a research project in class. In addition, results generated in the classroom were further validated in the research laboratory setting thereby providing a new model for faculty to engage in both course-based and individual undergraduate research experiences.
Keywords: course-based undergraduate research experience, protein purification, peptide pulldown assay, bromodomain
Introduction
Histones are nuclear proteins that play a fundamental role in the organization and packaging of eukaryotic DNA. An octamer of histone proteins (two copies each of H3, H4, H2A, H2B) wrap ~147 bp of DNA into nucleosomes, where the N- and C-terminal tails protrude out from the core nucleosome structure. These tails are decorated with a myriad of post-translational modifications (PTMs), such as acetylation, phosphorylation and methylation. Histone PTMs can act as binding platforms for proteins that regulate key cellular processes including transcription, DNA repair and DNA replication [1]. Currently, there is much interest in defining how histone PTMs influence both normal development and disease [2–4]. The cooperative nature of histone PTMs and recruitment of effector proteins that “read” or bind to these histone PTMs compose a complex and profound regulatory epigenetic mechanism.
Histone acetylation is a PTM associated with active transcription and serves as a recognition site for proteins that contain bromodomains. The bromodomain is a conserved acetyl-lysine binding motif found in 46 human proteins [5]. p300 is a key bromodomain-containing protein that interacts with at least 400 other proteins to regulate transcription [6] (Fig. 2A). Its acetyl-transferase domain catalyzes lysine acetylation of histone and non-histone proteins, which contributes to its role as a transcription coactivator [6–9]. The p300 bromodomain has also been shown to be critical for coactivator function and histone acetylation in vitro [9]. In cervical cancer cells, loss of the p300 bromodomain also compromises the transcriptional effects of p300 [10]. p300 is mutated in a significant number of endometrial cancers (www.tumorportal.org), but the functional effects of p300 bromodomain mutations have not been explored.
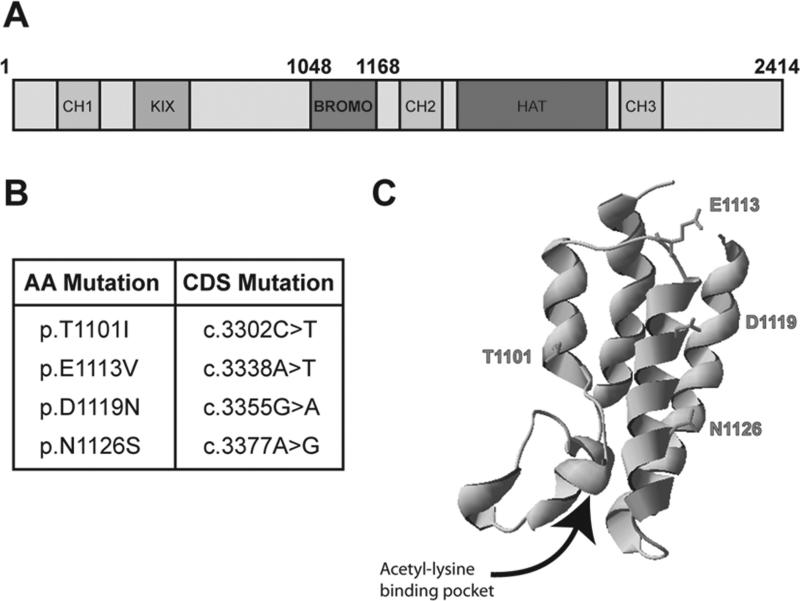
FIG 2.
Structural organization of p300 and bromodomain mutations identified in endometrial cancers. A) Schematic of the domain organization of p300. The bromodomain (BROMO) is shown in pink. B) Table of the bromodomain mutations identified in endometrial cancers using the COSMIC database. C) Crystal structure of the p300 bromodomain [5] showing the acetyl-lysine binding pocket. Mutated residues are highlighted in pink. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Here, we describe a course-based undergraduate research experience (CURE) exploring p300 bromodomain mutations in an upper-level undergraduate molecular biology course (Fig. 1). Undergraduate research experiences have been shown to improve understanding of science and lab techniques [11], and have been proposed as an important part of the undergraduate curriculum [12]. Indeed, independent undergraduate research experiences have been found to improve interest in pursuing a Ph.D. [13, 14] and increase analytic and technical skills [15, 16]. CUREs allow many students to participate in research in a course setting and can similarly improve students’ analytical and technical skills [11]. There are several defining features of CUREs, including the use of scientific practices such as developing hypotheses, the potential to discover something new, participation in a broader scientific context, collaboration and iteration [17]. A defining feature of CUREs is the participation in a research question with an outcome that is unknown to both instructor and student [17]. In addition, CUREs can bridge laboratory research and the classroom to engage students in experiments and projects that can be directly followed up in a research setting [18].
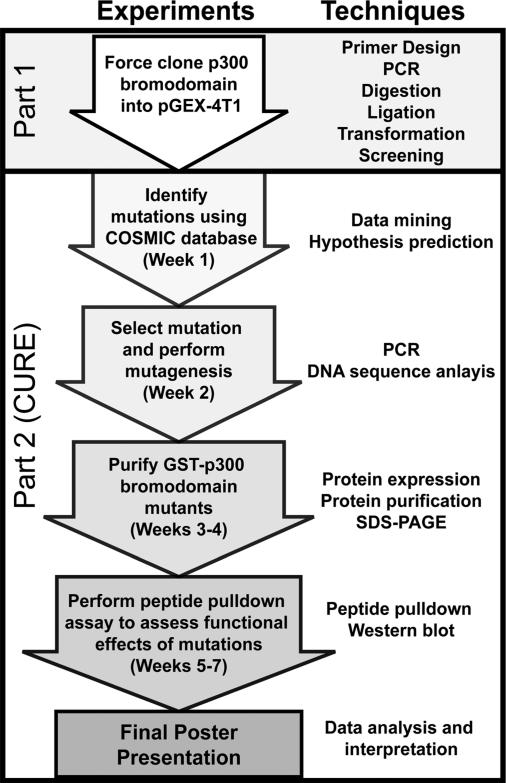
FIG 1.
Diagram showing the outline of the CURE. In Part 1 of the course, students used molecular cloning techniques to create a protein expression vector containing the wild-type p300 bromodomain. In Part 2 of the course (described in this report), students identified and create cancer-relevant mutations and tested the functional effects of these mutations using peptide pulldown experiments.
In this report, we describe a course module in which students identified and selected p300 bromodomain mutations identified from the Catalogue of Somatic Mutations in Cancer (COSMIC) database for further functional experiments. Overall, students in the CURE setting showed signifi-cant gains in confidence performing molecular techniques and positively reported on the course as a research experience. With hundreds of histone PTM-binding domains and thousands of cancer genomic sequences, this report provides a framework to build future CUREs aimed at determining the functional effects of cancer-relevant mutations on histone PTM binding domains. The students’ results were carefully validated after the course, demonstrating the transition of research from the course setting to the research laboratory.
Course Organization and Assessment
Students performed the experiments described here in BIO4130 Molecular Biology (an upper-level biology elective) at the University of North Carolina at Pembroke. Sixteen senior undergraduate students participated in the course, which was a blend of lab work and lectures consisting of one 75 min and one 115 min session each week for 16 weeks. Students were required to pass a pre-requisite genetics course (BIO3180 – Principles of Genetics) prior to enrolling in the course. Throughout the course, students worked in groups of two for all experiments. During the first seven weeks of the course (Part 1), students learned and practiced basic molecular biology techniques (i.e., PCR, DNA cloning and SDS-PAGE) to clone the p300 bromodo-main (amino acids 1020 to 1180) into a protein expression vector (pGEX) based on a previously published cloning course [19]. The second half of the semester consisted of the seven week CURE module described in this report (Fig. 1). In order to efficiently implement this research experience in the classroom, an individual mentored undergraduate researcher assisted in preparing materials and performing preliminary and validation experiments.
In order to address three key components of undergraduate science education (critical thinking, data interpretation, and communication) [20], the course was assessed using in-class and homework assignments that emphasized the interpretation of primary literature, exams focused on data analysis and critical thinking, a series of written lab reports and a final poster presentation. A detailed rubric for the poster is provided in Supporting Information File 1. Prior to each lab period, students had to prepare lab notebooks and take an on-line pre-laboratory test. In addition, students completed anonymous preand post-assessment surveys to assess changes in confidence performing molecular techniques and interest in pursuing research careers. Surveys were approved by the UNC-Pembroke Institutional Review Board under protocol #15-01-003.
Student Learning Objectives
The following learning objectives were designed for this CURE module:
Utilize cancer genome databases to identify cancer-relevant mutations.
Evaluate the effects of amino acid substitutions on protein–protein interaction.
Perform site-directed mutagenesis to create point mutations in a protein sequence.
Communicate findings, discuss pitfalls, and propose future experiments related to the laboratory project.
Laboratory Materials and Methods
Detailed protocols, including reagent lists and pre-laboratory questions, for each lab period are provided in Supporting Information File 2.
Identification of p300 Bromodomain Mutations Using COSMIC
The Catalogue of Somatic Mutations (COSMIC) database is a collection of cancer genome sequences that can be easily accessed and mined to identify mutations that occur in different types of cancer [21]. Students watched two video tutorials and completed a worksheet prior to searching the database for p300 bromodomain mutations in class. To perform the search, students identified and selected a cancer type in which p300 was found to be significantly mutated using www.tumorportal.org [22]. Students then used the COSMIC database to identify missense mutations in the p300 bromodomain that have been found in endome-trial cancers (see Supporting Information File 2, Week 1).
Site-Directed Mutagenesis
Site-directed mutagenesis was performed using the Quick-Change protocol (Stratagene) as detailed in Supporting Information File 2, Week 2. Primer sequences for each point mutant are provided in Supporting Information File 3. The template for the mutagenesis reactions was pGEX-4T1 containing the p300 bromodomain (residues 1020–1180) cloned into the EcoRI and SalI sites. Successful mutagenesis was confirmed by sequence analysis.
Protein Purification
Expression vectors were transformed into SoluBL21 competent E. coli cells (Genlantis) for protein expression and purification. Detailed protocols are provided in Supporting Information File 2 (Week 3 and 4). In brief, protein purification was performed using the MagneGST protein purification system (Promega) according to the manufacturer's protocol with slight modifications. Briefly, pellets were resuspended in lysis buffer supplemented with 1 mg/mL lysozyme (Sigma) and incubated with gentle rotation at 25°C for 30 min. An aliquot of the lysate was reserved for coomassie analysis, and the remaining lysate was added to pre-equilibrated MagneGST particles and incubated at 4°C for 1 hr. Beads were washed three times and the remaining protein was eluted two times with 150 μL of elution buffer. Samples of the flow through (supernatant collected after 4°C incubation) and the combined elutions were analyzed by 10% SDS-PAGE and Bio-Safe coomassie stain (BioRad) according to the manufacturer's protocol.
Peptide Pulldown Assay and Western Blot
Pull-down assays were performed in peptide binding buffer (PBB) [50mM Tris-HCl pH 8.0, 300mM NaCl, 0.1% NP-40 v/v] based on previously described protocols [23, 24]. Biotinylated peptides (500 pmol) were immobilized on Streptavidin MagneSphere® Paramagnetic Particles (Promega) for 30 min at 25°C. Peptide sequences are provided in Supporting Information File 2 (Week 5). Particles were washed twice with 1 mL PBB, and particles were incubated with 40 pmol of GST protein in PBB + 0.5% bovine serum albumin (BSA; Sigma Aldrich) for 1 hr at 4°C. A small aliquot of the input protein solution was reserved for analysis by western blotting. After three washes with PBB, bound proteins were eluted by boiling for 5 min in 1× SDS Laemmli buffer. Samples were analyzed for the presence of the GST-tagged bromodomain using 10% SDS-PAGE and transferred (BioRad PROTEAN system) to PVDF membrane (BioRad). Membranes were dried and stored at 4°C for one week. Rehydrated membranes were then incubated in GST antibody (Sigma) for 1 hr at 25°C, washed three times with phosphate buffered saline (PBS) + 0.1% Tween-20 and incubated with HRP-conjugated secondary antibody. The western blotting protocol is detailed in Supporting Information File 2, Weeks 6 and 7.
Results and Discussion
Identification and Selection of p300 Bromodomain Mutations (Weeks 1–2)
Four mutations found in endometrial cancers were identified using the COSMIC database (Fig. 2B). In class, these residues were mapped by the instructor to the p300 bromo-domain structure to stimulate discussion about how they may affect the function of the domain (Fig. 2C). Student pairs selected one mutation to study after developing a hypothesis to predict if the mutation would affect the structure and function of the domain. Through class discussion, two aspects of the mutation were decided to be the most important for predicting if the mutation would affect the function of the domain: 1) a change in the type of amino acid (hydrophobic to polar, for example); and 2) close proximity to the acetyl-lysine binding pocket. In the following lab report, students were assessed on their ability to communicate their hypothesis and the reasoning behind it. Students then designed primers to mutate the residue and performed site-directed mutagenesis to create the mutant bromodomain.
Protein Purification and Peptide Pulldown Assay (Weeks 3–7)
Students next performed protein purification to isolate the mutated protein domain. Prior to the purification, students were required to predict what the resulting coomassie after purification would look like. This helped them understand where the protein was throughout the purification process and ensured they understood which samples to keep for analysis. In all cases, protein purification and quantitation was successful (representative data shown in Fig. 3A). There was some apparent protein degradation, and it may be beneficial to add additional protease inhibitors in the future. Students successfully quantified the protein and calculated the amount of protein required for the peptide pulldown experiment.
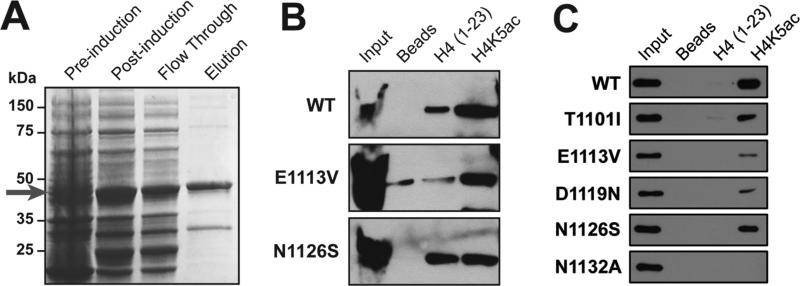
FIG 3.
Protein purification and peptide pulldown analysis performed by students in the molecular CURE module. A) Example of student-generated SDS-PAGE and coomassie stain analysis of protein purification. The elution lane contains the purified GST-p300 bromodomain as indicated by the red arrow. B) Example of student-generated western blot results from peptide pulldown experiments performed with the indicated p300 bromodomain mutation. C) Validation of student generated results in the research laboratory setting showing negligent effects of the p300 bromodomain mutations on the acetyl-lysine binding function of the domain. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Peptide pulldown experiments were performed using unmodified or acetylated histone H4 to assess functional affects of the mutations (Fig. 3B). There are many potential pitfalls with this assay, but all of the students successfully obtained the elutions. There are several critical steps in the peptide pulldown assay: 1) equal addition of protein to each tube; 2) careful washing of the beads after protein binding; 3) elution in equal volumes of SDS buffer. In order to mitigate potential failures, students made a large solution of protein and aliquoted a large volume of the mixture to each tube containing magnetic particles. Students were also carefully shown the proper washing technique and prompted to centrifuge each tube to collect all liquid before placing it on the magnetic rack to collect the particles. Finally, a large volume of elution buffer was added to minimize effects of pipetting error.
Overall, all of the groups successfully performed the assay, and representative data are shown in Fig. 3B. None of the mutations disrupted the interaction with histone H4 peptide that was acetylated at lysine 5 (H4K5ac, Fig. 3B). Some interaction with unmodified H4 peptide was observed, but this may have been due to technical error during washing. If any liquid remained in the tube after removing the unbound or wash buffer, some remaining unbound protein can be detected in the pulldown elution. One mutation (N1126S) seemed to strongly increase the interaction with unmodified H4 peptide (Fig. 3B). The interaction between the mutant p300 bromodomain proteins was then carefully validated by an undergraduate researcher independently in the research setting using an additional mutant (N1132A) that disrupts the acetyl interaction [25] (Fig. 3C). Indeed, mutating N1132 to alanine disrupted the bromodomain interaction with H4K5ac peptide, and two of the mutants identified in endometrial cancers (E1113V and D1119N) impaired the interaction with H4K5ac peptide (Fig. 3C). However, N1126S did not show increased interaction with unmodified H4, as demonstrated by students in the class, thereby suggesting that the binding observed in the class may likely have been due to improper washing technique. Collectively, these results suggest that further experiments to assess the binding affinities and coactivator functions of these mutants are warranted.
Student Gains and Feedback
Learning objectives for the CURE were assessed using exams, lab reports, assignments, and a final poster presentation. The lab reports and final poster presentation indicated that students were able to develop a hypothesis and interpret the results of the peptide pulldown assay. In addition, several students proposed compelling future directions, such as “testing if two mutations combined would affect the function of the bromodomain” or seeing “if the mutation affected interaction with other non-histone proteins”.
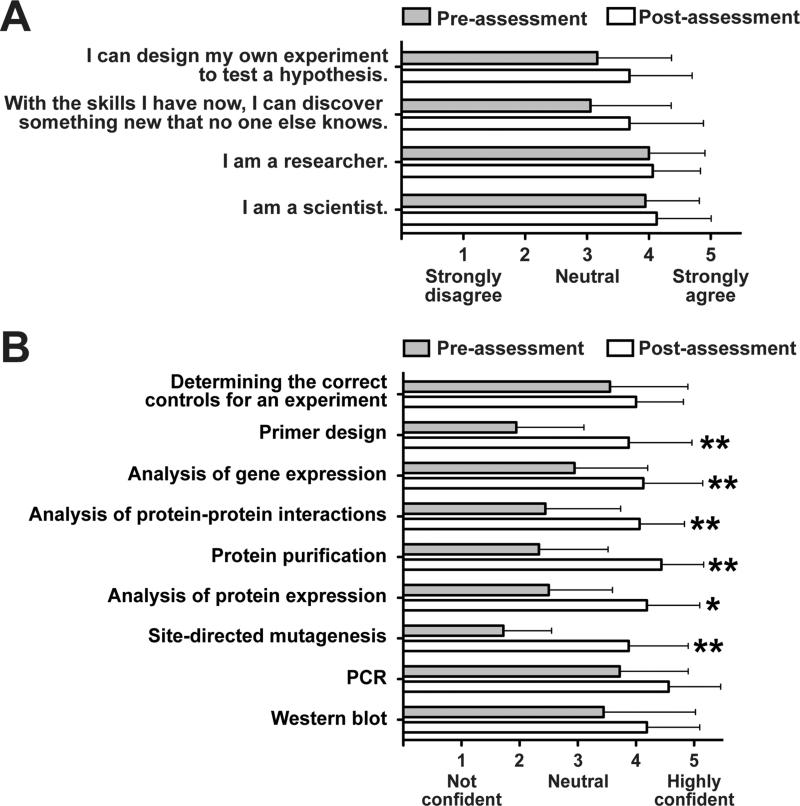
Students also performed anonymous, ungraded pre- and post-assessments to measure changes self-identification as a scientist. There were no significant changes in self-identification as a scientist or future plans to pursue graduate studies or a career in research, probably because most students already strongly identified as a scientist and researcher (Fig. 4A). However, there were significant gains in confidence performing molecular techniques that were used in the course (Fig. 4B). PCR, for example, was performed in pre-requisite labs, so most students already showed some confidence performing this technique. Finally, students responded favorably in an open ended-response regarding how the research project affected the experience in the course. Students felt the “project made the experience more realistic,” “helped me learn to do research, which I haven't really done before,” and the “research made it easier to understand the rest of the course”.
FIG 4.
Assessment of (A) self-identification as a scientist and (B) confidence performing molecular techniques before and after the CURE course. Students were asked to rate their agreement with a statement or confidence performing molecular techniques used throughout the CURE module on a scale of 1 to 5. * p < 0.05, ** p < 0.01 as determined by Chi-square tests.
Conclusions and Future Directions
The CURE described here provides a model for integrating epigenetics research in the classroom with basic research in the laboratory. Hundreds of histone PTMs have been detected in cells, and it is still unclear how these modifications function to recruit proteins, both individually and in combination [26]. Many ‘reader’ domains of histone modifications have been characterized, including bromodomains, tudor domains and PHD domains [27]. Mutations in reader domains could disrupt the function of a given protein to contribute to diseases, such as cancer. Given the ease at which researchers and students can access genetic data from cancers using databases such as COSMIC, we can start to ask questions and design experiments aimed at testing the effects of cancer mutations on the functions of epigenetic reader domains. This report provides a template for bringing these research questions to the classroom, and to allow students to engage in experiments with unknown outcomes. The positive feedback and confidence gains documented here demonstrate that this teaching strategy is an effective way to teach students fundamental molecular biology techniques and allow a large number of students to experience the thrill of scientific discovery in a classroom setting.
Supplementary Material
Acknowledgements
The authors thank the students and faculty of UNC-Pembroke for support in developing and implementing this course, especially Dr. Robert Poage and Takisha Bullard. In addition, Dr. Claire Gordy and Dr. Carlos Goller provided useful feedback during the development of the course. Dr. Krzysztof Krajewski provided aliquots of the peptides, and Dr. Jenny Kerschner provided feedback during the implementation and analysis of the course. This work was funded by a grant from the National Institute of General Medical Sciences, division of Training, Workforce Development, and Diversity under the Institutional Research and Academic Career Development Award, grant #K12-GM000678 and a grant from the National Science Foundation to B. D. S, grant # 1330320.
Abbreviations
- COSMIC
catalogue of somatic mutations in cancer
- CURE
course-based undergraduate research experience
- PBB
peptide binding buffer
- PBS
phosphate buffered saline
- PTM
post-translational modification
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Butler JS, Koutelou E, Schibler AC, Dent SYR. His-tone-modifying enzymes: Regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldmann T, Schneider R. Targeting histone modifications—Epigenetics in cancer. Curr. Opin. Cell. Biol. 2013;25:184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015;115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 8.Josling GA, Selvarajah SA, Petter M, Duffy MF. The role of bromodomain proteins in regulating gene expression. Genes (Basel) 2012;3:320–343. doi: 10.3390/genes3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohshima T, Suganuma T, Ikeda M. A novel mutation lacking the bromodomain of the transcriptional coactivator p300 in the SiHa cervical carcinoma cell line. Biochem. Biophys. Res. Commun. 2001;281:569–575. doi: 10.1006/bbrc.2001.4389. [DOI] [PubMed] [Google Scholar]
- 11.Corwin LA, Graham MJ, Dolan EL. Modeling course-based undergraduate research experiences: An agenda for future research and evaluation. CBE Life Sci. Educ. 2015;14:es1. doi: 10.1187/cbe.14-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Association for the Advancement of Sciences . Vision and change in undergraduate biology education: A call to action. Washington, DC: 2011. [May 23, 2015]. http://visionandchange.org/finalreport/. [Google Scholar]
- 13.Lopatto D. Undergraduate research experiences support science career decisions and active learning. CBE Life Sci. Educ. 2007;6:297–306. doi: 10.1187/cbe.07-06-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eagan MK, Hurtado S, Chang MJ, et al. Making a difference in science education: The impact of undergraduate research programs. Am. Educ. Res. J. 2013;50:683–713. doi: 10.3102/0002831213482038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopatto D. Survey of Undergraduate Research Experiences (SURE): First findings. Cell Biol. Educ. 2004;3:270–277. doi: 10.1187/cbe.04-07-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter A-B, Laursen SL, Seymour E. Becoming a scientist: The role of undergraduate research in students’ cognitive, personal, and professional development. Sci. Educ. 2007;91:36–74. [Google Scholar]
- 17.Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. Assessment of course-based undergraduate research experiences: A meeting report. CBE Life Sci. Educ. 2014;13:29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukami T. IBI* series winner. Integrating inquiry-based teaching with faculty research. Science. 2013;339:1536–1537. doi: 10.1126/science.1229850. [DOI] [PubMed] [Google Scholar]
- 19.Miller H, Witherow DS, Carson S. Molecular Biology Techniques: A Classroom Laboratory Manual. Academic Press; London, UK: 2011. [Google Scholar]
- 20.Coil D, Wenderoth MP, Cunningham M, Dirks C. Teaching the process of science: Faculty perceptions and an effective methodology. CBE Life Sci. Educ. 2010;9:524–535. doi: 10.1187/cbe.10-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malecek K, Ruthenburg A. Validation of histone-binding partners by peptide pull-downs and isothermal titration calorimetry. Methods Enzymol. 2012;512:187–220. doi: 10.1016/B978-0-12-391940-3.00009-3. [DOI] [PubMed] [Google Scholar]
- 24.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 26.Huang H, Lin S, Garcia BA, Zhao Y. Quantitative proteomic analysis of histone modifications. Chem. Rev. 2015;115:2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musselman CA, Lalonde M-E, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.