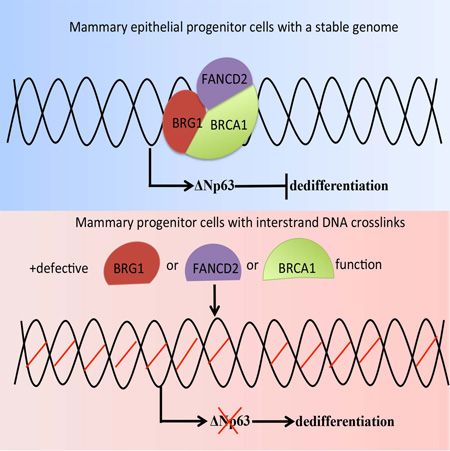
Summary
An abnormal differentiation state is common in BRCA1-deficient mammary epithelial cells, but the underlying mechanism is unclear. Here we report a convergence between DNA repair and normal, cultured human mammary epithelial (HME) cell differentiation. Surprisingly, depleting BRCA1 or FANCD2 (Fanconi Anemia [FA] proteins), or BRG1, a mSWI/SNF subunit, caused HME cells to undergo spontaneous epithelial-to-mesenchymal transition (EMT) and aberrant differentiation. This also occurred when WT HMEs were exposed to chemicals that generate interstrand DNA cross links (repaired by FA proteins) but not in response to double strand breaks. Suppressed expression of ΔNP63 also occurred in each of these settings, an effect that links DNA damage to the aberrant differentiation outcome. Taken together with somatic breast cancer genome data, these results point to a breakdown in a BRCA/FA--mSWI/SNF-ΔNP63- mediated DNA repair and differentiation maintenance process in mammary epithelial cells that may contribute to sporadic breast cancer development.
Keywords: Breast cancer, BRCA1, BRG1, FANCD2, Cisplatin, Crosslink Repair, EMT
Graphical Abstract
eTOC blurb (In Brief)
BRCA1 deficient mammary epithelial cells exhibit an abnormal differentiation state. Hua et al., show that maintenance of differentiation in human mammary epithelial cells depends upon BRCA1-dependent DNA interstrand crosslink repair and ΔNp63, which appears to be important for breast cancer suppression.
Introduction
The differentiation of stem/progenitor cells contributes to normal tissue homeostasis and to cancer suppression(Jopling et al., 2011; Santos et al., 2014; Tata et al., 2013; Tetteh et al., 2015; Wang et al., 2012). Most BRCA1 breast cancers are triple negative/basal-like (TNBC), while others are luminal(Mavaddat et al., 2012). Some of the cells of BRCA1-associated TNBC exhibit mesenchymal traits, suggesting that they have passed through an epithelial to mesenchymal transition (EMT)(Polyak and Weinberg, 2009). BRCA1 deficiency also disrupts normal mammary gland development in mice(Molyneux et al., 2010; Xu et al., 1999) and perturbs normal human mammary epithelial stem/progenitor-like cell differentiation(Lim et al., 2009; Liu et al., 2008; Proia et al., 2011). The mechanism underlying these phenomena has been unclear.
BRCA1 suppresses breast and ovarian cancer development, at least in part, by maintaining genome integrity(Huen et al., 2010). Previous work indicates that endogenous or exogenous DNA damage elicits differentiation defects in melanocyte stem cells(Inomata et al., 2009), hematopoietic stem cells(Wang et al., 2012), B cells(Sherman et al., 2010), myeloid progenitor cells(Santos et al., 2014), and intestinal progenitor cells(van Es et al., 2012), indicating a link between differentiation maintenance and a proper response to DNA damage. Others have reported a link between defective BRCA1 function and a breakdown in normal mammary epithelial cell differentiation, Thus, we asked whether DNA damage arising from the loss of BRCA1–driven DNA repair is sufficient to promote an aberrant differentiation state in mammary epithelial cells.
The ATPase-dependent chromatin remodeling protein, BRG1, is a BRCA1-interacting protein of unexplained value in a BRCA1 disease-suppressing context(Bochar et al., 2000), although it has been proposed as a breast and ovarian cancer suppressor(Bultman et al., 2008; Jelinic et al., 2014; Ramos et al., 2014). BRG1 is a key component of the differentiation-promoting mammalian (m)SWI/SNF chromatin remodeling complex(Ho and Crabtree, 2010). Similar to BRCA1, one possible mechanism by which a mSWI/SNF complex might suppress malignancy is through the maintenance of genome integrity, given that the loss of BRG1 or other mSWI/SNF components generates spontaneous anaphase bridges(Dykhuizen et al., 2013). Hence, we explored the possibility that BRCA1 and BRG1 cooperate to support proper mammary epithelial cell differentiation by contributing to efficient DNA damage repair.
Here we report that BRCA1/BRG1/FANCD2-dependent crosslink repair stabilizes the differentiated state of primary, immortalized, diploid human mammary epithelial (HME) cells. Similarly, exposure of otherwise naive HME cells to inter-strand crosslink-inducing agents, elicited aberrant differentiation in HME cells. In addition, BRCA1/BRG1/FANCD2-driven crosslink repair-dependent differentiation was stabilized by the action of the p53 family member, ΔNp63. Together, these results and abundant genomic sequencing data from sporadic human breast cancers imply that this mechanism can serve as a mediator of breast cancer suppression.
Results
BRCA1 is required for differentiation maintenance in human mammary epithelial cells
In light of prior findings(Huen et al., 2010; Liu et al., 2008; Mani et al., 2008; Proia et al., 2011; Tetteh et al., 2015) and to test whether BRCA1 is essential for the maintenance of mammary epithelial cell differentiation, we assessed the differentiated state before and after loss of BRCA1 function (Figure 1A) in the FACS-purified CD44low (an epithelial phenotype) cells from a strain of wtBRCA1 hTERT-immortalized, primary human mammary epithelial cells. We then infected the cells with a tetracycline-inducible shScramble (control) or shBRCA1-encoding lentiviral vector (targeting the BRCA1 3’-UTR). Doxycycline (Dox) exposure resulted in marked BRCA1 (p220) depletion without a change in proliferation (Figures 1A-1B and Figure S1A–S1D).
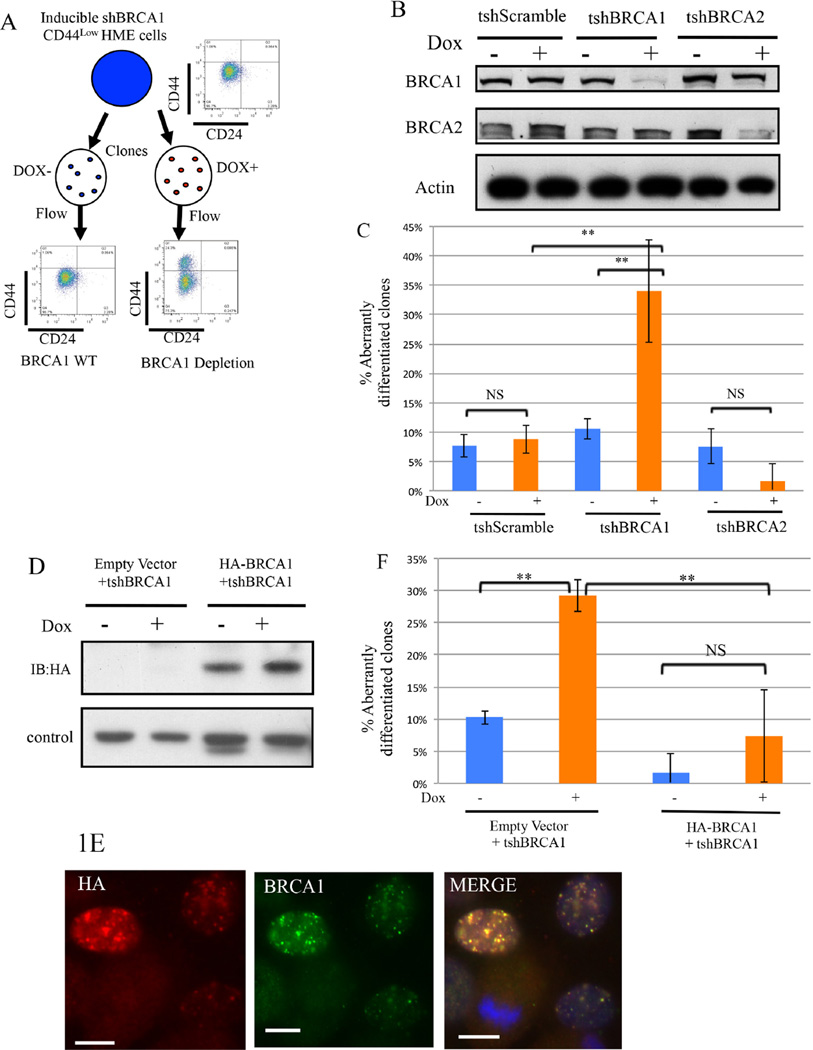
Figure 1. BRCA1 contributes to the maintenance of differentiation in CD44low human mammary epithelial (HME) cells (Also see Figure S1).
1A) Experimental scheme to search for a differentiation marker expression change in CD44low HME cell clones after BRCA1 depletion.
1B) Immunoblotting for BRCA1 or BRCA2 in CD44low HME cells infected with the indicated tet-on shRNA vectors before and after doxycycline exposure.
1C) Summary of the analysis of aberrantly differentiated CD44high clones (that contain ≥5% CD44high cells) emerging after BRCA1 or BRCA2 depletion in CD44low HME cells. The data represented are mean values ±S.D. **P<0.01 and NS: Not significant.
1D) Immunoblotting for shRNA-resistant HA-BRCA1 in CD44low HME cells before and after endogenous BRCA1 depletion. A nonspecific band serves as the loading control.
1E) Formation of nuclear foci by shRNA-resistant HA-BRCA1 in CD44low cells after depleting endogenous BRCA1. Cells were treated with cisplatin (1µM, 24hours). Scale bar: 10µm. 1F) Prevention of the emergence of aberrantly differentiated clones by shRNA-resistant BRCA1 in CD44low HME cells following shBRCA1 expression. The data represented are mean values ±S.D. **P<0.01.
Then single, CD44low HME cells from the shScramble- and shBRCA1-transduced populations were seeded on individual plastic disks and incubated in the continuous presence or absence of Dox. Individual colonies were isolated and analyzed for cell-surface expression of CD24 (an epithelial-like marker) and CD44 (a CD44high state represents a mesenchymal and a CD44low state an epithelial one). Clones containing > 5% CD44high cells, were classified as aberrantly differentiated, since the vast majority of clones derived from shScramble– transduced or untreated parental cells contained less than this level of CD44high cells.
In CD44low HME cells inducing an shScramble revealed no effect on BRCA1 expression or the abundance of CD44high cells (Figure 1B-1C and Supplemental table 1). However, following Dox- induced BRCA1 depletion, 30% of newly isolated clones contained significant numbers of CD44high cells (Figure 1B-1C and Supplemental table 1). The values associated with BRCA1 depletion represent a ~3-fold increase when compared with uninduced controls (P=0.002) (Figure 1C). The presence of CD44high cells in the uninduced control setting is likely a result of spontaneous conversion and/or leaky hairpin expression.
BRCA2 is a BRCA1- interacting protein, and some of its DNA damage repair properties are complemented by BRCA1 function. However, depletion of BRCA2 in tet-on inducible shBRCA2 CD44low HME cells did not cause aberrant differentiation (Figure 1B-1C). This appeared to be the result of a failure of the CD44low cells to clone and to proliferate robustly after BRCA2 depletion (Figure S1A–S1D).
To determine whether the shBRCA1-driven change in mammary epithelial differentiation was BRCA1 depletion-specific, HA-tagged shRNA-resistant BRCA1 and an empty vector were introduced, in parallel, into inducible CD44low HME cells before Dox exposure. Constitutive expression of shRNA-resistant BRCA1 was confirmed by western blotting (Figure 1D). This protein also formed discrete nuclear foci after exogenous DNA damage (Figure 1E), consistent with its ability to perform certain normal, post-damage BRCA1 functions. Unlike the control, it prevented conversion of CD44low to CD44high HME cells following BRCA1 depletion (Figures 1F and Supplemental table 1). Thus, the shBRCA1-driven, CD44 phenotype change represents an on-target, BRCA1 depletion-associated effect.
Since BRCA1 depletion had no effect on CD44low cell colony formation (Figure S1A–S1D), selection against this property did not contribute to the BRCA1 depletion-driven expansion of CD44high cells. Moreover, the fraction of CD44high cells arising from each BRCA1-depleted clone did not strictly correlate with the efficiency of BRCA1 depletion (Figure 2A). This suggested the existence of a threshold BRCA1 concentration below which cells may exhibit an aberrant CD44high phenotype. This aside, these results indicate that sufficient BRCA1 is necessary for the maintenance of a normal state of HME cell differentiation.
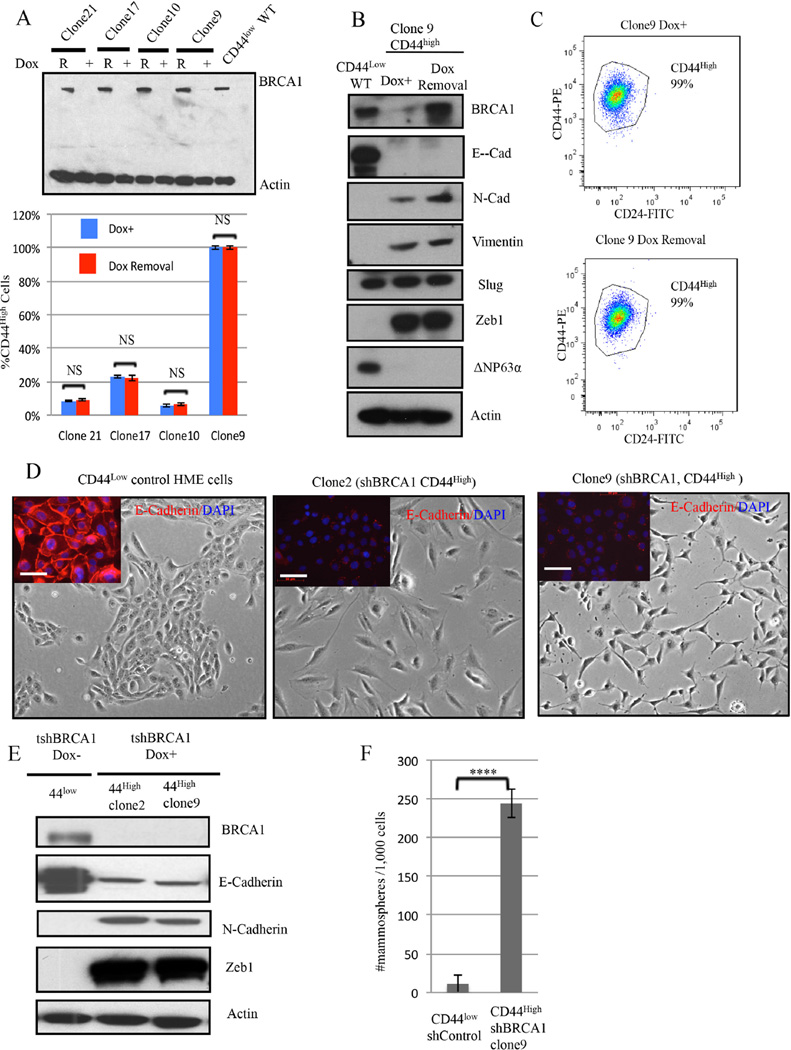
Figure 2. Aberrant differentiation of HME cells following BRCA1 depletion.
2A) (Top panel) Immunoblotting for BRCA1 after Doxycycline-induced BRCA1 depletion (DOX +) and after BRCA1 re-accumulation (R) for 4 weeks after Dox removal. (Lower panel)The abundance of CD44high cells was measured before and 4 weeks after doxycycline removal in four, independent, BRCA1- depleted clones. The data represented are mean values ±S.D. NS: Not significant.
2B) Immunoblotting for BRCA1 and EMT markers in CD44high HME cells (clone9) after BRCA1 depletion or reexpression.
2C) Representative CD24 and CD44 FACS profiles of clone 9 derived after BRCA1 depletion (clone 9 Dox+) and 4 weeks after subsequent Doxcycline removal.
2D) Respective phase-contrast images of CD44low parental and of CD44high cells (clones 2 and 9) isolated after Dox-induced BRCA1 depletion in CD44low cells. The inserts depict the results of anti-E-cadherin (red) and DAPI (blue) staining. Scale bar: 50 µm
2E) Immunoblotting for EMT marker proteins in two, independent, aberrantly differentiated CD44high clones (clones 2 and 9) that appeared after BRCA1 depletion in CD44low cells. 2F) Mammosphere assays were performed on a clone (clone9) containing uniformly CD44high HME cells derived after BRCA1 depletion of CD44low cells or on shluc CD44low control cells. The data are represented as mean values ±S.D. ****P<0.0001.
To determine whether re-expression of BRCA1 can revert an otherwise stable BRCA1 depletion-induced CD44high phenotype, Dox was removed; and nearly pre-depletion level BRCA1 expression reappeared at ~5 days (Figure 2A-2B). However, the CD44high phenotype remained stable in four, different clones for at least 4 weeks after Dox washout (Figure 2A-2C). Thus, the BRCA1 depletion associated differentiation change was not reversible following re-expression of the protein in formerly depleted cells.
Since changes in mammary differentiation have been associated with EMT induction in HME cells (Mani et al., 2008), we asked whether the change from a CD44low to CD44high phenotype was associated with the appearance of an EMT phenotype. Indeed, CD44high cells arising after BRCA1 depletion exhibited mesenchymal morphology (Figure 2D-middle and right vs left panel). Similarly, expression of E-cadherin, an epithelial marker, decreased while that of the mesenchymal markers, N-cadherin and ZEB1, increased in these cells (Figure 2E). In addition, mammary epithelial stem/progenitor-like cell activity, as reflected by mammosphere formation(Dontu et al., 2003), was significantly higher in the emerging BRCA1-depleted CD44high cells when compared with their CD44low counterparts (Figure 2F). These results are consistent with earlier findings(Liu et al., 2008; Proia et al., 2011).
Collectively, these results indicate that BRCA1 depletion in mammary epithelial cells can result in newly appearing CD44high cells that exhibit traits associated with a more primitive stem/progenitor-like state.
DNA Damage and Mammary Cell Differentiation
We then asked whether a defect in BRCA1-driven DNA damage responsiveness, a central BRCA1 function, accompanied the EMT observed in BRCA1-depleted CD44high HME cells. Importantly, BRCA1 is known to engage in the repair of both DSBs and ICLs, albeit by only partially overlapping mechanisms(Bunting et al., 2012; Bunting et al., 2010; Cao et al., 2009; Scully et al., 1997). Therefore, we challenged these cells with two, different DNA damaging agents: etoposide, which generates double strand breaks (DSB), and cisplatin, which largely generates interstrand DNA crosslinks (ICLs).
Etoposide exposure (1µM for 48 hours) significantly reduced colony formation but failed to promote the appearance of CD44high cells (Supplemental table 2). When CD44low cells were independently exposed to cisplatin at different concentrations (0.5µM for 8days, 1µM for 8days), we detected multiple clones containing CD44high cells in both cases (Figure 3A and Supplemental table 2). CD44high cell-containing clones also appeared at the highest cisplatin dose (2 µM for 48 hours). After plating the same number of cells, the number of surviving clones that was detected (N=28) was comparable to those that appeared after etoposide exposure (1µM for 48 hours, N=35). Under these conditions, only cisplatin exposure was associated with increased CD44high expression in any of the surviving clones. Thus, after eliciting comparable cell toxicity, DNA damage induced by cisplatin but not by etoposide triggered the development of a mesenchymal-like CD44high phenotype in the previously CD44low cells.
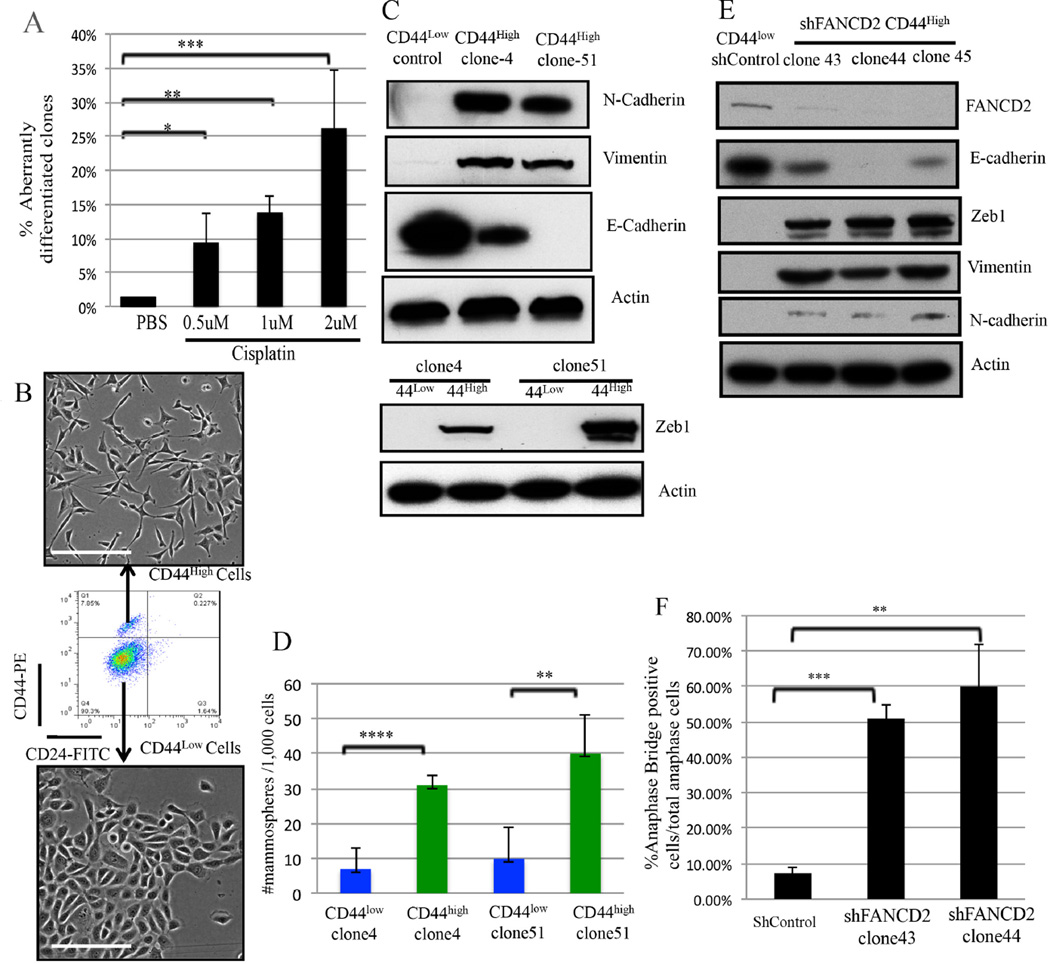
Figure 3. DNA Crosslink damage affects the differentiation of CD44low HME cells (Also see Figures S2).
3A) Summary of the effect of Cisplatin exposure upon the state of HME cell differentiation as reflected by the abundance of clones containing CD44high cells (≥5%). The data are represented as mean values ±S.D. *P<0.05, **P<0.01 and ***P<0.001.
3B) Phase-contrast images of FACS-purified CD44high and CD44low cells derived from an HME clone (Clone 4) following cisplatin-treatment. Scale Bar: 100µm
3C) Immunoblotting for EMT markers in FACS-purified CD44low and CD44high cells (clones 4 and 51) following cisplatin treatment of CD44low (control) cells.
3D) Mammosphere assays performed on cisplatin-treated, FACS-purified CD44high cells compared with their CD44low counterparts in two independent clones. The data represented are mean values ±S.D. **P<0.01 and ****P<0.0001.
3E) Immunoblotting for FANCD2 and EMT markers in CD44high cells from three, independent, FANCD2-depleted clones.
3F) Statistical analysis of anaphase bridge-positive cells in two, independent FANCD2-depleted clones (clone43 and clone44). The data represented are mean values ±S.D. **P<0.01 and ***P<0.001
Similar to the effects of BRCA1 depletion, when CD44high and CD44low HME cells from each of two, independent cisplatin-treated clones were sorted by FACS, the CD44high HME cells exhibited elongated mesenchymal morphology. The mesenchymal morphology was accompanied by a lack of cell-cell junctions while the CD44low cells formed well-defined islands via direct cell-cell contact and exhibited ‘cobblestone’ epithelial morphology (Figure 3B). Unlike CD44low cells, the CD44high cells revealed lower/absent E-cadherin expression and relatively high N-cadherin, Vimentin, and ZEB1 abundance (Figure 3C). Moreover, the CD44high cells generated mammospheres more efficiently than CD44low cells derived from the same clones (Figure 3D).
Taken together with the previously described results, these data imply that unrepaired ICL damage, on its own, can trigger the development of an aberrant state of mammary epithelial cell differentiation.
FANCD2 and BRG1 are essential for differentiation maintenance by promoting crosslink repair
FANCD2, a BRCA1- interacting protein(Garcia-Higuera et al., 2001), plays a key role in ICL repair (Kim and D’Andrea, 2012) , and BRCA1 deficiency is known to impair post-damage FANCD2 nuclear focus formation(Bunting et al., 2012; Garcia-Higuera et al., 2001). Consistent with defective ICL repair underlying conversion of mammary epithelial cells into mesenchymal stem-like cells, FANCD2 depletion from CD44low HME cells resulted in the emergence of multiple (5/45) CD44high cell–containing clones. In contrast, all 64 shControl-infected clones remained CD44low (Figure S2A–S2D). Western blotting confirmed the depletion of FANCD2 in the relevant clones (Figure 3E), and these CD44high cells exhibited a mesenchymal morphology and EMT-like protein expression while CD44low cells retained their epithelial properties (Figures 3E and Figure S2D).
Importantly, these FANCD2-deficient CD44high cells also exhibited signs of spontaneous DNA damage in the form of anaphase bridges (Figure 3F and Figure S2E). This result is consistent with failure of FANCD2 to contribute to the repair of the DNA lesions that give rise to these structures(Naim et al., 2013). These data again imply that failed DNA repair can elicit a change in the differentiation state of CD44low HME cells.
Endogenous BRCA1 interacts with the endogenous SWI/SNF protein, BRG1(Bochar et al., 2000), as was confirmed in 293T and MCF7 cells (Figure S3A–S3C), implying that endogenous BRCA1/BRG1 complexes also exist in human mammary epithelial cells. Similarly, endogenous/endogenous BRG1 and FANCD2 co-immunoprecipitate (Figure S3D), leading to speculation that BRG1, FANCD2, and BRCA1 co-contribute to certain common DNA repair functions(Dykhuizen et al., 2013).
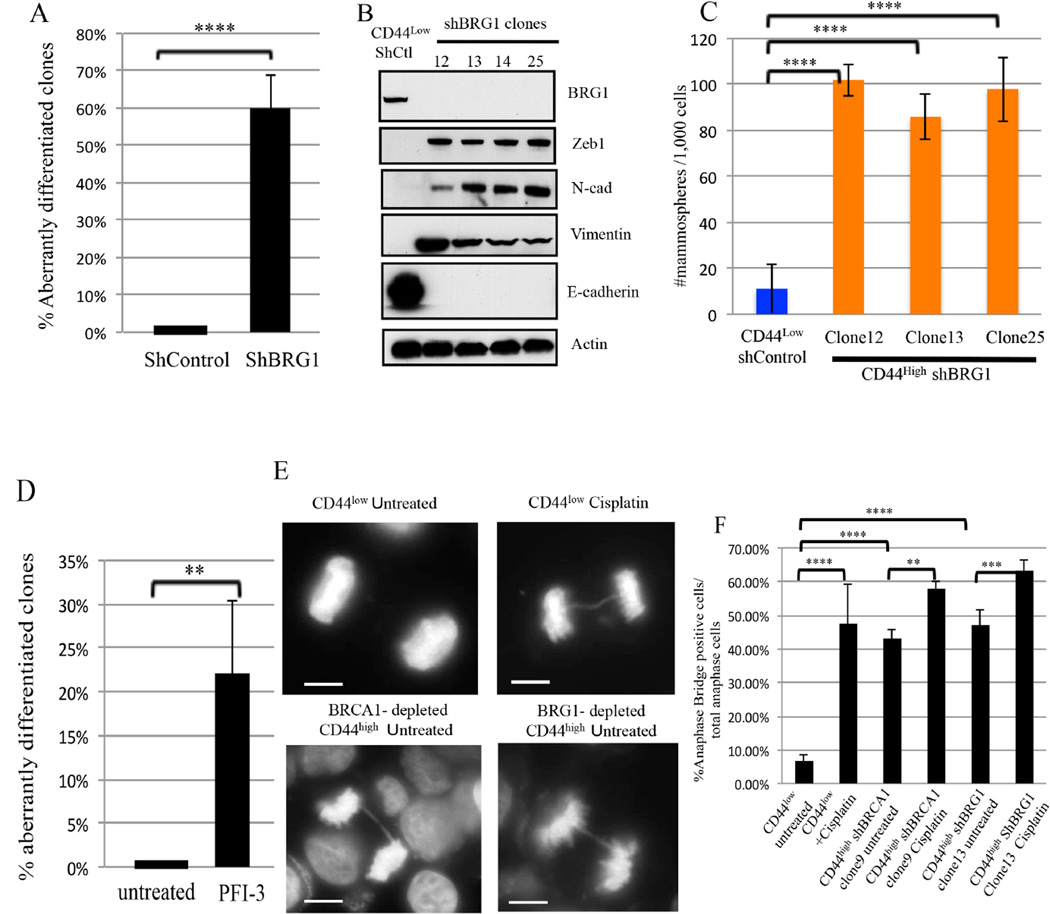
Like BRCA1 or FANCD2 depletion, BRG1 depletion also elicited an aberrant differentiation effect in CD44low HME cells (Figure 4A, and Figure S4A–S4B). BRG1-depleted CD44high cells exhibited EMT properties (Figure 4B and Figure S4C) and formed mammospheres more efficiently than controls (Figure 4C). Similar to BRG1 depletion, expression of a dominant negative ATPase-defective BRG1 mutant cDNA [K785R(Sif et al., 2001)] in CD44low HME cells led to a CD44high phenotype in numerous cells (Figure S4E) while WT BRG1 expression had no effect. Exposure of a BRG1 inhibitor (Vangamudi et al., 2015), PFI-3 (50µM), also led to an accumulation of CD44high cells compared with untreated controls (Figure 4D and Figure S4F–S4G). Thus, BRG1, like BRCA1 and FANCD2, participates in maintaining a stably differentiated HME cell state
Figure 4. BRG1 suppresses DNA damage-associated aberrant differentiation (Also see Figures S3 and S4).
4A) Summary of the effect of BRG1 depletion on the differentiation state of CD44low HME cells. ****P<0.0001.
4B) Immunoblotting for EMT markers from four, independent CD44high clones following BRG1 -depletion.
4C) Mammosphere assays were performed on CD44high HME cells that appeared after BRG1 depletion and on Shluc CD44low control cells. The data represented are mean±S.D. ****P<0.0001.
4D) Summary of the BRG1 inhibitor, PFI-3 (50 µM), exposure on the differentiation state in CD44low HME cells. . The data represented are mean values ±S.D. **P<0.01. **P<0.01. 4E) Representative images of normal anaphase chromosomes and anaphase bridges in untreated and cisplatin-treated (1µM, 24hr) CD44low cells, respectively. Other panels depict drug-free CD44high cells following long term, hairpin-mediated BRCA1 (clone 9) or BRG1 (Clone 13) depletion. Scale Bar: 10 µM.
4F) Analysis of anaphase bridge-positive cells in drug-free CD44low, shBRCA1 clone 9, and shBRG1 clone 13 CD44high cells and after exposure of these cells to cisplatin (1 µM, 24 hours). The data represented are mean±S.D.
Protein C receptor (PROCR)-expressing mammary epithelial cells represent multipotent mammary stem cells in mice (Wang et al., 2015) and in humans(Shipitsin et al., 2007). We detected the overexpression of PROCR (Figure S5A) in BRCA1 or BRG1- depleted CD44high cells by comparison with CD44low cells. This suggests aberrant CD44high HME cells after BRG1 or BRCA1 depletion may reflect a more primitive stem-/progenitor-like state. These results are analogous to Drosophila neuroblast dedifferentiation following the loss of mSWI/SNF complex function(Eroglu et al., 2014; Koe et al., 2014).
Consistent with the fact that both BRCA1 and BRG1 facilitate DNA decatenation (Dykhuizen et al., 2013; Lou et al., 2005), the BRCA1- or BRG1-depleted CD44high HME cells exhibited greater spontaneous anaphase bridge development, a manifestation of spontaneous DNA damage, than control CD44low cells (Figure 4E-4F). Thus, attenuating BRCA1 or BRG1 or FANCD2 function in CD44low HME cells led to aberrant differentiation and a parallel defect in resolving sister chromatids, an effect that can generate chromosome instability (Chan et al., 2009). Importantly, these results again imply that the control of genome integrity is linked to the maintenance of mammary epithelial cell differentiation.
BRCA1/BRG1/FancD2 depletion effects in other MEC lines
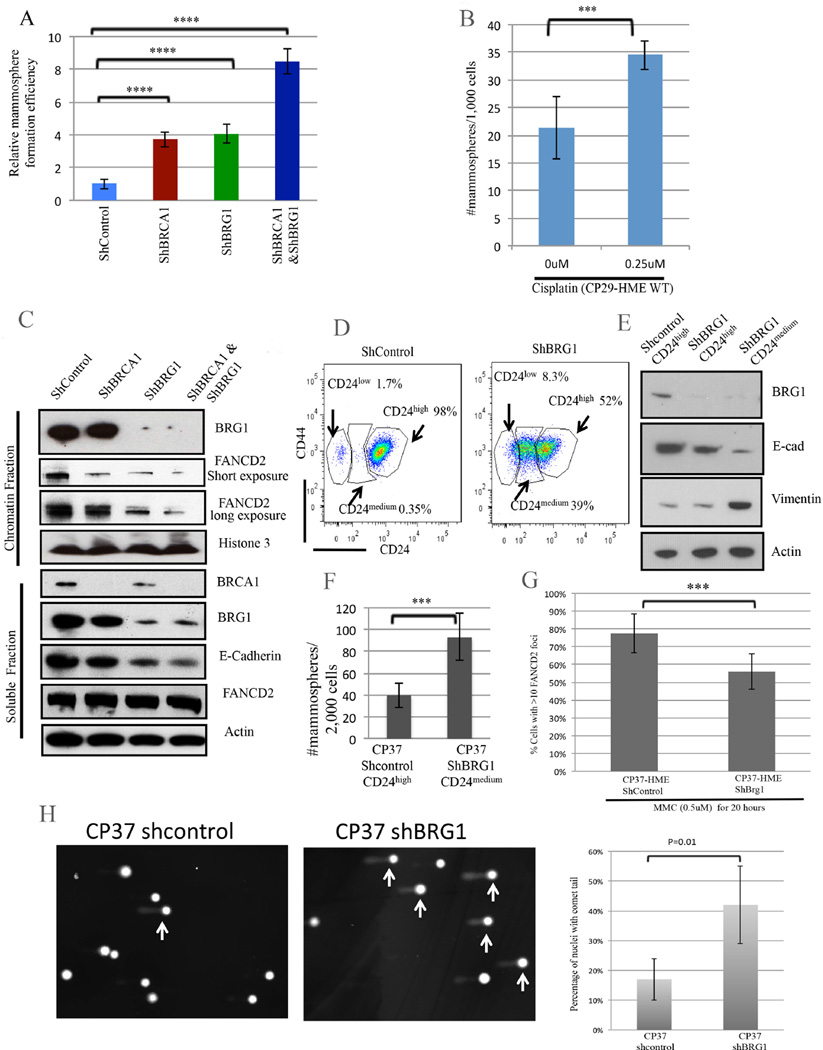
To assess the generality of BRCA1/BRG1 regulation of mammary epithelial cell differentiation, we found that loss of BRCA1 or BRG1, or both together, resulted in increased mammosphere-forming ability (Figure 5A) in another BRCA1+/+ HME line (CP29-HME) (Pathania et al., 2014). In addition, each of these perturbations as well as FancD2 depletion enhanced mitomycin C (MMC-another ICL generator) sensitivity in CP29-HME (Figure S5B–S5C). Exposure of naive CP29-HME to cisplatin (0.25µM) also led to enhanced mammosphere formation (Figure 5B). We further observed that depletion of BRCA1, BRG1, or both proteins together, triggered a loss of chromatin- bound FANCD2 in CP29-HME cells (Figure 5C). These data may well represent another indication of a close functional relationship among these proteins.
Figure 5. BRCA1 and BRG1 cooperate in crosslink repair and differentiation maintenance. (Also see Figures S5 and S6).
5A) Effect of BRCA1, BRG1, and BRCA1+BRG1 co-depletion on mammosphere-forming efficiency in CP29-HME cells. The mammosphere number was corrected for clonogenic survival efficiency on plastic to measure the relative mammosphere formation efficiency. The data represented are mean±S.D. **** P<0.0001.
5B) A mammosphere assay was performed on wt CP29-HME cells before and after cisplatin treatment. The data represented are mean±S.D. *** P<0.001
5C) Immunoblotting for chromatin-associated and soluble FANCD2 and E-cadherin following hairpin depletion of BRCA1, BRG1, or both in primary, telomerase-immortalized BRCA1+/+: p53 +/+ CP29-HME cells.
5D) CD24 and CD44 expression profiles following BRG1 depletion in CP37-HME cells.
5E) Immunoblotting for BRG1 and for EMT markers following BRG1 depletion in CP37- HME cells. FACS- purified CD24high and CD24medium cells were analyzed here.
5F) A mammospshere assay of CP37-HME cells following BRG1 depletion. The data represented are mean±S.D. *** P<0.001
5G) Quantification of FANCD2-containing DNA damage foci in CP37-HME cells after mitomycin C exposure. The data are represented as mean values ±S.D. *** P<0.001.
5H) Percent of cells with comet tails in BRG1-depleted CP37-HME cells compared to shcontrol cells. The data represented are mean values±S.D.
In yet a third HME line (CP37-HME(Pathania et al., 2014)), we again observed that BRG1 depletion promoted aberrant differentiation (Figure 5D-5F). After BRG1 depletion, 39% of these cells acquired a CD24medium mesenchymal phenotype (Figure 5D) while 98% of shcontrol-treated cells remained CD24high. Despite the lack of a CD44 change, the cells, nonetheless, revealed traits consistent with a change in differentiation state (Figure 5E-5F). Of note, others have observed a relationship between reduced CD24 expression and EMT in mammary epithelial cells (Al-Hajj et al., 2003; Mani et al., 2008).
In keeping with the aforementioned results pointing to BRG1/BRCA1/FANCD2 functional interactivity (Figure 5C), BRG1 depletion in CP37-HME cells also impaired the formation of FANCD2-containing nuclear foci after mitomycin C treatment (Figure 5G). Finally, in a related manifestation, CP37 HME cells revealed a major increase in comet formation after BRG1 depletion (Figure 5H), supporting a proposed role for BRG1 in the repair of spontaneously arising chromatid breaks.
Fanconi Anemia and mSWI/SNF genes are frequently mutated in breast cancer
Since BRCA1 is an established Fanconi Anemia (FA) gene (FANCS) (Sawyer et al., 2015), and BRCA1, FANCD2, and BRG1 all support ICL repair and a stable mammary epithelial differentiation state, we asked whether a breakdown of the FA or mSWI/SNF machinery leads to development of somatic breast cancer by analyzing the breast cancer genomic sequences in the 974 patient, TCGA Breast Cancer (BrCa) database (Cancer Genome Atlas, 2012; Cerami et al., 2012; Gao et al., 2013).
Collectively, the breast cancer genomes of 28% (264/974) of the TCGA patients contain mutations in 17 BRCA1/FA and 17 mSWI/SNF complex genes (Figure S6A). Moreover, each of the 264 cancers contained at least one BRCA/FA and/or one SWI/SNF mutation. This is consistent with the knowledge that FA and mSWI/SNF genes can serve as breast cancer suppressors (D’Andrea, 2010; Houghtaling et al., 2005; Kadoch et al., 2013).
In addition, this 264 breast tumor subset contains all of the FA and SWI/SNF mutations in the N= 974 breast cancer collection. All of these FA and SWI/SNF mutations are somatic, and these 264 tumors include abundant luminal and triple negative cancers (Cancer Genome Atlas, 2012). Given the role of FA gene products and a key SWI/SNF complex component in sustaining normal mammary epithelial differentiation, these FA- and/or SWI/SNF- mutant tumors represent a large and novel BrCa subset, defined with respect to defective DNA repair-centered processes that may have contributed to their development as cancers.
We also found that FANCD2 was preferentially mutated in the sporadic hormone- negative breast cancers while FANCA was mutated in the sporadic hormone- positive breast cancers among this 264 case subset (Figure S6B). Similarly ARID1A was more frequently mutated in hormone- positive breast cancers while ARID1B is more frequently mutated in hormone negative tumors (data not shown). Perhaps, certain defective differentiation control functions dominated in these cancers depending upon the nature of their defective FA or mSWI/SNF subunit function.
In addition, mSWI/SNF mutations were identified in approximately half of the patient tumor DNAs that contained BRCA1 mutations (N=15, 15/32, 47%). However, many more that contained mSWI/SNF mutations lacked BRCA1 mutations (N=135, 135/942, 14%; P<0.0001; Fig S6C). Our results also indicate that BRCA1 and BRG1 cooperate in both in mammary epithelial differentiation maintenance (mammosphere suppression) and participate in ICL damage repair (Figure 5A-5B). Therefore, since BRCA1/FA and mSWI/SNF mutations were co-identified in multiple, albeit a minority of breast cancers, one might also hypothesize that BRCA1/FA and mSWI/SNF functions cooperate to suppress breast cancer development.
ΔNp63 links DNA damage responsiveness and differentiation control in mammary epithelial cells
p63, a multifunctional p53 family member, can suppress the development of mammary epithelial EMT, likely by inhibiting TGFβ function(Adorno et al., 2009; Lindsay et al., 2011) . And BRCA1 (Buckley et al., 2011) and FANCD2(Park et al., 2013) positively regulate p63 transcription.
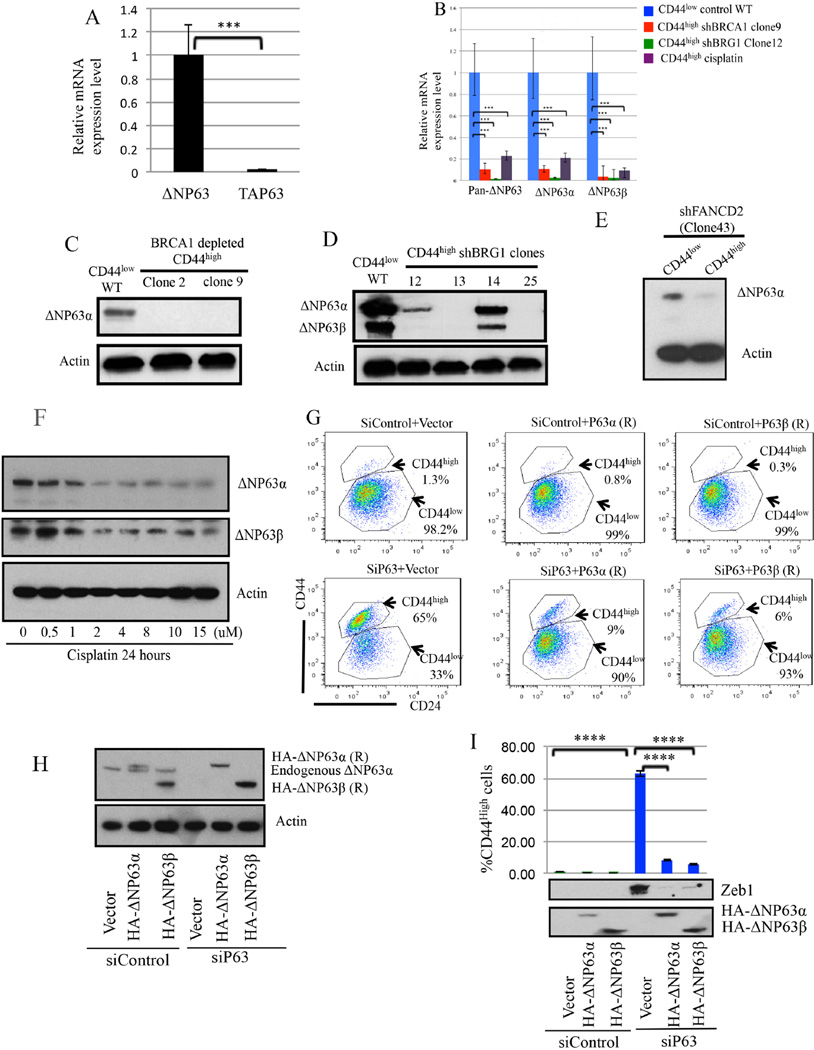
HME cells contain ΔNp63 and not TAP63 p63 isoforms (Figure 6A) (Lindsay et al., 2011). In BRCA1-, BRG1-, or FANCD2- depleted CD44high HME cells, ΔNp63 mRNA and protein expression were diminished when compared with their expression in naive CD44low HME (Figure 6B-6E). Similarly, BRCA1 depletion reduced ΔNp63 expression in HME cells which remained CD44low during the assay (Figure S5D). Depleting another FA protein, PALB2, also reduced ΔNp63 expression in CD44low cells (Figure S5E). Consistent with the results of others(Fomenkov et al., 2004), there was a concomitant reduction in expression of the two ΔNp63 isoforms, α and β after 24 hour exposure of CD44low HME cells to cisplatin (Figure 6F). These findings suggest that ICLs trigger the suppression of ΔNp63 expression (Fomenkov et al., 2004).
Figure 6. ΔNp63 depletion is associated with aberrant HME cell differentiation (Also see Figure S7).
6A) ΔNp63 is the primary p63 isoform in CD44low HME cells. The data represented are mean values±S.D. *** P<0.001
6B) ΔNP63α and ΔNP63β mRNA expression were significantly reduced in aberrantly differentiated CD44high cells after BRCA1 or BRG1 depletion or after Cisplatin treatment. The data represented are mean values±S.D. ***P<0.001.
6C-6E) Immunoblotting for ΔNp63 in CD44high cells after BRCA1/BRG1/FANCD2 depletion.
6F) Cisplatin suppresses endogenous ΔNP63 expression in CD44low HME cells.
6G-6I) ΔNP63 is necessary and sufficient to suppress the aberrant differentiation of CD44low cells. After siP63 transfection, the percentage of CD44high cells was measured, and expression of the EMT marker, Zeb1, and of the two HA-tagged ΔNP63 vectors was analyzed. ****P<0.0001.
To determine if ΔNP63 depletion affects HME cell differentiation, we sought and detected evidence of a conversion of CD44low to CD44high HME cells after siRNA-mediated ΔNp63 depletion (Figure 6G-6I). This abnormal differentiation-associated phenotype was prevented by co-expression of HA-tagged, siP63-resistant ΔNp63α or ΔNp63β but not by relevant controls (Figure 6G-6I). Thus, ΔNp63 expression suppressed the aberrant differentiation of HME cells.
siRNA-mediated ΔNp63 depletion also led to increased expression of the mesenchymal marker and EMT driver, ZEB1, in HME cells, which was suppressed by siP63-resistant HA-ΔNp63α or HA-ΔNp63β expression (Figure 6I). Thus, maintenance of ΔNP63 expression sufficed to prevent EMT development in these cells (Adorno et al., 2009).
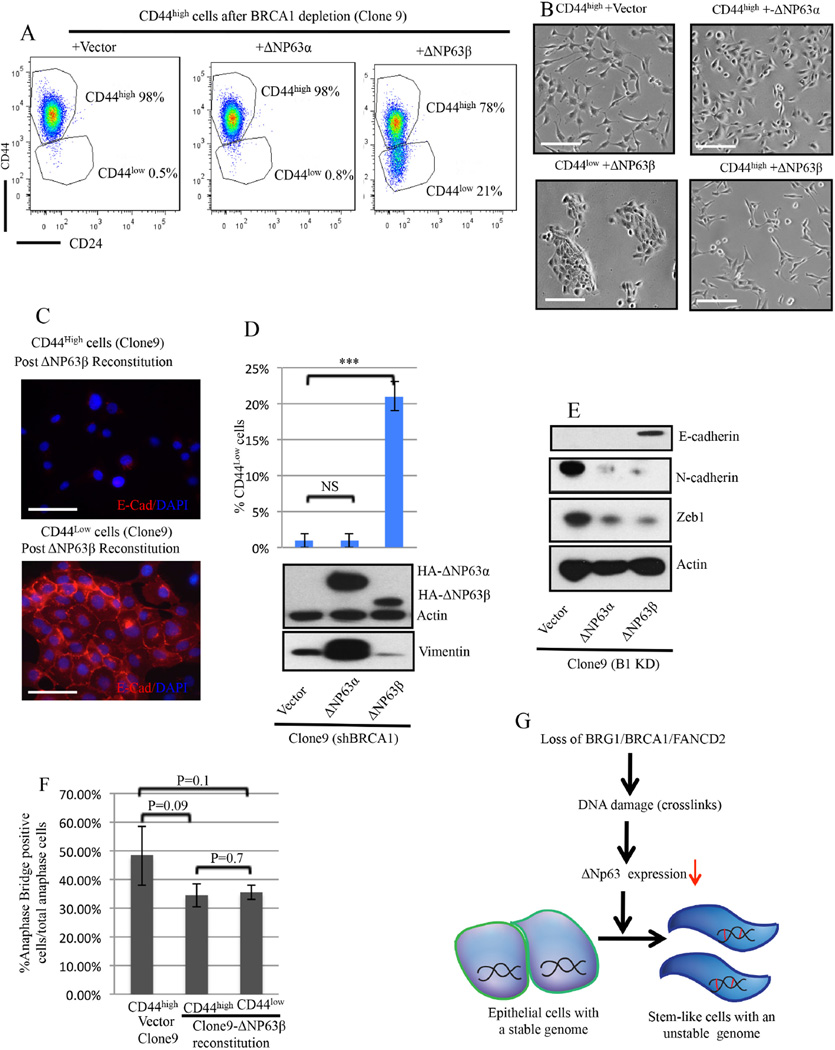
We also attempted to reconstitute BRCA1 -depleted CD44high cells in two ways. First, allowing the BRCA1 concentration in HME cells to rise to nearly what it was before Dox-triggered depletion failed to alter EMT protein expression signature or their ΔNp63-depleted state (Figure 2B). By contrast, in cells that had become ΔNp63α/β co-depleted after Dox induced BRCA1 depletion, subsequent ectopic ΔNp63β re-expression led to the development of a mesenchymal to epithelial transition (MET) phenotype (Figure 7A-7E) and to conversion of significant numbers of CD44high to CD44low cells (Figure 7A-right FACS panel). Indeed, the fraction of ΔNp63β -generated CD44low cells increased from 3% after 2 weeks to 20% after 6 weeks. After FACS purification, these CD44low cells revealed an epithelial phenotype, while their residual CD44high counterparts retained mesenchymal morphology (Figure 7B-7E). ΔNp63β re-expression also led to reduced N-cadherin and Zeb1 and increased E-cadherin expression (Figure 7E).
Figure 7. ΔNP63β expression promotes differentiation of BRCA1-deficient CD44high HME cells.
7A) Representative CD24 and CD44 profiles after ΔNP63 α vs β reconstitution of a BRCA1-depleted clone (clone9) composed, almost uniformly, of CD44high cells when the experiment was initiated.
7B) Phase-contrast images of CD44high cells after empty vector or HA-ΔNP63α transduction of CD44high cells (clone9) and phase contrast images of CD44high and CD44low cells isolated by FACS after HA-ΔNP63β transduction of CD44high cells (clone9). Scale bar: 100µm.
7C) CD44low cells arising after HA-ΔNP63β reconstitution were positive for E-Cadherin while CD44high cells remained E-cadherin negative. Scale bar: 50µm.
7D) Percentage of CD44low cells after ΔNP63β, ΔNP63α,β or empty vector reconstitution of CD44high cells (clone9).
7E) Immuoblotting for EMT markers after ΔNP63β or ΔNP63αβ or empty vector reconstitution in CD44high cells (clone9).
7F) Percentage of anaphase bridge-positive cells in ΔNp63β- reconstituted CD44high and CD44low BRCA-depleted clone 9 cells. The data represented are mean values±S.D..
7G) A model of the mechanism underlying BRCA1/BRG1/FANCD2 dependent differentiation maintenance in HME cells in response to DNA damage. The short red bars represent interstrand DNA cross links.
While reconstitution with ΔNp63α failed to promote a marked CD44high to CD44low HME cell change or dramatic EMT reversal (Figure 7A), it did lead to a major reduction in N-cadherin and ZEB1 expression (Figure 7E). The basis for the differing effects of the two isoforms is unknown.
Thus, after BRCA1 depletion and the development of an aberrantly differentiated/ EMT phenotype, reversal of these characteristics was not achievable by eliminating the initial insult, i.e. BRCA1 depletion. It did, however, occur after re-expression of a downstream effector protein, ΔNp63β. The latter strongly implies that the aberrant differentiation of these cells arising after DNA damage is a product of the dedifferentiation of more differentiated derivative cells.
Notably, while ΔNp63β reconstitution led to the appearance of a sizeable subpopulation of CD44low, MET-appearing cells, it failed to reduce the abundance of their anaphase bridges (Figure 7F). Thus, the dedifferentiated phenotype can be eliminated while the offending damage remains.
Our data suggest a model in which BRCA1, BRG1, and FANCD2 together contribute to the maintenance of HME cell differentiation by sustaining ΔNp63 expression in the face of ongoing DNA damage (Figure 7G).
Effects on HME cells of simple aldehyde exposure
Simple, naturally occurring aldehydes have been shown by others to elicit DNA crosslink damage that requires FA pathway function to repair. In FA-deficient mice lacking a suitable aldehyde dehydrogenase species, they also promoted leukemogenesis(Langevin et al., 2011; Pontel et al., 2015).
In an effort to assess their potential to perturb mammary differentiation control, we exposed HME cells to acetaldehyde and, independently, to formaldehyde(Langevin et al., 2011; Pontel et al., 2015). Both promoted mammosphere formation, a manifestation of early progenitor/stem cell activity (Figure S7A and S7B). Interestingly, formaldehyde exposure also induced a change to a more mesenchymal morphology (Figure S7C) and suppressed E-cadherin and increased Vimentin expression in CD44low cells after 24 hours exposure, suggesting a role in promoting EMT (Figure S7D). Taken together, these data strongly suggest that certain simple aldehydes in sufficient concentration can promote aberrant differentiation of HME cells.
Similar to cisplatin exposure, we also found that exposure to acetaldehyde or formaldehyde suppressed ΔNp63 expression in CD44low cells (Figure S7E–S7F). Furthermore, 0.2mM formaldehyde led to incomplete and 1uM disulfiram, an ALDH inhibitor, led to no ΔNp63 depletion. However, when combined, these two perturbations led to greater depletion than was detected after formaldehyde exposure, alone (Figure S7G).
These results are consistent with sufficient formaldehyde or acetaldehyde exposure triggering a key step on the road to MEC dedifferentiation, at least in part, by suppressing ΔNp63 expression. The ΔNp63 depletion effect was further enhanced by what is presumed to be greater accumulation of formaldehyde that had been inefficiently metabolized because a key metabolizing enzyme(s) was inhibited by Disulfiram (Figure S7G).
Discussion
We have demonstrated a convergence of two, important, tumor suppressing processes in mammary epithelial cells, genome stability control and differentiation maintenance. Both are governed by BRCA1/FANCS, by its known partner proteins, BRG1 and FANCD2, and likely by other FA and mSWI/SNF gene products, as well. Using derivative clonal analysis as a lineage-tracing tool, we have confirmed that BRCA1 is necessary for differentiation maintenance in human mammary epithelial cells.
Loss of BRCA1, FANCD2, or BRG1 expression triggered differentiation-related CD44 or CD24 cell surface expression changes in ostensibly normal mammary epithelial cells. Where tested, such changes were accompanied by increased PROCR expression, acquisition of EMT morphology and an associated protein expression profile, enhanced mammosphere formation, and clear evidence of spontaneously arising chromosomal instability (e.g. anaphase bridge formation and comets). Importantly, exposure to an interstrand crosslinking agent, alone, elicited most of these outcomes. These results and the human breast cancer genomic evidence, discussed above, suggest the existence of a BRCA1/FA-mSWI/SNF-ΔNp63- driven DNA repair- dependent epithelial differentiation control system operating as a major component of breast cancer suppression.
We also observed that BRCA1, FANCD2, or BRG1 deficiency elicited an aberrant differentiation change from an epithelial to a more primitive, stem/progenitor-like phenotype. An analogous EMT-associated phenotype has been observed in BRCA1-defective mouse models(Ye et al., 2015). Moreover, others have shown that primary mammary epithelial cells from BRCA1 mutation carriers contain abnormally high numbers of CD44high or p27+ progenitor-like cells by comparison with their non-isogenic, wt counterparts(Choudhury et al., 2013).
Importantly, cisplatin, simple aldehydes, BRG1, FancD2, or BRCA1 depletion, alone, led to reduced ΔNp63 expression and spontaneously arising DNA damage in CD44low HME cells. Each of these conditions also led to an aberrant differentiation state. The aberrant differentiation phenotype following BRCA1 depletion was reversed, albeit incompletely, by ectopic expression of ΔNp63β. While it promoted the re-differentiation of BRCA1-deficient CD44high/ EMT positive cells to CD44low /MET derivatives, it failed to reduce anaphase bridges in these cells. Thus, not surprisingly, the DNA damage remained despite the re-differentiation effect.
These findings show that the results of BRCA1 depletion in CD44low HME cells are not a product of overgrowth by a pre-existing, ΔNp63-negative CD44high cell population. They also argue strongly that the development of an aberrant mammary epithelial phenotype in such a setting represents a form of dedifferentiation to a more stem/progenitor-like state.
This putative disease-generating effect is compatible with genomic data obtained from a sizeable subset of a large breast cancer group. This subset was targeted by a combination of somatic BRCA1/FA and mSWI/SNF mutations that, we propose, reflect a new, disease-contributing pathophysiological process. In one scenario, defective ICL repair or a related repair mechanism(s) may lead to: 1) the development of replication stress (Pathania et al., 2014; Pathania et al., 2011), an established characteristic of BRCA1 +/− mammary epithelial cells and of numerous epithelial cancers(Gorgoulis et al., 2005), and/or 2) transcription-associated damage (Aguilera and Garcia-Muse, 2012; Hatchi et al., 2015; Hill et al., 2014; Schwab et al., 2015) that may also underlie at least part of this disease-producing process.
Stimulated by the work of Patel and co-workers, we tested two, small aldehydes (formaldehyde and acetaldehyde) and observed that they, too, elicited a dedifferentiating effect in mammary epithelial progenitor cells (Langevin et al., 2011; Pontel et al., 2015). An aldehyde oxidase inhibitor significantly exacerbated the effect of a limiting concentration of formaldehyde, suggesting that aldehyde-driven DNA damage leads to defective FANC-mSWI/SNF-ΔNp63 differentiation control, which may contribute to the widely reported breast cancer risk-elevating effect of ethanol ingestion (Jayasekara et al., 2015). A test of this notion will be needed in the future.
Finally, these results reinforce the argument that genome caretaking sustains not only chromosomal integrity but also normal cellular differentiation, organ development, and tumor suppression (Eroglu et al., 2014; Park et al., 2013; Santos et al., 2014; Wang et al., 2012).
Experimental Procedures
Cell culture
All human mammary epithelial (HME) cells were immortalized with hTERT and maintained in MEGM medium as reported previously (Pathania et al., 2014). 293T and MCF7 cells were cultivated in DMEM containing 10% FBS. All cells were cultivated at 37 °C in a 10% CO2-containing atmosphere.
FACS and Single clone generation
HME cells were stained with CD24-FITC and CD44-PE antibodies and sorted in a BD FACSAria II instrument.
Single mammary epithelial cell clone were generated by plating ~500 single CD44low in a 10cm plate. After 8–10 days, individual clones were retrieved, and each was expanded in a single well of a 24 well plate for flow cytometry analysis.
Western blotting and antibodies
Cells were lysed for 2 hours on ice in NETN-250 lysis buffer. To separate the soluble and chromatin fractions, cells were lysed for 30 minutes on ice in NETN-250 lysis buffer. Soluble and chromatin fractions were separated by centrifugation at 13,200g at 4°C.
Immunoprecipitation was performed by incubation of NTEN 250-based soluble extract with anti-BRCA1 or anti-BRG1 antibodies and protein A-coupled Sepharose beads (GE). The beads were washed in NTEN100 buffer and boiled for 5 min at 95°C in Laemmli buffer and centrifuged. The supernatants were subjected to western analysis.
The antibodies were listed in the supplemental information.
Clonogenic and cell growth assays
To measure cell growth, ~ 300 cells/well were seeded in 6-well plastic plates and treated with drugs and cultivated for 10 days. Cells were fixed and visualized with crystal violet.
To measure colony-forming efficiency before vs after BRCA1/BRCA2 depletion, one or two cells from that line were seeded in each well of a 96-well plate with or without doxycycline. After 8 days, cells were fixed and stained with crystal violet to visualize individual clones.
Cell cycle was analyzed with a FITC Mouse Anti-BrdU kit according to the manufacturer’s protocol.
ShRNAs and SiRNA
The shRNA and siRNA sequences were listed in the supplemental information. The packaging of lentivirus was described as before as described (Sheng et al., 2010).
Anaphase bridges
Proliferating cells were plated at ~ 60% confluence, fixed with 4% paraformaldehyde for 15 minutes, and stained with 4’,6-diamidino-2-phenylindole (DAPI; Vector Labs). A total of at least 100 anaphase or early telophase cells were scored in each experiment.
Comet Assay
Alkaline comet assay were performed on CP37-HME cells before and after BRG1 depleting using the Single-Cell Gel Electrophoresis Assay kit (Trevigen) according to the manufacturer’s instructions. About 400 cells were accounted in each experiment.
Quantitative RT-PCR
RNA from relevant cell lines was extracted using an RNeasy Plus Mini Kit (Qiagen). cDNA was prepared using a SuperScript III kit (Invitrogen). cDNA was analyzed by qRT-PCR with iQ SYBR green supermix (Bio-Rad), and the results were analyzed using the delta-delta threshold cycle method. Primer sequences are listed in supplemental information.
Vectors and Cloning
ShRNA-resistant HA-BRCA1 was described earlier(Pathania et al., 2011). Flag-tagged BRG1 WT- and K785R- encoding plasmids were gifts of Dr. Robert Kingston (Massachusetts General Hospital). Full-length ΔNP63β and ΔNP63β cDNAs were amplified with specific primers (listed in supplemental item 4) and cloned into a POZ-FH-N retro-vector as previously described(Nakatani and Ogryzko, 2003).
TCGA Mutational analysis of Fanconi Anemia(FA) genes and mSWI/SNF genes
TCGA-collected mutations in BRCA1/ Fanconi Anemia (FA) and 17 core mSWI/SNF genes were identified at the cbioport website using TCGA 2015 Cell(Cerami et al., 2012; Gao et al., 2013) data (974 tumors with sequencing and CNA data). The analysis results can be retrieved using the link (http://bit.ly/1MpaUnB). Concurrent relationships between mutations in BRCA1/FA, and mSWI/SNF genes were assessed using two-sided Fisher’s exact tests.
Statistic Analysis
Otherwise stated, all statistic analysis was performed by a two-sided student’s t-test.
Supplementary Material
Highlights.
BRCA1 is necessary for the maintenance of mammary epithelial cell differentiation
Interstrand crosslink repair stabilizes mammary epithelial cell differentiation
FA and mSWI/SNF genes are frequent mutated in sporadic breast cancers
ΔNP63 links DNA repair and differentiation maintenance in HME cells
Acknowledgments
We thank all Livingston laboratory members for numerous helpful discussions. We are also grateful to Drs. Robert Weinberg and Qiufu Ma for important suggestions and other valuable input along the way. We also thank Dr A. D’Andrea for the generous gifts of valuable reagents and helpful insights, and the DFCI flow cytometry facility for excellent technical support. This work was supported by grants from the National Cancer Institute (NCI)-Mechanisms of Breast Development and Carcinogenesis (2PO1CA80111-16), BRCA1 Function in Post Damage Foci (5R01CA136512-05), and from the Susan B Komen Foundation for the Cure (to DML) and from the Breast Cancer Research Foundation (to DML).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.W and D.M.L conceptualized and designed the study. H.W conducted most of the experiments. B.B. provided advice relevant to the p63 and other HME-associated analyses. A.G.L, K.T, S.D and B.L provided advice and helped with certain experiments, and S.P. and J.F. provided immortalized HMEC strains and advice on how to study them. R.G and A.G helped TCGA data analysis. K.P advised on the design and means of analysis of certain experiments. D.M.L supervised the project. H.W and D.M.L wrote the manuscript, and all authors read, edited, and/or commented upon the manuscript.
References
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Buckley NE, Conlon SJ, Jirstrom K, Kay EW, Crawford NT, O’Grady A, Sheehan K, Mc Dade SS, Wang CW, McCance DJ, et al. The DeltaNp63 proteins are key allies of BRCA1 in the prevention of basal-like breast cancer. Cancer research. 2011;71:1933–1944. doi: 10.1158/0008-5472.CAN-10-2717. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Molecular cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Molecular cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature cell biology. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Choudhury S, Almendro V, Merino VF, Wu Z, Maruyama R, Su Y, Martins FC, Fackler MJ, Bessarabova M, Kowalczyk A, et al. Molecular profiling of human mammary gland links breast cancer risk to a p27(+) cell population with progenitor characteristics. Cell stem cell. 2013;13:117–130. doi: 10.1016/j.stem.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & development. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho YJ, Zhao K, Crabtree GR. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu E, Burkard TR, Jiang Y, Saini N, Homem CC, Reichert H, Knoblich JA. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- Fomenkov A, Zangen R, Huang YP, Osada M, Guo Z, Fomenkov T, Trink B, Sidransky D, Ratovitski EA. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell cycle. 2004;3:1285–1295. doi: 10.4161/cc.3.10.1155. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Molecular cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Molecular cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SJ, Rolland T, Adelmant G, Xia X, Owen MS, Dricot A, Zack TI, Sahni N, Jacob Y, Hao T, et al. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes & development. 2014;28:1957–1975. doi: 10.1101/gad.241620.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nature reviews. Molecular cell biology. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- Jayasekara H, MacInnis RJ, Room R, English DR. Long-Term Alcohol Consumption and Breast, Upper Aero-Digestive Tract and Colorectal Cancer Risk: A Systematic Review and Meta-Analysis. Alcohol and alcoholism. 2015 doi: 10.1093/alcalc/agv110. [DOI] [PubMed] [Google Scholar]
- Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, Gao J, Schultz N, Gonen M, Soslow RA, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nature genetics. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nature reviews. Molecular cell biology. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes & development. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koe CT, Li S, Rossi F, Wong JJ, Wang Y, Zhang Z, Chen K, Aw SS, Richardson HE, Robson P, et al. The Brm-HDAC3-Erm repressor complex suppresses dedifferentiation in Drosophila type II neuroblast lineages. eLife. 2014;3:e01906. doi: 10.7554/eLife.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Lindsay J, McDade SS, Pickard A, McCloskey KD, McCance DJ. Role of DeltaNp63gamma in epithelial to mesenchymal transition. The Journal of biological chemistry. 2011;286:3915–3924. doi: 10.1074/jbc.M110.162511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS. BRCA1 regulates human mammary stem/progenitor cell fate. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nature structural & molecular biology. 2005;12:589–593. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, Ramus SJ, Spurdle A, Robson M, Sherman M, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell stem cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nature cell biology. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods in enzymology. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Park E, Kim H, Kim JM, Primack B, Vidal-Cardenas S, Xu Y, Price BD, Mills AA, D’Andrea AD. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Molecular cell. 2013;50:908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, Su Y, Ting DT, Polyak K, Richardson AL, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nature communications. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, Feunteun J, Livingston DM. BRCA1 is required for postreplication repair after UV-induced DNA damage. Molecular cell. 2011;44:235–251. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature reviews. Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, et al. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Molecular cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell stem cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P, Karnezis AN, Craig DW, Sekulic A, Russell ML, Hendricks WP, Corneveaux JJ, Barrett MT, Shumansky K, Yang Y, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nature genetics. 2014;46:427–429. doi: 10.1038/ng.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA, Faryabi RB, Ergen AV, Day AM, Malhowski A, Canela A, Onozawa M, Lee JE, Callen E, Gutierrez-Martinez P, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian G, Consortium FC, Majewski J, Dyment DA, Innes AM, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer discovery. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Molecular cell. 2015;60:351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Sheng Q, Liu X, Fleming E, Yuan K, Piao H, Chen J, Moustafa Z, Thomas RK, Greulich H, Schinzel A, et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer cell. 2010;17:298–310. doi: 10.1016/j.ccr.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MH, Kuraishy AI, Deshpande C, Hong JS, Cacalano NA, Gatti RA, Manis JP, Damore MA, Pellegrini M, Teitell MA. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Molecular cell. 2010;39:873–885. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A–containing Brg1 and hBrm chromatin remodeling complexes. Genes & development. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends in cell biology. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature cell biology. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangamudi B, Paul TA, Shah PK, Kost-Alimova M, Nottebaum L, Shi X, Zhan Y, Leo E, Mahadeshwar HS, Protopopov A, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer research. 2015 doi: 10.1158/0008-5472.CAN-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, Zeng YA. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nature genetics. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.