Abstract
The Kelch-like ECH-associated protein 1 (Keap1)/nuclear factor erythroid 2-related factor 2 (Nrf2)/antioxidant response elements pathway enables cells to survive oxidative stress conditions through regulating the expression of cytoprotective enzymes such as NAD(P)H:quinone oxidoreductase 1 (NQO1). This work presents the design and synthesis of novel anilinoquinazoline derivatives (2–16a) and evaluation of their NQO1 inducer activity in murine cells. Molecular docking of the new compounds was performed to assess their ability to inhibit Keap1–Nrf2 protein–protein interaction through occupying the Keap1–Nrf2-binding domain, which leads to Nrf2 accumulation and enhanced gene expression of NQO1. Docking results showed that all compounds can potentially interact with Keap1; however, 1,5-dimethyl-2-phenyl-4-(2-phenylquinazolin-4-ylamino)-1,2-dihydropyrazol-3-one (9), the most potent inducer, showed the largest number of interactions with key amino acids in the binding pocket (Arg483, Tyr525, and Phe478) compared to the native ligand or any other compound in this series.
Keywords: Kelch domain, molecular modeling, Keap1/Nrf2, cytoprotection, NQO1 induction
Introduction
Antioxidants, from a chemical viewpoint, are considered electron donors to free radicals, molecular centers that tend to lose electrons initiating oxidations, and thus protecting the cells against oxidative stress.1–3 Such oxidative stress situations, generated from imbalanced production of reactive oxygen species, for example, superoxide anion radical and hydrogen peroxide, are unfortunately associated with many pathological conditions including stroke, diabetes, Alzheimer’s disease, cancer, and chronic inflammation.4 As a natural mechanism to counteract oxidative stress, aerobic cells express superoxide dismutase converting superoxide to hydrogen peroxide, which is subsequently disposed of catalase and peroxidases. In addition to these enzymatic defenses, there are indirect antioxidants, namely, the cytoprotective enzymes, that catalyze a wide variety of chemical reactions, protecting cells and organisms and allowing their adaptation to many types of stress.5 Among the most critical cytoprotective enzymes is the Kelch-like ECH-associated protein 1 (Keap1)–nuclear factor erythroid 2-related factor 2 (Nrf2)–antioxidant response elements (AREs) pathway.6,7 The Keap1/Nrf2/ARE pathway enables the cells to adapt and survive oxidative and inflammatory stress conditions through regulating the expression of a network of >100 genes. This pathway is inducible by various stress stimuli and small molecules (termed inducers), whereby the inducers react with specific cysteine residues of the protein sensor Keap1, which loses its ability to target Nrf2 for ubiquitination and proteasomal degradation, resulting in its stabilization, followed by binding to the ARE and transcriptional activation of downstream target genes, such as NAD(P)H:quinone oxidoreductase 1 (NQO1).6,8–11 Recently, the Keap1–Nrf2 protein–protein interaction has been viewed as a critical target for intervention and potential management of a variety of oxidative stress-related pathologies, including cancer, Parkinson’s and Alzheimer’s disease, and diabetes.12–15 Design of noncovalent small-molecule modulators of the Keap–Nrf2 interaction has been intensively explored.16,17 Quinazoline derivatives are considered excellent bioactive substances where a number of biological activities have been associated with antioxidant activity.18–20
In continuation of our work toward identification of NQO1 inducers,21,22 we herein report the synthesis, NQO1 inducer activity, and Keap1 binding in silico screening results for a novel class of anilinoquinazolines.
Materials and methods
Chemistry
Melting points (uncorrected) were determined in open capillaries on a Gallen Kamp melting point apparatus (Gallemkamp, MSE Ltd., London, UK). Precoated silica gel plates (Kieselgel 0.25 mm, 60 G F254; EMD Millipore, Billerica, MA, USA) were used for thin layer chromatography. A solvent system of chloroform/methanol mixture (8:2 mL) was used as a developing system, and the spots were detected by ultraviolet light. Infrared (IR) spectra (KBr disk) were recorded using a Perkin Elmer FT-IR spectrophotometer (PerkinElmer Inc., Waltham, MA, USA). Nuclear magnetic resonance (NMR) spectra were scanned on a Bruker NMR spectrophotometer (Bruker AXS Inc., Madison, WI, USA), operating at 500 MHz for 1H spectra and 125.76 MHz for 13C spectra. Chemical shifts are expressed in δ-values (ppm) relative to tetramethylsilane as an internal standard, using dimethyl sulfoxide (DMSO)-d6 as a solvent. Elemental analyses were done on a model 2400 Perkin Elmer CHNSO analyzer (PerkinElmer Inc.). All the values were within ±0.4% of the theoretical values. All reagents used were of analytical reagent grades. The starting material 4-chloro-2-phenylquinazoline 1 was purchased from Sigma-Aldrich Co. (St Louis, MO, USA) and was directly used for preparation of the target compounds.
General procedure for the synthesis of 2-phenyl-quinazoline-4-amine derivatives (2–16a)
Compound 1 (2.40 g, 0.01 mol) and different amines (0.012 mol) were added to dry dimethylformamide (10 mL) containing three drops trimethylamine. Reaction mixture was refluxed for 24 hours, and solid product formed was filtrated and recrystallized from acetic acid to give 2–16a.
N-Heptyl-2-phenylquinazolin-4-amine (2)
Yield, 87%; melting point (mp) 141°C–143°C. IR (cm−1): 3,278 (NH), 3,074 (aromatic [arom]), 2,924, 2,950, 2,851 (aliphatic [aliph]), 1,639 (CN). 1H NMR: δ 0.9 (t, 3H, CH3), 1.2 [m, 10H, 5CH2], 3.8 [t, 2H, CH2-NH], 7.6–8.7 [m, 9H, Ar-H], 10.8 [s, 1H, NH]. 13C NMR: δ 14.3, 22.5, 26.8, 28.9, 31.5, 31.6, 42.1, 112.6, 120.3, 124.8 (2), 128.2, 129.3, 129.5, 131.7, 133.8, 135.5, 139.5, 157.3, 160.2. Mass spectroscopy (MS) m/z (%): 319 (M+) (3.23), 204 (100). Calculated for C21H25N3 (319): C, 78.96; H, 7.89; N, 13.15. Found: C, 78.59; H, 8.13; N, 12.81.
N-(Octan-2-yl)-2-phenylquinazolin-4-amine (3)
Yield, 80%; mp 192°C–193°C. IR (cm−1): 3,191 (NH), 3,100 (arom), 2,954, 2,925, 2,850 (aliph), 1,629 (CN). 1H NMR: δ 0.7 [t, 3H, CH3], 0.8 [d, 3H, CH3, J=7.2 Hz], 1.2–1.8 [m, 10H, 5CH2], 2.8 [m, 1H, CH], 7.6–8.8 [m, 9H, Ar-H], 10.0 [s, 1H, NH]. 13C NMR: δ 14.3, 20.3, 21.2, 25.8, 28.8, 31.6, 36.4, 48.6, 112.5, 120.4, 124.9 (2), 128.2, 129.4, 129.6 (2), 131.9, 133.8, 136.0, 157.5, 159.8, 160.6. MS m/z (%): 333 (M+) (12.7), 255 (100). Calculated for C22H27N3 (333.47): C, 79.24; H, 8.16; N, 12.60. Found: C, 79.50; H, 7.84; N, 12.25.
2-Phenyl-N-(2-(pyrrolidin-1-yl)ethyl)quinazolin-4-amine (4)
Yield, 90%; mp 193°C–194°C. IR (cm−1): 3,325 (NH), 358 (arom), 2,935, 2,846 (aliph), 1,617 (CN). 1H NMR: δ 1.4–1.5 [m, 4H, CH2-CH2 Cyclo], 2.4–2.5 [m, 4H, CH2-N-CH2 Cyclo], 2.6 [t, 2H, N-CH2], 3.8 [t, 2H, CH2-NH], 7.8–8.5 [m, 10H, Ar-H + NH]. 13C NMR: δ 24.5 (2), 40.5, 54.7, 57.7 (2), 114.3, 123.0, 125.6 (2), 128.3, 128.6, 128.7 (2), 130.4, 133.0, 139.2, 150.3, 159.7, 160.1. MS m/z (%): 318 (M+) (22.5), 247 (100). Calculated for C20H22N4 (318): C, 75.44; H, 6.96; N, 17.60. Found: C, 75.09; H, 6.63; N, 17.92.
N-(2-(1-Methylpyrrolidin-2-yl)ethyl)-2-phenylquinqzolin-4-amine (5)
Yield, 85%; mp >360°C. IR (cm−1): 3,308 (NH), 3,060 (arom), 2,950, 2,819, (aliph), 1,618 (CN). 1H-NMR: δ 1.7–2.3 [m, 6H, 3CH2 Cyclo], 1.9 [m, 2H, CH2-CH], 2.4 [m, 1H, CH Cyclo], 2.5 [s, 3H, N-CH3], 3.0 [m, 2H, CH2-NH], 7.4–8.6 [m, 10H, Ar-H + NH]. 13C NMR: δ 21.4, 29.4, 29.5, 40.4, 40.5, 55.2, 66.0, 114.3, 123.4, 125.7 (2), 128.2, 128.3, 128.7 (2), 130.5, 133.2, 139.1, 150.3, 159.7, 160.2. MS m/z (%): 332 (M+) (21.6), 316 (100). Calculated for C21H24N4 (332): C, 75.87; H, 7.28; N, 16.85. Found: C, 76.11; H, 7.57; N, 17.20.
N-(2-(1-Methyl-1H-pyrrol-2-yl)ethyl)-2-phenylquinqzolin-4-amine (6)
Yield, 82%; mp 172°C–173°C. IR (cm−1): 3,334 (NH), 3,059 (arom), 2,930, 2,825 (aliph), 1,618 (CN). 1H NMR: δ 1.9 [s, 3H, CH3], 2.8–3.9 [m, 4H, 2CH2], 5.6–6.6 [m, 3H, 3CH Pyrrole], 7.5–8.5 [m, 10H, Ar-H + NH]. 13C NMR: δ 25.9, 33.6, 41.0, 106.4, 106.7, 114.3, 121.8, 123.0, 125.5, 125.7 (2), 128.1, 128.3, 128.6 (2), 130.4, 133.1, 133.4, 150.4, 159.8, 162.7. MS m/z (%): 328 (M+) (3.26), 248 (100). Calculated for C21H20N4 (328): C, 76.80; H, 6.14; N, 17.06. Found: C, 76.55; H, 6.47; N, 16.81.
1-(3-(2-Phenylquinazolin-4-ylamino)propyl)pyrrolidin-2-one (7)
Yield, 88%; mp >360°C. IR (cm−1): 3,370 (NH), 3,100 (arom), 2,932, 2,847, (aliph), 1,654 (CO), 1,572 (CN). 1H NMR: δ 1.5–3.2 [m, 6H, 3CH2 Cyclo], 1.9 [m, 2H, CH2-CH2-CH2], 3.3–3.5 [m, 4H, NH-CH2 + N-CH2], 7.3–8.8 [m, 10H, Ar-H + NH]. 13C NMR: δ 18.5, 20.5, 29.1, 40.3, 40.4, 56.5, 114.3, 123.1, 125.8 (2), 128.3, 128.6, 130.5 (2), 133.1, 135.7, 139.1, 150.3, 159.6, 160.1, 183.2. MS m/z (%): 346 (M+) (15.38), 317 (100). Calculated for C21H22N4O (346): C, 72.81; H, 6.40; N, 16.17. Found: C, 72.54; H, 6.08; N, 16.46.
N-(1-ethyl-1H-pyrazol-5-yl)-2-phenylquinazolin-4-amine (8)
Yield, 92%; mp 243°C–244°C. IR (cm−1): 3,414 (NH), 3,062 (arom), 2,956, 2,854 (aliph), 1,617 (CN). 1H NMR: δ 1.3 [t, 3H, CH3], 4.3 [q, 2H, CH2], 7.4–8.5 [m, 11H, Ar-H], 12.5 [s, 1H, NH]. 13C NMR: δ 16.2, 40.4, 93.8, 114.6, 121.4, 126.3 (2), 127.0, 127.8, 128.2 (2), 129.0, 131.8, 133.2, 135.0, 149.1, 152.9, 162.8 (2). MS m/z (%): 315 (M+) (9.54), 286 (100). Calculated for C19H17N5 (315): C, 72.36; H, 5.43; N, 22.21. Found: C, 72.69; H, 5.16; N, 22.51.
1,5-Dimethyl-2-phenyl-4-(2-phenylquinazolin-4-ylamino)-1,2-dihydropyrazol-3-one (9)
Yield, 84%; mp 149°C–150°C. IR (cm−1): 3,413 (NH), 3,060 (arom), 2,923, 2,839, (aliph), 1,654 (CO), 1,618 (CN). 1H NMR: δ 2.3 [s, 3H, CH3], 3.1 [s, 3H, N-CH3], 7.3–8.5 [m, 14H, Ar-H], 9.4 [s, 1H, NH]. 13C NMR: δ 11.6, 31.2, 114.2 (2), 123.6, 124.0 (2), 126.2, 126.7 (2), 128.2, 128.4, 128.7 (2), 129.6 (2), 130.6 (2), 133.5, 135.9, 138.8, 150.8, 159.5, 160.1, 162.7. MS m/z (%): 407 (M+) (5.98), 331 (100). Calculated for C25H21N5O (407): C, 73.69; H, 5.19; N, 17.19. Found: C, 73.44; H, 5.50; N, 17.56.
N-(3-(1H-Imidazol-1-yl)propyl)-2-phenylquinazolin-4-amine (10)
Yield, 79%; mp 174°C–175°C. IR (cm−1): 3,231 (NH), 3,058 (arom), 2,927, 2,866 (aliph), 1,617 (CN). 1H NMR: δ 2.1–4.1 [m, 6H, CH2-CH2-CH2-N], 7.2–8.4 [m, 12H, Ar-H], 8.5 [s, 1H, NH]. 13C NMR: δ 30.6, 40.5, 44.4, 114.3, 119.9, 123.1, 125.7 (2), 128.3, 128.6, 128.8, 130.5 (2), 133.1 (2), 137.8, 139.0, 150.3, 159.6, 160.1. MS m/z (%): 329 (M+) (17.23), 288 (100). Calculated for C20H19N5 (329): C, 72.93; H, 5.81; N, 21.26. Found: C, 72.71; H, 5.49; N, 20.93.
N-(3-(2-Methylpiperidin-1-yl)propyl)-2-phenylquinazolin-4-amine (11)
Yield, 84%; mp 254°C–256°C. IR (cm−1): 3,434 (NH), 3,089 (arom), 2,957, 2,779, (aliph), 1,633 (CN). 1H NMR: δ 1.2 [s, 3H, CH3], 1.3–2.6 [m, 9H, 4CH2 + CH Cyclo], 1.6–3.8 [m, 6H, 3CH2], 7.4–8.9 [m, 9H, Ar-H], 10.8 [s, 1H, NH]. 13C NMR: δ 12.3, 22.2, 23.5, 27.9, 34.4, 40.4, 50.1, 55.1, 59.0, 112.9, 120.9, 125.0 (2), 128.2 (2), 129.3 (2), 129.8, 133.7, 135.8, 157.6 (2), 160.6. MS m/z (%): 360 (M+) (33.85), 344 (100). Calculated for C23H28N4 (360): C, 76.63; H, 7.83; N, 15.54. Found: C, 76.91; H, 7.49; N, 15.22.
2-Phenyl-N-(2-piperidin-1-yl)ethylquinazolin-4-amine (12)
Yield, 90%; mp 317°C–319°C. IR (cm−1): 3,401 (NH), 3,100 (arom), 2,936, 2,713 (aliph), 1,630 (CN). 1H NMR: δ 2.2–2.7 [m, 10H, 5CH2 Cyclo], 2.8 [t, 2H, N-CH2], 4.3 [s, 2H, CH2NH], 7.4–9.1 [m, 9H, Ar-H], 10.2 [s, 1H, NH]. 13C NMR: δ 24.5, 28.0 (2), 47.4, 50.7, 59.4 (2), 112.6, 120.5, 125.0 (2), 128.1, 128.2, 129.4 (2), 130.4, 131.5, 133.9, 157.4, 160.2, 162.8. MS m/z (%): 332 (M+) (45.11), 219 (100). Calculated for C21H24N4 (332.20): C, 75.87; H, 7.28; N, 16.85. Found: C, 76.11; H, 7.55; N, 17.10.
N-(2-Morpholinoethyl)-2-phenylquinazolin-4-amine (13)
Yield, 76%; mp 273°C–274°C. IR (cm−1): 3,413 (NH), 3,076 (arom), 2,927, 2,836 (aliph), 1,599 (CN). 1H NMR: δ 2.3–2.4 [m, 4H, CH2-N-CH2 morpholino], 2.5–3.2 [m, 4H, 2CH2], 3.6–4.3 [m, 4H, CH2-O-CH2], 7.3–8.8 [m, 9H, Ar-H], 10.3 [s, 1H, NH]. 13C NMR: δ 51.5, 54.9, 55.4 (2), 63.7 (2), 113.3, 126.9, 127.8 (2), 129.6 (2), 129.8 (2), 133.1 (2), 135.5, 159.1, 160.2, 163.4. MS m/z (%): 334 (M+) (22.17), 256 (100). Calculated for C20H22N4O (334): C, 71.83; H, 6.63; N, 16.75. Found: C, 71.50; H, 6.30; N, 16.45.
N-(3-Morpholinopropyl)-2-phenylquinazolin-4-amine (14)
Yield, 80%; mp 235°C–236°C. IR (cm−1): 3,324 (NH), 3,088 (arom), 2,954, 2,864 (aliph), 1,610 (CN). 1H NMR: δ 1.8–1.9 [m, 2H, NH-CH2-CH2-CH2], 2.2–2.3 [m, 4H, CH2-N-CH2 morpholino], 3.1 [t, 2H, N-CH2], 3.2 [t, 2H, NH-CH2], 3.8–3.9 [m, 4H, CH2-O-CH2], 7.5–8.5 [m, 9H, Ar-H], 11.2 [s, 1H, NH]. 13C NMR: δ 23.7, 40.5, 51.4, 54.3 (2), 63.6 (2), 113.9, 123.8, 128.8 (2), 128.9 (2), 129.6 (2), 132.7, 133.1, 135.6, 152.2, 160.3, 161.7. MS m/z (%): 348 (M+) (10.62), 221 (100). Calculated for C21H24N4O (348): C, 72.39; H, 6.94; N, 16.08. Found: C, 72.08; H, 6.60; N, 16.35.
N-(1-Benzylpiperidin-2-yl)-2-phenylquinazolin-4-amine (15)
Yield, 83%; mp 250°C–251°C. IR (cm−1): 3,380 (NH), 3,077 (arom), 2,988, 2,867 (aliph), 1,630 (CN). 1H NMR: δ 2.1–2.8 [m, 9H, 4CH2 + CH Cyclo], 4.3 [s, 2H, CH2-Ph], 7.4–8.9 [m, 14H, Ar-H], 10.2 [s, 1H, NH]. 13C NMR: δ 22.6, 28.0, 34.5, 50.7, 59.4, 83.2, 112.6, 125.0, 128.2, 129.2 (2), 129.4 (3), 129.9 (3), 130.4 (2), 131.5, 132.0, 133.9, 136.1, 157.5, 160.2, 162.8. MS m/z (%): 394 (M+) (5.88), 314 (100). Calculated for C26H26N4 (394.22): C, 79.16; H, 6.64; N, 14.20. Found: C, 79.48; H, 6.36; N, 13.83.
6-(2-Penylquinazolin-4-ylamino)hexanoic acid (16a)
Yield, 76%; mp 163°C–165°C. IR (cm−1): 3,438 (OH), 3,311 (NH), 3,078 (arom), 2,939, 2,854 (aliph), 1,687 (CO), 1,613 (CN). 1H NMR: δ 1.4–1.8 [m, 6H, 3CH2], 2.2 [t, 2H, CH2CO], 3.8 [t, 2H, NH-CH2], 7.3–8.8 [m, 9H, Ar-H], 10.5 [s, 1H, NH], 14.9 [s, 1H, OH]. 13C NMR: δ 24.6, 26.3, 28.3, 34.0, 41.9, 112.6, 126.8, 128.2 (2), 129.4 (2), 129.6 (2), 133.7 (2), 135.8, 157.4 (2), 160.2, 174.8. MS m/z (%): 335 (M+) (39.45), 290 (100). Calculated for C20H21N3O2 (335): C, 71.62; H, 6.31; N, 12.53. Found: C, 71.29; H, 6.60; N, 12.19.
Biological evaluation
Hepa1c1c7 murine hepatoma cells were grown in a humidified atmosphere at 37°C, 5% CO2. The cell culture medium was α-minimum essential medium (α-MEM) and was supplemented with 10% (v/v) heat- and charcoal-inactivated fetal bovine serum. For evaluation of the potential NQO1 inducer activity, cells (104 per well) were grown in 96-well plates for 24 hours, after which the cell culture medium was replaced with fresh medium containing each inducer (dissolved in DMSO and diluted in the medium 1:1,000), and the cells were grown for a further 48 hours. There were eight replicates of each treatment of serial dilutions of inducers. The final DMSO concentration in the cell culture medium was maintained at 0.1% (v/v) in all wells. At the end of the treatment period, cell lysates were prepared in digitonin and the specific activity of NQO1 was determined using menadione as a substrate as described.23,24 The concentration that doubles the specific activity of NQO1 (CD value) was used as a measure of inducer potency. Mean values for the eight replicate wells are shown for each data point. The standard deviation for each data point was within 5% of the mean value. The classical NQO1 inducer sulforaphane was included as a positive control in each bioassay and was consistently giving a CD value of 0.2 µM.
Molecular modeling study
The molecular model of all the new anilinoquinazoline derivatives was built using molecular operating environment (MOE) software suite Version 10.2008 maintaining all the default parameters. The structures’ geometry was optimized, and a systematic conformational search was carried out to a root mean square gradient of 0.01 Å using the ConfSearch module implemented in MOE. Computations were set to be performed with the Merck Force Field (MMFF94s). The new compounds’ ability to access and block the Nrf2-binding site of Keap1 was evaluated by performing a molecular docking study using the crystallographic structure of Keap1 obtained from the Protein Data Bank (PDB ID: 4IQK). Following addition of the missing hydrogens and calculating the partial charges of the receptor, validation of the docking has been carried out by docking of the native ligand. Afterward, the ligands were docked where they were left free to explore all conformations possible inside the enzyme. Multiple separate docking simulations using default parameters were performed followed by choosing the best conformations based on the combination of S score data, E conformation, and appropriate fitting with the relevant amino acids in the binding pocket.
Results and discussion
Chemistry
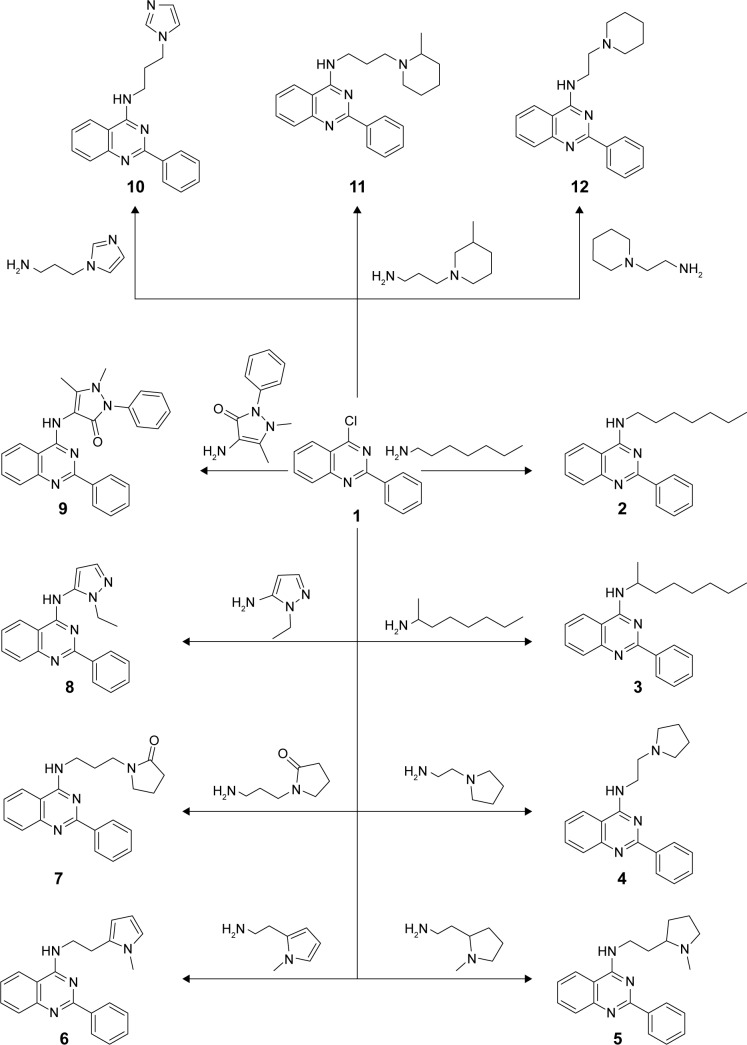
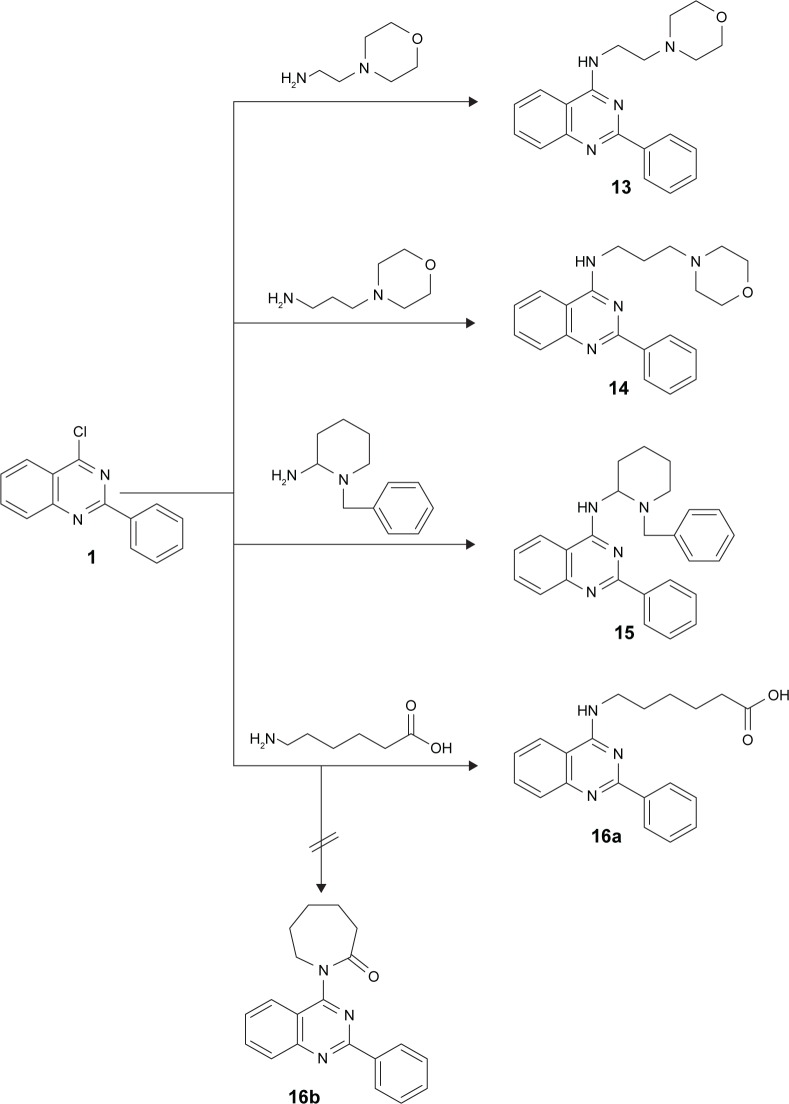
Novel anilinoquinazoline derivatives 2–16a were prepared from the key starting material 4-chloro-2-phenylquinazoline 1 via reaction with primary amines having side chains bearing aliphatic groups (eg, heptane, octane), aromatic groups (eg, pyrrolidine, imidazole, piperidine), or substituted aromatic rings (eg, methyl pyrrolidine, benzyl piperidine, methyl piperidine). Refluxing of 1 with the appropriate amine in dry dimethylformamide in the presence of triethylamine as a base catalyst furnished the desired corresponding compounds 2–16a (Figures 1 and 2). The proposed structures of the products were confirmed by their analytical and spectral data. First, the IR of the reaction products showed in all compounds an absorption band corresponding to their NH function in the region 3,434–3,191 cm−1, in addition to a CN band in the region 1,639–1,572 cm−1. Moreover, compounds 7, 9, and 16a showed an extra carbonyl absorption band in the region 1,687–1,654 cm−1. Finally, compounds 4–16a bearing aliphatic carbon chains showed absorption peaks around 2,988–2,713 cm−1. Moreover, 1H NMR of 2–16a revealed a singlet signal in the region 9.4–11.2 ppm for the NH groups. With regard to compounds 16, 1 was reacted with 6-aminocaproic acid with the expected product to be the azepan-2-one derivative 16b; however, the hexanoic acid derivative 16a was obtained instead. That was revealed on the basis of elemental analysis and IR, which showed the presence of OH absorption band at 3,438 cm−1, NH band at 3,311 cm−1, and a carbonyl band at 1,687 cm−1. That was further confirmed using spectral analysis where 1H NMR showed a triplet at 3.8 ppm for the NH-CH2, signal at 10.5 ppm for the NH group, and another signal at 14.9 ppm for the OH group. Moreover, 13C NMR of 16a revealed signals at 24.6 ppm for CH2-CH2-COOH, 26.3 ppm for CH2-CH2-CH2-COOH, 28.3 ppm for NH-CH2-CH2, 34.0 ppm for CH2-COOH, 41.9 ppm for NH-CH2, and 174.8 ppm for CO group.
Figure 1.
Synthesis of derivatives 2–11.
Notes: Reagents and conditions: a mixture of 1 (0.01 mol) and different amines (0.012 mol) in dry DMF (10 mL) containing three drops of Et3N was refluxed for 24 hours and then left to cool. The solid product formed was collected by filtration and recrystallized from acetic acid.
Abbreviation: DMF, dimethylformamide.
Figure 2.
Synthesis of derivatives 13–16.
Notes: Reagents and conditions: a mixture of 1 (0.01 mol) and different amines (0.012 mol) in dry DMF (10 mL) containing three drops of Et3N was refluxed for 24 hours and then left to cool. The solid product formed was collected by filtration and recrystallized from acetic acid.
Abbreviation: DMF, dimethylformamide.
Biological activity
The ability of the novel compounds to double the activity of NQO1 was used as a measure of their inducer potency (CD values), and results obtained are presented in Table 1. Evaluation of the NQO1 inducer activity showed that compounds 2, 5, and 14 were inactive, whereas compounds 4, 6, 11, 12, 13, and 16a had weak activity; however, CD value was not reached (Figure 3). On the other hand, compounds 3 (CD =14 µM), 7 (CD =26 µM), and 15 (CD =19 µM) had moderate inducer activity. Compounds 8 (CD =5.2 µM) and 10 (CD =5.5 µM) were of approximately equal potency. However, the cell responses to them were very different where the dose–response dependency was very clear for compound 8, whereas it was completely absent for compound 10. The most potent inducer in this series was compound 9 (CD =3.9 µM), which also showed a very clear dose response with a magnitude of induction of approximately sixfold at a concentration of 50 µM.
Table 1.
NQO1 inducer activity and CD values of the test compounds
| Conc (µM) | Compound #
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16a | |
| 0.1563 | NR | 1.05 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 0.3125 | NR | 1.08 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 0.625 | NR | 1.07 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 0.781 | 1.25 | NR | 1.24 | 1.09 | 0.97 | 1.01 | 1.23 | 1.34 | 1.66 | 1.11 | 1.01 | 1.12 | 1.01 | 1.04 | 1.12 |
| 1.25 | NR | 1.19 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 1.563 | 1.25 | NR | 1.3 | 1.1 | 0.96 | 1.01 | 1.33 | 1.56 | 1.78 | 1.1 | 1.12 | 1.17 | 1.04 | 1.04 | 1.14 |
| 2.5 | NR | 1.28 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 3.125 | 1.21 | NR | 1.49 | 1.12 | 1.08 | 1.1 | 1.57 | 1.86 | 1.89 | 1.2 | 1.26 | 1.16 | 1.06 | 1.1 | 1.23 |
| 5 | NR | 1.56 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 6.25 | 1.2 | NR | 1.68 | 1.2 | 1.31 | 1.25 | 2.12 | 2.41 | 2.03 | 1.3 | 1.36 | 1.24 | 1.11 | 1.23 | 1.38 |
| 10 | NR | 1.93 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 12.5 | NR | NR | 1.76 | 1.28 | 1.54 | 1.48 | 2.63 | 3.23 | 2.1 | 1.5 | NR | 1.39 | 1.1 | 1.45 | 1.67 |
| 20 | NR | 2.13 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 25 | NR | NR | NR | NR | NR | 1.98 | 3.34 | 4.74 | NR | NR | NR | 1.41 | 1.06 | 1.53 | 2.22 |
| 50 | NR | NR | NR | NR | NR | 2.73 | 4.16 | 6.45 | NR | NR | NR | NR | NR | NR | 3.19 |
| 100 | NR | NR | NR | NR | NR | 3.63 | 4.25 | NR | NR | NR | NR | NR | NR | NR | 4.02 |
| CDa | NR | 14 | NR | NR | NR | 26 | 5.2 | 3.9 | 5.5 | NR | NR | NR | NR | NR | 19 |
Note:
CD data presented are the averages of three independent experiments, each with eight replicate wells of cells, and SD for each data point was within 5% of the value.
Abbreviations: CD, concentration of the test compound that doubles NQO1 activity; NQO1, NAD(P)H:quinone oxidoreductase 1; NR, not recorded; SD, standard deviation.
Figure 3.
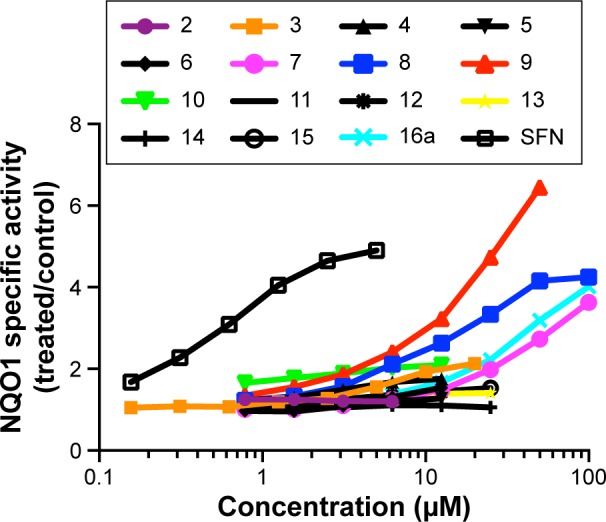
Concentration dependence of NQO1 inducer activity of the test compounds.
Notes: Hepa1c1c7 cells (104 per well) were grown in 96-well plates for 24 hours. The cell culture medium was then replaced with fresh medium containing serial dilutions of each inducer, the cells were grown for a further 48 hours, and the specific enzyme activity of NQO1 was determined using menadione as a substrate in cell lysates. Mean values for eight replicate wells are shown for each data point. The standard deviation for each data point was within 5% of the mean value. The classical NQO1 inducer SFN was used as a positive control.
Abbreviations: NQO1, NAD(P)H:quinone oxidoreductase 1; SFN, sulforaphane.
The anilinoquinazolines represent a new chemical class of NQO1 inducers, thus adding to the existing knowledge of the diversity of the many chemical scaffolds that have been reported to induce this enzyme. The classical NQO1 inducers are primarily oxidants and electrophiles or other compounds that react (or are metabolized to products that react) and chemically modify cysteine sensors of Keap1. They belong to ten distinct chemical classes: 1) oxidizable diphenols, phenylenediamines, and quinones, 2) Michael reaction acceptors (olefins or acetylenes conjugated with electron-withdrawing groups), 3) isothiocyanates and sulfoxythiocarbamates, 4) thiocarbamates, 5) dithiolethiones, 6) conjugated polyenes, 7) hydroperoxides, 8) trivalent arsenicals, 9) heavy metals, and 10) vicinal dimercaptans.11 A new generation of NQO1 inducers is also emerging, that of noncovalent small-molecule modulators of the Keap–Nrf2 protein–protein interaction.16,17,25–27 Although we cannot exclude the possibility that the anilinoquinazolines could be metabolized to electrophilic species in cells, we hypothesize that the anilinoquinazolines themselves may have a more unusual mechanism of action, namely, the ability to directly disrupt the binding of Keap1 to Nrf2, and thus belong to the category of noncovalent inducers. To begin testing this hypothesis, we next performed molecular modeling studies.
Molecular modeling
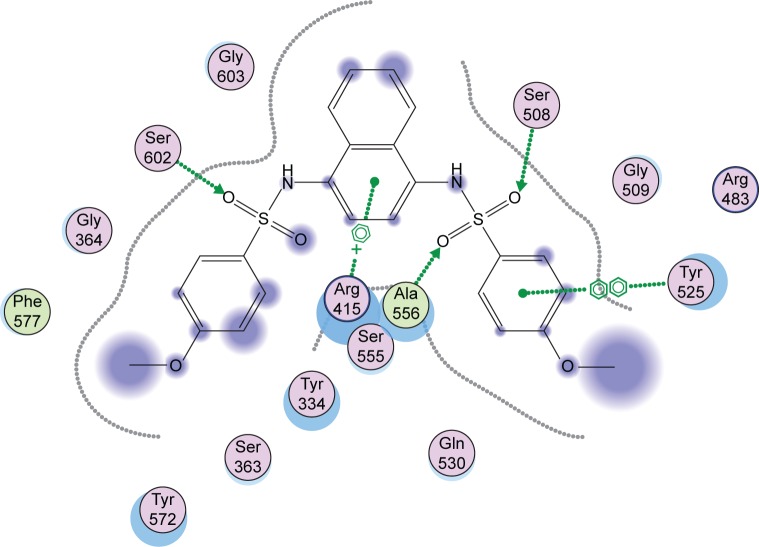
It has been established that binding of Keap1 to Nrf2 promotes its degradation, thus maintaining low levels of expression of downstream target genes. Several small-molecule compounds have been reported to have binding affinity to the Keap1 Kelch domain, therefore antagonizing its activity.25–27 In order to assess the ability of the newly synthesized compounds to access and block the Kelch domain of Keap1, a molecular docking study was performed using the crystal structure obtained from the PDB ID: 4IQK using MOE software suite Version 10.2008. The key interactions detected between the validated native ligand and the receptor are found to be arene–cation interaction with Arg415, arene–arene interaction with Tyr525, and three hydrogen bonds with Ser602, Ser508, and Ser555 with S=−13.306 Kcal/mol with a root mean square deviation of 0.6635 Kcal/mol/Å (Figure 4). Docking of the synthesized compounds revealed that binding to Arg415 through an arene–cation interaction is an important common interaction among all compounds. Moreover, by observing the interactions of compounds 4–16a, it was found that compounds showing activity have a larger number of interactions with the binding site of the sensor protein via their side chain or via their main skeleton. That may emphasize the role of variation of the side chain in either making its own interactions or in pushing the phenylquinazoline moiety to more interactions with more amino acids.
Figure 4.
Interactions of the native ligand with the Kelch domain of Keap1 (PDB ID: 41QK).
Note: The green circles denote lipophilic amino acids and the purple ones denote hydrophilic amino acids.
Abbreviations: Keap1, Kelch-like ECH-associated protein 1; PDB, Protein Data Bank.
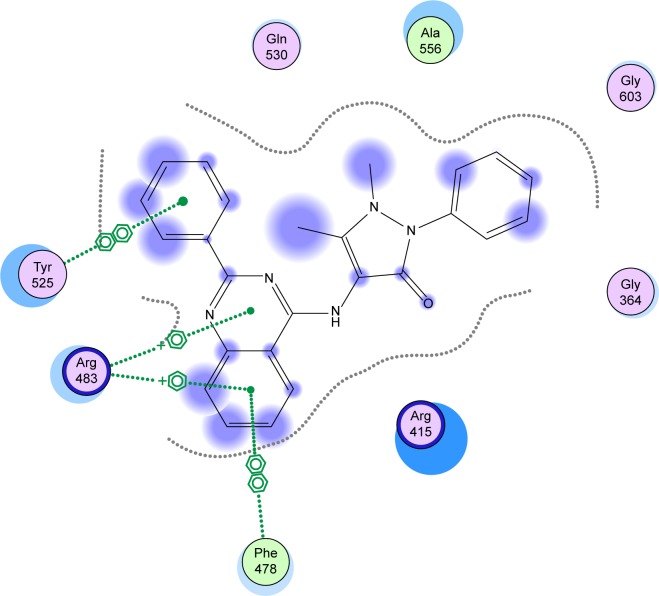
Finally, it was observed that compound 9 showed the best activity (CD =3.9 µM) as well as the best binding affinity with S=−11.7347 Kcal/mol. Compound 9 was able to make two arene–cation interactions with Arg483. Although the Arg involved in compound 9 interactions is different from Arg415 that the native ligand interacts with, the overlap of 9 over the native ligand showed a change in the orientation of the compound to ensure better fitting in the pocket (Figure 5). Moreover, an arene–arene interaction is noticed with Tyr525 similar to the native ligand. Finally an additional arene–arene interaction is noticed with Phe478; that additional interaction may be related to the high activity of that compound since it is only noticed in the interactions of compound 9 with the sensor protein (Figure 6).
Figure 5.
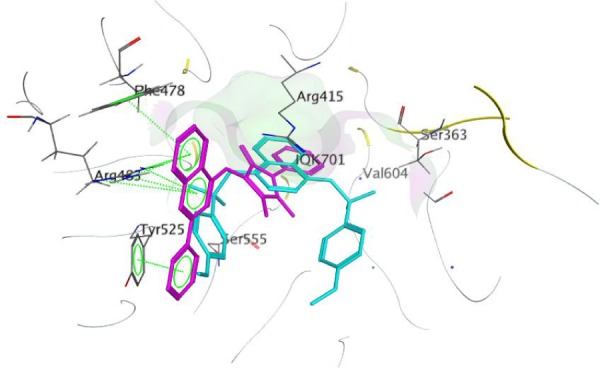
Overlap of compound 9 (magenta) over native ligand (cyan) with the Kelch domain of Keap1.
Abbreviation: Keap1, Kelch-like ECH-associated protein 1.
Figure 6.
Interactions of compound 9 with the Kelch domain amino acids of Keap1.
Abbreviation: Keap1, Kelch-like ECH-associated protein 1.
Conclusion
This study deals with the synthesis of novel anilinoquinazoline derivatives with potential NQO1 inducer activity. Among the derivatives 2–16a, 12 compounds showed activity, with six of them showing CD values ranging between 3.9 µM and 25 µM. Finally, the molecular docking study has shown that the most active derivative, compound 9 (CD =3.9 µM), showed arene–arene interactions with key amino acids in the Nrf2-binding site of Keap1. The obtained results introduce compound 9 as a lead for anilinoquinazoline scaffold-based Nrf2 activators, thus serving as a starting point for lead optimization of new molecules based on the chemotype described herein.
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding of this research through the Research Group Project number RGP-302. Maureen Higgins and Albena T Dinkova-Kostova are grateful to Cancer Research UK (C20953/A10270 and C20953/A18644) for financial support.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Harman D. Antioxidant supplements: effects on disease and aging in the United States population. J Am Aging Assoc. 2000;23(1):25–31. doi: 10.1007/s11357-000-0004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64(1):97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 3.Sies H. Oxidative stress: from basic research to clinical application. Am J Med. 1991;91(3C):31S–38S. doi: 10.1016/0002-9343(91)90281-2. [DOI] [PubMed] [Google Scholar]
- 4.Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501(1):116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47(1):89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 7.Ross D. Quinone reductases multitasking in the metabolic world. Drug Metab Rev. 2004;36(3–4):639–654. doi: 10.1081/dmr-200033465. [DOI] [PubMed] [Google Scholar]
- 8.Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A. 1988;85(21):8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13(11):1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 10.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 11.Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85(4):241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Ichikawa T, Janicki JS, Cui T. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin Ther Targets. 2009;13(7):785–794. doi: 10.1517/14728220903025762. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Redell JB, Moore AN, Dash PK. A novel strategy to activate cytoprotective genes in the injured brain. Biochem Biophys Res Commun. 2011;407(3):501–506. doi: 10.1016/j.bbrc.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steel R, Cowan J, Payerne E, O’Connell MA, Searcey M. Anti-inflammatory effect of a cell-penetrating peptide targeting the Nrf2/Keap1 interaction. ACS Med Chem Lett. 2012;3(5):407–410. doi: 10.1021/ml300041g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson TP, Amirahmadi S, Joshi G, et al. Discovery of potent, novel Nrf2 inducers via quantum modeling, virtual screening, and in vitro experimental validation. Chem Biol Drug Des. 2012;80(6):810–820. doi: 10.1111/cbdd.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L, Magesh S, Chen L, et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1-Nrf2 protein-protein interaction. Bioorg Med Chem Lett. 2013;23(10):3039–3043. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang C, Miao Z, Sheng C, Zhang W. Updated research and applications of small molecule inhibitors of Keap1-Nrf2 protein-protein interaction: a review. Curr Med Chem. 2014;21(16):1861–1870. doi: 10.2174/0929867321666140217104648. [DOI] [PubMed] [Google Scholar]
- 18.Nesterova NA, Kovalenko SI, Belenichev IF, Karpenkos OV, Sidorova IV. Formation of combinational library of quinazoline-4-yl-hydrazones with antioxidant activity. Ukraine Med Khim. 2004;6:14–21. [Google Scholar]
- 19.Nesterova NA, Kovalenko SI, Karpenkos OV, Belenichev IF. Synthesis and antioxidant activity of 4-arylidenehydrazinoquinazolines. Ukr Farmatsevtichnii Zhurnal (Kiev) 2004;22:5–10. [Google Scholar]
- 20.Al-Omar MA, Al-Rashood ST, El-Subbagh HI, Abdel Hamide SG. Interaction of 2-thio-4-oxo-quinazoline derivatives with guinea pig liver molybdenum hydroxylases, xanthine oxidase and aldehyde oxidase. J Biol Sci. 2005;5(3):370–378. [Google Scholar]
- 21.Ghorab MM, Higgins M, Alsaid MS, Arafa RK, Shahat AA, Dinkova-Kostova AT. Synthesis, molecular modeling and NAD(P)H:quinone oxidoreductase 1 inducer activity of novel cyanoenone and enone benzenesulfonamides. J Enzyme Inhib Med Chem. 2014;29(6):840–845. doi: 10.3109/14756366.2013.858146. [DOI] [PubMed] [Google Scholar]
- 22.Ghorab MM, Higgins M, Dinkova-Kostova AT, Alsaid MS, Shahat AA. Novel thioureido derivatives carrying thione and sulfonamide moieties induce the cytoprotective enzyme NAD(P)H:Quinone oxidoreductase 1. Asian J Chem. 2014;26(24):8501–8504. [Google Scholar]
- 23.Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal Biochem. 1988;169(2):328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 24.Fahey JW, Dinkova-Kostova AT, Stephenson KK, Talalay P. The “Prochaska” microtiter plate bioassay for inducers of NQO1. Methods Enzymol. 2004;382:243–258. doi: 10.1016/S0076-6879(04)82014-7. [DOI] [PubMed] [Google Scholar]
- 25.Marcotte D, Zeng W, Hus JC, et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a noncovalent mechanism. Bioorg Med Chem. 2013;21(14):4011–4019. doi: 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev. 2012;32(4):687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertrand HC, Schaap M, Baird L, et al. Design, synthesis, and evaluation of triazole derivatives that induce Nrf2 dependent gene products and inhibit the Keap1-Nrf2 protein-protein interaction. J Med Chem. 2015;58(18):7186–7194. doi: 10.1021/acs.jmedchem.5b00602. [DOI] [PubMed] [Google Scholar]