Abstract
6-(Methylsulfinyl)hexyl isothiocyanate (6-MSITC), 6-(methylthio)hexyl isothiocyanate (6-MTITC), and 4-(methylsulfinyl)butyl isothiocyanate (4-MSITC) are isothiocyanate (ITC) bioactive compounds from Japanese Wasabi. Previous in vivo studies highlighted the neuroprotective potential of ITCs since ITCs enhance the production of antioxidant-related enzymes. Thus, in this present study, a genome-wide DNA microarray analysis was designed to profile gene expression changes in a neuron cell line, IMR-32, stimulated by these ITCs. Among these ITCs, 6-MSITC caused the expression changes of most genes (263), of which 100 genes were upregulated and 163 genes were downregulated. Gene categorization showed that most of the differentially expressed genes are involved in oxidative stress response, and pathway analysis further revealed that Nrf2-mediated oxidative stress pathway is the top of the ITC-modulated signaling pathway. Finally, real-time polymerase chain reaction (PCR) and Western blotting confirmed the gene expression and protein products of the major targets by ITCs. Taken together, Wasabi-derived ITCs might target the Nrf2-mediated oxidative stress pathway to exert neuroprotective effects.
Keywords: Wasabi, isothiocyanates, sulforaphane, gene profile analysis, Nrf2 pathway
Introduction
Wasabi (Wasabi japonica (Miq.) Matsumura), commonly known as Japanese horseradish, is a member of the Brassi-caceae vegetables. Its rhizome has a pungent flavor, which is popularly used as a spice among Japanese household. Studies have shown that Wasabi has multifarious functions such as antimicrobial, anticoagulation, anti-inflammatory, anti-obesity, and anticancer.1–5 These activities can be attributed to a group of bioactive compounds identified as isothio-cyanates (ITCs).6 They include 4-(methylsulfinyl)butyl isothiocyanate (4-MSITC, usually called sulforaphane, SFN), 6-(methylsulfinyl)hexyl isothiocyanate (6-MSITC), and 6-(methylthio)hexyl isothiocyanate (6-MTITC; Fig. 1). Our previous study revealed that a structure–activity relationship of Wasabi ITCs was present for the inhibition of cyclooxygenase-2 expression with a dependence on the methyl chain length of Wasabi ITCs.7 The longer the methyl chain length of Wasabi ITCs, the stronger the inhibition of cyclooxygenase-2 expression.
Figure 1.
Chemical structures of Wasabi-derived ITCs used in the study: (A) 4-(methylsulfinyl)butyl isothiocyanate (4-MSITC, usually called sulforaphane, SFN), (B) 6-(methylsufinyl)hexyl isothiocyanate (6-MSITC), and (C) 6-(methylthio)hexyl isothiocyanate (6-MTITC).
Recently, Tarozzi et al have provided a review highlighting the potential of SFN against neurodegenerative diseases by implicating the activation of nuclear factor E2-related factor 2/cis-antioxidant response element (Nrf2-ARE) pathway.8 SFN pretreatment could protect disruption of blood–cerebrospinal fluid barrier and shield astrocytes and neuron cells from toxic effects caused by various oxidants through the increase of intracellular glutathione (GSH) and the induction of nicotinamide adenine dinucleotide phosphate (NAD[P]H):quinone oxidoreductase 1 (NQO1) via the activation of Nrf2 pathway.9–11 Similarly, in quinone-induced dopaminergic cell death model, SFN exerted protective function by mediating the toxic accumulation of quinones via induction of NQO1 expression.12 Other than providing long-term protection against oxidative damage via upregulation of the antioxidant redox system, SFN can also downregulate the expressions of inflammation-associated genes.13 Indeed, SFN is a budding neuroprotective agent but little is known yet about its analogs found in Japanese Wasabi. To date, animal study, following Parkinson’s disease mouse model, demonstrated that 6-MSITC reduced motor dysfunction induced by 6-hydroxydopamine via reducing oxidative stress and apoptotic cell death.14 Also, 6-MSITC prevented oxidative stress cytotoxicity by raising the intracellular GSH content through the increase of γ-glutamylcysteine synthetase induced by the activation of Nrf2/ARE pathway in an oxidative stress-induced animal model.11 However, the exact molecular mechanism of interaction of Wasabi-derived ITCs toward neuroprotection at the cellular level has not yet been ascertained.
Nrf2 is a basic region leucine zipper transcription factor that activates the Nrf2/ARE pathway. It acts as the master regulator of the cellular antioxidant response via modulating the expressions of over 250 genes.15 Impaired Nrf2 leads to a dysfunctional Nrf2 pathway that decreases cellular defense against oxidative stress.16 The overproduction of oxidative stress could contribute to cell death, which is associated with the progression of neurodegenerative diseases. In tert-butyl hydroquinone-induced astrocytes, the cells from Nrf2- deficient mice were more sensitive to oxidative stress than the cells from wild-type mice.17 Hence, upregulation of Nrf2 activity is an attractive approach to combat the increase of oxidative stress during the development of neurodegeneration. Interestingly, in vivo studies revealed that Nrf2 inducers reduced toxic-induced cellular damage in the brain of wild-type Nrf2 mice but not in Nrf2 knockout mice.18,19 For instance, SFN administration in rats exposed to traumatic brain injury attenuated oxidative stress and neuronal damage via upregulation of Nrf2-dependent antioxidant enzymes such as heme oxygenase 1 (HO-1) and NQO1.20 HO-1 catalyzes heme degradation to form CO, free iron, and biliverdin that immediately undergoes enzymatic reduction to form bilirubin, a potent antioxidant and protector of neuron cells against oxidative stress even at minute concentration.21 NQO1 catalyzes the two-electron reduction of quinones and diverts the participation of these agents from one-electron oxidoreduction and oxidative stress.22 Therefore, further understanding of how Nrf2/ARE pathway prevents the progress of neurodegenerative diseases through the use of these bioactive agents is important.
DNA microarray can investigate the expressions of thousands of genes simultaneously in a given cell type or tissue sample.23,24 In our previous investigation, the anti-inflammatory genes and associated signaling pathways targeted by 6-MSITC were successfully clarified by employing DNA microarray technology to macrophages.25 In this present study, to clarify the molecular mechanism of Wasabi-derived ITCs on neuroprotection at the cellular level, we carried out DNA microarray analysis to profile gene expression changes in a neuronal model cell line, IMR-32, stimulated by these ITCs. Moreover, Ingenuity Pathway Analysis (IPA) was used to map out cellular signaling pathways for these ITC-regulated gene expressions.
Materials and Methods
Materials
ITCs (SFN, 6-MSITC, and 6-MTITC) were purified from Wasabi by reversed-phase high performance liquid chromatography (HPLC) to >99.3% purity26 and dissolved in dimethyl sulfoxide for cell culture experiments. The antibodies against Nrf2 (C-20), Keap1 (E-20), NQO1 (C-19), HSP70 (D69), GAPDH, rabbit IgG, and horseradish peroxidase (HRP)-conjugated anti-goat secondary antibody were purchased from Santa Cruz Biotechnology. AKR1C1, AKR1C3, and TXNRD1 antibodies were obtained from Abcam. HRP-conjugated anti-rabbit and anti-mouse secondary antibodies were from Cell Signaling Technology.
IMR-32 cell culture. Human neuroblastoma IMR-32 cells (cell no. TKG0207) were obtained from Riken Bioresource Center Cell Bank. IMR-32 cells were grown in Eagle’s Minimum Essential Medium (Nissui Seiyaku) supplemented with 2 mM l-glutamine (Nacalai Tesque), 1% v/v MEM nonessential amino acid solution (Nacalai Tesque), and 10% v/v fetal bovine serum (Equitech-Bio) under a humidified 5% CO2 atmosphere at 37 °C.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Toxicity of ITCs on IMR-32 cells was checked by incubating the cells with 0–20 μM concentrations of ITCs and then assessed the viability using MTT assay. In brief, IMR-32 cells were seeded onto the 96-well plate (1 ×104 cells/well). After 24-hour preculture, the cells were treated with 0–20 μM concentration of ITCs for 12 hours. Then, 5 mg/mL of MTT was added to each well and incubated for another 4 hours. After incubation, 100 μL of stop solution was then added to each well and the absorbance at 595 nm was then measured after thorough pipetting to disperse the generated blue formazan.
Total RNA extraction
IMR-32 cells were precultured in 10 cm dishes for 24 hours and then treated by 10 μM of ITCs (SFN, 6-MSITC, and 6-MTITC) in 0.2% dimethyl sulfoxide for another 9 hours. Total RNA was extracted using RNeasy Mini Kit (Qiagen™), following the manufacturer’s instructions. RNA integrity was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies).
Microarray hybridization and transcript analyses
Four hundred nanograms of total RNA were used to generate cDNA with Eukaryotic Poly-A RNA Control Kit (Affymetrix) and GeneChip® One-cycle cDNA Synthesis Kit (Affymetrix), following the manufacturer’s protocol. After cleanup of double-stranded cDNA by Sample Cleanup Module Kit (Affymetrix), biotin-labeled cRNAs were synthesized at 37 °C for 16 hours by GeneChip IVT Labeling Kit (Affymetrix). Following the cleanup and quantification, the fragmented and biotin-labeled cRNAs were hybridized at 45 °C for 16 hours to the Affymetrix GeneChip (Human Genome U133 Plus 2.0 oligonucleotide arrays) containing approximately more than 54,000 probe sets. The GeneChip was washed and stained by GeneChip Hybridization, Wash, and Stain Kit in Fluidics Station (Affymetrix). The hybridized fluorescence was scanned using Affymetrix Launcher. The images were processed for visualization and normalization of each probe set to a common baseline using GeneSpring GX 10.1 (Agilent Technologies). Gene products of fold change greater than 2 were further analyzed using Gene Ontology (GO) software (http://www.geneontology.org) for biological processes, molecular functions, and signaling pathways.
Pathway analyses
Pathway and global functional analysis were performed using IPA (Ingenuity® Systems; www.ingenuity.com). A data set containing gene accession numbers and the corresponding fold change of ITC-treated cells versus control was uploaded into the software and were mapped out using GO.
GO analysis generated the biological functions as well as pathways from the IPA library that is significant to the data set. Genes from the data sets associated with biological functions or canonical pathway with level of significance less than 0.05 were used to map out molecular networks. Resulting networks were ranked based on the scores generated from Fisher’s exact test to indicate the probability of each biological function and/or canonical pathway was not due to chance alone.
Real-time PCR
The primers (Table 1) used for real-time PCR in the present study, including AKR1C1, AKR1C3, NQO1, GSR, TXNRD1, and GCLM, were designed according to the NCBI sequence database using the software Primer3. Reverse transcription and real-time PCR were performed with DyNAmo™ SYBR® Green 2-Step qRT-PCR Kit (Finnzymes Oy) according to manufacturer’s manual. Briefly, RNA (200 ng) was reversed to cDNA using Oligo dT and M-MuLV RNase at 37 °C for 30 minutes, and the reaction was then terminated at 85 °C for 5 minutes. Real-time PCR was performed with the Roter-Gene-3000 AKAA (Corbett Research) in triplicates using the standard curve. The Tm of PCR was determined according to each primer sequence (https://www.finnzymes.fi/tm.determination.html). Each PCR contained 250 ng of reversed transcripts, 75 ng of each primer, and 10 μL of Master Mix (Finnzymes Oy). The thermal cycling condition was held at 95 °C for 15 minutes followed by 55 cycles of 30 seconds at 94 °C, 30 seconds at corresponding Tm (Table 1), and 30 seconds at 72 °C. The result was represented by relative expression level normalized with control cells.
Table 1.
Primer sequences used for real-time PCR.
| GENE SYMBOL | DIRECTION | PRIMER SEQUENCES | TM (°C) |
|---|---|---|---|
| AKR1C1 | Fw | 5′-ATC CCT CCG AGA AGA ACC AT-3′ | 59 |
| Re | 5′-ACA CCT GCA CGT TCT GTC TG-3′ | ||
| AKC1C3 | Fw | 5′-AAG TAA AGC TTT GGA GGT CAC A-3′ | 59 |
| Re | 5′-GGA CCA ACT CTC GTC GAT GAA-3′ | ||
| NQO1 | Fw | 5′-AGT GCA GTG GTG TGA TCT CG-3′ | 59 |
| Re | 5′-GGT GGA GTC ACG CCT GTA AT-3′ | ||
| GSR | Fw | 5′-GAT CCC AAG CCC ACA ATA GA-3′ | 59 |
| Re | 5′-CTT AGA ACC CAG GGC TGA CA-3′ | ||
| TXNRD1 | Fw | 5′-ATC AGG AGG GCA GAC TTC AA-3′ | 61 |
| Re | 5′-CCC ACA TTC ACA CAT GTT CC-3′ | ||
| GCLM | Fw | 5′-GGG AAC CTG CTG AAC TGG-3′ | 61 |
| Re | 5′-GCA TGA GAT ACA GTG CAT TCC-3′ |
Western blot analyses
IMR-32 cells were seeded into a 10-cm dish and precultured for 24 hours. Ten micromolars of ITC (SFN, 6-MSITC, or 6-MTITC) were added and cocultured for another 12 hours. Cells were harvested by lysis buffer, and then homogenized in an ultrasonicator for 10 seconds twice and incubated on ice for 30 minutes.26 After the homogenates were centrifuged at 14,000 × g for 15 minutes at 4 °C, the protein concentration was determined by protein assay kit (Bio-Rad Laboratories). Forty micrograms of protein lysates were run on 10%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to polyvinylidene diflouride (PVDF) membrane (Amersham Pharmacia Biotech). The membrane was incubated with specific antibody overnight at 4 °C and further incubated for 1 hour with HRP-conjugated secondary antibody. Bound antibodies were detected using the ECL system and the relative amounts of proteins associated with specific antibody were quantified using Lumi Vision Imager software (TAITEC Co.).
Statistical analyses
All experiments were done for at least three trials. The differences between the sample treatment and the control were statistically analyzed using Student’s t-test. A statistical probability of P < 0.05 was considered significant.
Results
Gene profile analysis in IMR-32 cells treated by Wasabi-derived ITCs
IMR-32 cells were treated with 0–20 μM of SFN, 6-MSITC, or 6-MTITC for 12 hours.25,27 The cyto-toxicity assay results showed that there was no significant decrease in the number of viable cells in such treatment (Supplementary Fig. 1), indicating that less than 20 μM of these ITCs is the safe concentration for treating IMR-32 cells. Thus, we performed gene profiling from the cells treated by 10 μM of SFN, 6-MSITC, or 6-MTITC for 9 hours. Cell mRNA was prepared, and mRNA profiling was carried out using Affymetrix HG UG133 plus 2.0 oligonucleotide arrays containing approximately over 54,000 probe sets as elaborated in the “Materials and Methods” section. As summarized in Table 2, the total number of gene expression mediated by 6-MSITC (263 genes) and 6-MTITC (233 genes) were more than twice higher than SFN (108 genes). Detailed evaluation manifested that 6-MSITC had the strongest regulation on gene expression (100 upregulated and 163 downregulated genes), followed by 6-MTITC (98 upregulated and 135 downregulated genes) and SFN (67 upregulated and 41 downregulated genes). Moreover, total number of the genes downregulated by 6-MSITC and 6-MTITC were more than that of the genes upregulated while SFN acted by contrast.
Table 2.
Number of genes regulated by Wasabi-derived ITCs.
| FOLD CHANGE | SFN | 6-MSITC | 6-MTITC |
|---|---|---|---|
| ≥4 | 16 | 21 | 22 |
| ≥3 to <4 | 7 | 12 | 12 |
| ≥2 to <3 | 44 | 67 | 64 |
| Subtotal | 67 | 100 | 98 |
| ≥−4 | 40 | 3 | 2 |
| ≥−3 to <−4 | 1 | 13 | 10 |
| ≥−2 to <−3 | 0 | 147 | 123 |
| Subtotal | 41 | 163 | 135 |
| Total | 108 | 263 | 233 |
Grouping of genes targeted by Wasabi-derived ITCs
To understand the function of the genes targeted by SFN, 6-MSITC, and 6-MTITC, all genes that were differentially expressed (at least 2 folds) were subjected to GO analyses for classification based on biological processes, molecular functions, or cellular component (data not shown). Majority of the upregulated genes with known annotation information was associated with various categories such as metabolic processes, transcription, transport, oxidoreductase, signal transduction, and stress response. For downregulated genes, most of the subsets (≥ 2-fold change) were related to binding, CNS-specific function, signal transduction, transcription, transferase activity, and transport. We identified that 14, 17, and 18 genes of the total number of differentially expressed genes from SFN, 6-MSITC, and 6-MTITC treatments, respectively, were associated with oxidative stress response. Consequently, gene profile analyses revealed that most of the differentially expressed genes induced by SFN, 6-MSITC, and 6-MTITC are linked to the oxidative stress response.
Identification of the biological pathways by IPA
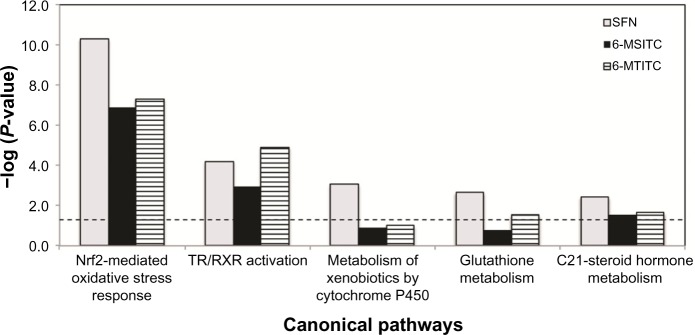
To identify the biologically relevant networks and pathways within the differentially expressed genes of IMR-32 cells, pathway analyses were done using Ingenuity Pathways Knowledge Base on the datasets. Numerous pathways with significant threshold (P < 0.05) were obtained from these analyses. The first five most statistically significant canonical pathways with respect to these ITCs are illustrated in Figure 2. Interestingly, gene profiles of SFN, 6-MSITC, and 6-MTITC treatment shared identical top two canonical pathways including Nrf2-mediated oxidative stress response and TR/RXR activation (Fig. 2). Furthermore, we summarized the genes associated with each pathway that are modulated by ITCs in Table 3. We found that 6-MSITC and 6-MTITC caused greater number of differentially expressed genes (≥2-fold change) than SFN. These results imply that 6-MSITC and 6-MTITC have greater influence in the gene expression regulation of IMR-32 cells than SFN.
Figure 2.
Comparative canonical pathway analyses of differentially expressed genes in IMR-32 neuron cells stimulated with Wasabi ITCs. Differentially upregulated and downregulated genes were evaluated for canonical pathway analyses using IPA software as elaborated in the “Materials and Methods” section. Only five of the top most significant pathways with respect to Wasabi ITCs are shown here.
Note: List of corresponding significant pathways is indicated below and their respective level of significance (P < 0.05) denoted by the length of the bars.
Table 3.
List of genes involved in canonical pathways modulated significantly by SFN, 6-MSITC, and 6-MTITC in IMR-32 cells.
| ITCS | CANONICAL PATHWAYS | P-VALUE | REGULATION | NO. OF GENES | REGULATED GENES |
|---|---|---|---|---|---|
| SFN | Nrf2-mediated oxidative stress response | 5.01E-11 | ↑ | 12 | BACH1, DNAJB4, FTH1, FTL, GCLM, GSR, GSTM3, HMOX1, MAFF, NQO1, SQSTM1, TXNRD1 |
| TR/RXR activation | 6.03E-05 | ↑ | 5 | AKR1C1, AKR1C2, AKR1C3, ME1, SYT2 | |
| Metabolism of xenobiotics by cytochrome P450 | 8.71E-04 | ↑ | 4 | AKR1C1, AKR1C2, AKR1C3, GSTM3 | |
| Glutathione metabolism | 2.19E-03 | ↑ | 3 | GCLM, GSR, GSTM3 | |
| C21-steroid hormone metabolism | 3.72E-03 | – | 2 | AKR1C1, AKR1C3 | |
| 6-MSITC | Nrf2-mediated oxidative stress response | 1.23E-07 | ↑ | 14 | BACH1, DNAJB1, DNAJB4, FTH1, FTL, GCLM, GSR, HMOX1, KEAP1, MAFF, MAFG, NQO1, SQSTM1, TXNRD1 |
| TR/RXR activation | 1.12E-03 | ↑ | 6 | ACACA, AKR1C1, AKR1C2, AKR1C3, ME1, SYT2 | |
| Wnt/β-catenin signaling | 6.61E-03 | ↑ | 7 | GJA1, KREMEN1, PPP2R2B, PPP2R2C, SRC, WNT5A, WNT5B | |
| PTEN signaling | 8.91E-03 | ↑ | 5 | FOXO6, IKBKB, ITGA4, MAGI1, PDGFRA | |
| Ephrin receptor signaling | 1.05E-02 | ↑ | 7 | ANGPT1, CXCR4, FIGF, GNG12, ITGA4, NGEF, SRC | |
| 6-MTITC | Nrf2-mediated oxidative stress response | 4.68E-08 | ↑ | 14 | BACH1, DNAJB1, DNAJB4, FTH1, FTL, GCLM, GSR, HMOX1, KEAP1, MAFF, MAFG, NQO1, SQSTM1, TXNRD1 |
| TR/RXR activation | 1.23E-05 | ↑ | 8 | ACACA, AKR1C1, AKR1C2, AKR1C3, ME1, PPARGC1A, RXRA, SYT2 | |
| Hepatic Fibrosis/Hepatic stellate cell activation | 2.51E-04 | – | 8 | COL1A1, FGFR2, FIGF, FLT4, IGFBP5, PDGFRA, TNFRSF1A, VEGFA | |
| Axonal guidance signaling | 1.00E-03 | ↑ | 13 | CXCR4, FIGF, ITGA4, PLXNA2, PRKACB, SEMA6B, SHANK2, SHC1, SLIT2, SLIT3, VEGFA, WNT5A, WNT5B | |
| Estrogen receptor signaling | 3.24E-03 | ↑ | 6 | HNRNPD, PCK2, PPARGC1A, SHC1, SRC, TAF4B |
Expression profiling of Nrf2-mediated genes by Wasabi-derived ITCs
To investigate the neuroprotective effects of SFN, 6-MSITC, and 6-MTITC, we investigated the genes coding for proteins involved in apoptosis regulations. Microarray data revealed that the expressions of proapoptosis genes were unaltered by ITC treatment (data not shown). Pre-experiment data also showed that ITCs could protect IMR-32 cells from oxidative stress induced by H2O2 (data not shown). This implies that ITCs do not induce neuronal cell death. Thus, we profiled the effects of SFN, 6-MSITC, and 6-MTITC on the expression of genes mediated by Nrf2 pathway (Table 4). They were classified into five categories including (a) the genes coding upstream regulators, (b) the genes coding antioxidant proteins, (c) the genes coding metabolizing enzymes and detoxifying protein genes, (d) the genes coding chaperone and stress response protein genes, and (e) the genes coding ubiquitination and proteosomal degradation protein. We found that the gene expression of most of the anti oxidant proteins and metabolizing enzymes from 6-MSITC and 6-MTITC treatment was higher than SFN. These are ferritin heavy polypeptide 1 (FTH1), ferritin light polypeptide (FTL), glutathione reductase (GSR), HO-1, NQO1, sequestosome 1 (SQSTM1), and thioredoxin reductase 1 (TXNRD1) for the antioxidant proteins and enzymes. We identified aldo–keto reductase family 1 member C1, C2, C3 (AKR1C1, AKR1C2, AKR1C3), and glutamate-cysteine ligase modifier subunit (GCLM) for metabolizing enzymes. We found that 6-MSITC and 6-MTITC have greater capacity to induce the expressions of genes associated with the Nrf2 pathway in neuron cells.
Table 4.
List of genes involved in the Nrf2-mediated oxidative stress pathway by SFN, 6-MSITC, and 6-MTITC in IMR-32 cells.
| GENE INVOLVED | REPRESENTATIVE PUBLIC ID | SFN | 6-MSITC | 6-MTITC |
|---|---|---|---|---|
| (A) Upstream regulators | ||||
| ACTIN (ACTA1) | NM_001100 | 1.27 | 1.05 | 1.09 |
| ATF4 | NM_001675 | 1.32 | 1.63 | 1.57 |
| BACH | NM_001186 | 1.94 | 2.08 | 2.24 |
| c-FOS (FIGF) | NM_004469 | −1.66 | −2.36 | −2.64 |
| c-MAF (MAF) | NM_005360 | −1.07 | −1.05 | 1.07 |
| CBP (CREBBP) | NM_004380 | 1.05 | 1.21 | 1.24 |
| ERK1/2 (MAP2K1) | AI571419 | 1.07 | 1.18 | 1.26 |
| FRA1 (FOSL1) | BG251266 | 1.20 | 1.39 | 1.09 |
| JUN | NM_002228 | −1.13 | −1.11 | 1.08 |
| MAFF | AL021977 | 2.21 | 2.90 | 2.78 |
| P300 (EP300) | AI459462 | −1.20 | −1.34 | −1.61 |
| (B) Genes coding antioxidant protein | ||||
| CAT | NM_001752 | 1.04 | 1.05 | 1.01 |
| FTH1 | AA083483 | 2.74 | 3.41 | 3.48 |
| FTL | BG538564 | 2.59 | 2.88 | 2.86 |
| GPX2 | NM_002083 | 1.08 | 1.07 | 1.15 |
| GSR | AI888037 | 3.63 | 3.50 | 3.45 |
| HO1 (HMOX1) | NM_002133 | 11.50 | 59.14 | 54.63 |
| PRDX1 | L19184 | 1.58 | 1.82 | 1.74 |
| SOD | NM_000454 | 1.23 | 1.43 | 1.38 |
| SQSTM1 | AW293441 | 3.36 | 4.66 | 5.00 |
| TRXD1 (TXNRD1) | NM_003330 | 4.82 | 4.82 | 4.77 |
| TXN | AF065241 | 1.35 | 1.32 | 1.16 |
| (C) Genes coding metabolizing enzymes | ||||
| AFAR (AKR7A2) | NM_003689 | −1.18 | −1.26 | −1.24 |
| AKR1C1 | S68290 | 10.60 | 11.02 | 12.02 |
| AKR1C2 | M33376 | 11.00 | 11.96 | 12.23 |
| AKR1C3 | AB018580 | 103.64 | 103.85 | 109.70 |
| AOX1 | NM_001159 | 1.09 | 1.01 | 1.03 |
| CBR1 | BC002511 | −1.09 | −1.04 | −1.12 |
| CYP1A | NM_000499 | −1.24 | −1.22 | −1.07 |
| CYP2A (CYP2A13) | NM_000766 | −1.16 | −1.03 | −1.24 |
| CYP2C (CYP2C18) | NM_000772 | −1.06 | 1.05 | 1.07 |
| CYP3A (CYP3A4) | NM_017460 | 1.10 | 1.10 | 1.20 |
| CYP4A (CYP4A11) | NM_000778 | −1.26 | −1.39 | −1.10 |
| EPHX1 | NM_000120 | 1.51 | 1.89 | 1.67 |
| FMO1 | NM_002021 | −1.16 | 1.07 | −1.00 |
| GCLC | NM_001498 | 1.62 | 1.48 | 1.54 |
| GCLM | NM_002061 | 8.44 | 11.88 | 5.39 |
| GST (GSTA1) | NM_000846 | −1.11 | −1.11 | −1.05 |
| NQO1 | NM_000903 | 27.37 | 27.67 | 28.41 |
| UGT | NM_001072 | 1.30 | 1.00 | 1.11 |
| MRP1 (ABCC1) | NM_004996 | −1.01 | 1.04 | 1.03 |
| SR-BI (SCARB1) | NM_005505 | −1.06 | −1.03 | −1.02 |
| (D) Genes coding chaperones and stress response proteins | ||||
| HSP22 (HSPB8) | AF133207 | −1.39 | −1.07 | −1.09 |
| CCT7 | NM_006429 | 1.05 | 1.09 | 1.11 |
| CLPP | NM_006012 | −1.04 | 1.00 | −1.03 |
| ERP29 | NM_006817 | −1.01 | 1.10 | 1.09 |
| FKBP5 | NM_004117 | 1.10 | −1.32 | −1.16 |
| HERPUD1 | AF217990 | 1.21 | 1.28 | 1.31 |
| HSP40 (DNAJB4) | NM_007034 | 4.30 | 5.34 | 5.39 |
| HSP90 (HSP90B1) | AK025862 | 1.59 | 1.48 | 1.59 |
| PPIB | NM_000942 | 1.20 | 1.26 | 1.33 |
| PTPLAD1 | NM_016395 | −1.01 | 1.07 | 1.05 |
| (E) Genes coding ubiquitination-related factors | ||||
| HIP2 (UBE2K) | NM_005339 | 1.17 | 1.23 | 1.33 |
| PSM (PSMA1) | NM_002786 | 1.04 | 1.11 | 1.07 |
| UB2R1 (UBE2R2) | BE221883 | 1.05 | 1.11 | 1.06 |
| UBB (UBA52) | AF348700 | −1.09 | −1.06 | −1.06 |
| USP14 | NM_005151 | 1.12 | 1.18 | 1.14 |
| VCP | AF100752 | 1.29 | 1.51 | 1.41 |
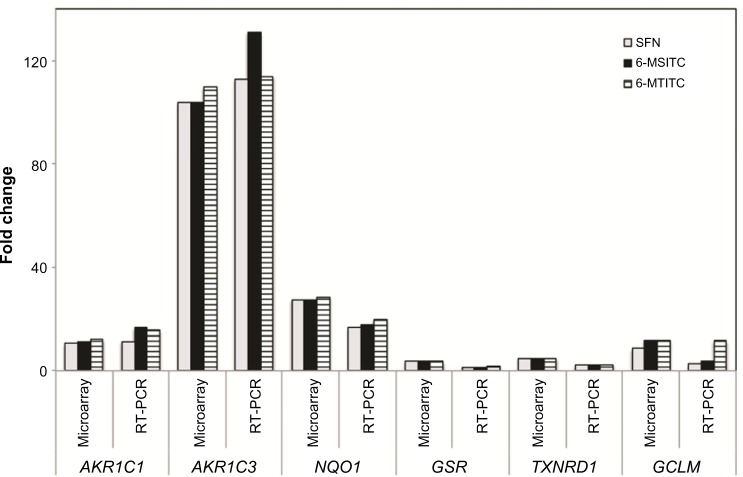
To confirm the results of microarray analyses, the expression levels of six selected genes were further detected by real-time PCR (Fig. 3). Most of these genes exhibited a similar expression pattern between the microarray and real-time PCR data. For instance, 6-MSITC induced gene expression of AKR1C1 by 16 folds in the real-time PCR experiment, whereas 11 folds in microarray analysis. The 6-MSITC-induced AKR1C3 gene expression was 131 folds in real-time PCR while 104 folds in microarray analysis. The effect of 6-MSITC on NQO1 induction level was found higher in microarray analysis (27 folds) than in real-time PCR (18 folds).
Figure 3.
Validation of differentially expressed genes in Wasabi ITC-treated IMR-32 cells from DNA microarray analysis by real-time PCR. DNA microarray results were compared to real-time PCR results for selected genes. Real-time PCR was performed using DyNAmo™ SYBR® Green 2-Step qRT-PCR Kit as described in the “Materials and Methods” section. Fold changes represented the ratio between the treated samples values to that of the untreated samples. Expression changes are depicted as fold change (y-axis). Gene symbols are shown below.
Influence of 6-MSITC on Nrf2-mediated protein levels
To verify the involvement of ITCs in Nrf2-mediated oxida-tive stress response pathway, we chose 6-MSITC, the highest bioactive ITC among the three, to investigate the levels of Nrf2-mediated proteins by Western blotting. Dose (Fig. 4A) and time (Fig. 4B) experiments showed that treatment with 10 μM of 6-MSITC for 12 hours could effectively induce the production of Nrf2 and Nrf2-mediated proteins including NQO1, TXNRD1, AKR1C1, and AKR1C3, but no signifi-cant effect on the expression of Keap1. The products of housekeeping gene, GAPDH, was unaltered during such treatment. Moreover, we found that the present results are in agreement with our previous study on 6-MSITC using human hepatoblastoma HepG2 cells.29 These data suggest that 6-MSITC induced the expressions of antioxidant enzymes through the activation of Nrf2/Keap1 system.
Figure 4.
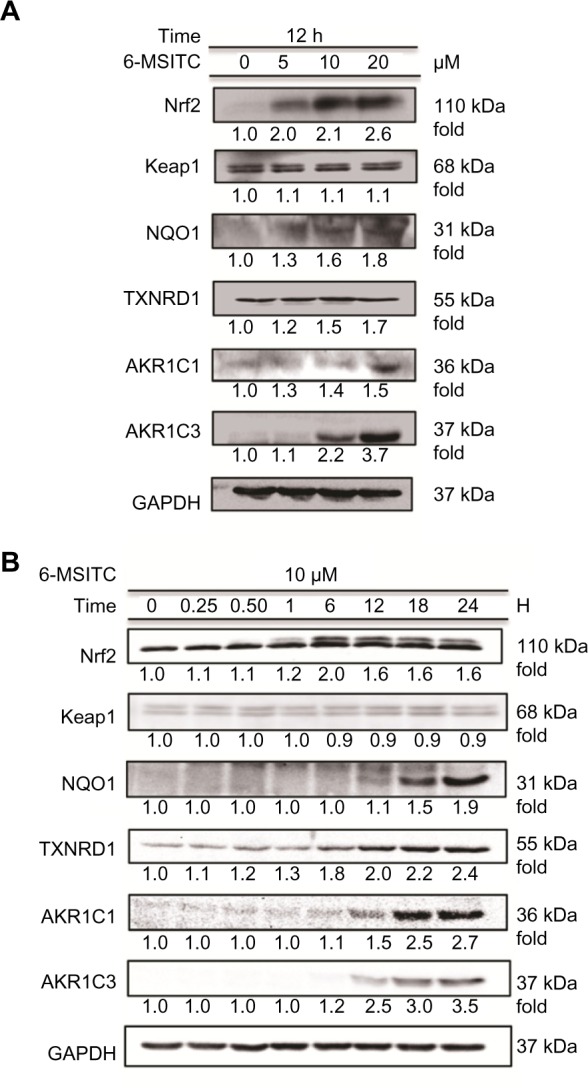
Effect of 6-MSITC on Nrf2 level and Nrf2-mediated induction of typical proteins. (A) IMR-32 cells were treated with 0−20 μM of 6-MSITC for 12 hours. (B) IMR-32 cells were treated with 10 μM of 6-MSITC for 0−24 hours. Nrf2, Keap1, NQO1, TXNRD1, AKR1C1, AKR1C3, and GAPDH were detected using Western blot analysis with their respective antibodies. The induction fold of the protein was calculated as the intensity of the treatment relative to that of control normalized to GAPDH by densitometry. The blots shown are the examples of three separate experiments.
Influence of 6-MSITC on Nrf2 protein level at transcription and posttranscription
The increase in the level of Nrf2 protein by 6-MSITC is possible due to transcriptional and posttranscriptional regulation. Thus, we first examined the microarray data. Cells treated with 10 μM of SFN, 6-MSITC, or 6-MTITC for 9 hours upregulated only 1.06-, 1.03-, and 1.07-fold of Nrf2, respectively, compared to that without treatment, indicating that ITCs did not regulate the expression Nrf2 gene in such treatment. Next, we investigated the influence of these ITCs on the stability of Nrf2 protein by adding cycloheximide (CHX), a protein synthesis inhibitor. IMR-32 cells were pretreated with 6-MSITC (10 μM) for 3 hours and then treated with CHX (5 μg/mL) for 15–60 minutes. As shown in Figure 5, the level of Nrf2 protein was decreased to 58% after 60 minutes after stopping protein synthesis with CHX. On the other hand, pretreatment with 6-MSITC extended Nrf2 stability, showing no degradation even after 60 minutes. The results indicated that 6-MSITC might increase Nrf2 protein level by inhibiting the turnover of Nrf2 protein, rather than by stimulating Nrf2 gene expression at transcriptional level.
Figure 5.
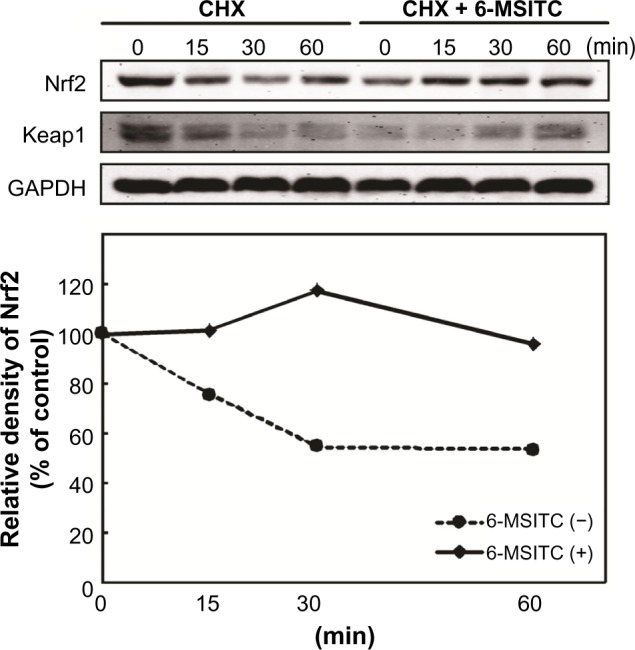
Effect of 6-MSITC on the stability of Nrf2. IMR-32 cells were pretreated with or without 10 μM of 6-MSITC for 3 hours, followed by exposure with 5 mg/mL of CHX for 0–1 hours. Nrf2, Keap1 and GAPDH were detected by Western blot analysis with their respective antibodies. Histograms show the densitometric analysis of Nrf2 compared with the control.
Discussion
ITCs have been reported to exhibit protective effects against oxidative stress in astrocytes, dopaminergic cell death, and traumatic brain injury by inducing the transcriptional factor Nrf2 that activates endogenous defenses of the cell via a battery of cytoprotective genes.10,12,30 Thus, this implies that the ITCs may possess neuroprotection, although the exact molecular mechanisms are not fully clarified. In the present study, we investigated, for the first time, the gene expression profiles of IMR-32 neuron cells treated by three ITCs (SFN, 6-MSITC, and 6-MTITC) to study the neuroprotective mechanisms on a genome-wide level using microarray technology. IMR-32 cell line is an ideal cell model for molecular study of distinct patterns of antioxidant-related pathway because it contains functional ARE capable of inducing endogenous cytoprotective genes.31 SFN is a major component of broccoli.32 6-MSITC and 6-MTITC are SFN analogs found to be the major bioac-tive compounds of Japanese Wasabi.2 DNA microarray analysis data revealed that SFN, 6-MSITC, and 6-MTITC could significantly affect the gene expressions of IMR-32 neuron cells. With over 54,000 gene probes on the array, 6-MSITC treatment at 10 μM for 9 hours regulated the expression of a total of 263 genes by greater than or equal to 2 folds (Fig. 1). Of the total number of genes regulated by 6-MSITC, 100 are upregulated and 163 are downregulated. The number of genes regulated by 6-MSITC is twice higher than that regulated by the treatment with SFN, suggesting that 6-MSITC elicited a stronger stimulation on gene expression than SFN in the IMR-32 cells. On the other hand, the number of genes regulated by 6-MTITC is close to the number of genes regulated by 6-MSITC, indicating that removal of oxygen atom on the sulfinyl sulfur of the methyl group has no significant influence on gene regulation. Both 6-MSITC and 6-MTITC have the same number of carbon atoms, but differ on the sulfur substituent attached to the methyl group. 6-MSITC is a methylthioalkyl ITC containing S = O substituent in the methyl group, whereas, 6-MTITC is a methylsulfinylalkyl ITC without oxygen atom attached to the sulfur atom of the methyl group.33,34 On the other hand, SFN is two-carbon atom less than 6-MSITC and 6-MTITC. It looks that the capacity of Wasabi ITCs to regulate gene expression depends on the carbon chain length between the ITC group and the sulfinyl sulfur. However, it will be interesting to investigate the effect of long carbon chain or aromatic ITCs on the gene expression profile of IMR-32 cells. Furthermore, it will also be valuable to explore the influence of Wasabi-derived ITCs on gene regulation of other types of brain cells by DNA microarray.
It is noticed that most of the genes targeted by SFN, 6-MSITC, and 6-MTITC belonged to oxidative stress response cluster. We found that 14, 17, and 18 genes of the total number of differentially expressed genes by SFN, 6-MSITC, and 6-MTITC, respectively, were associated with oxidative stress response. The upregulation of phase 2 metabolizing enzymes such as AKR1C3 and GCLM and antioxidant proteins such as GSR, HO-1, and TXNRD1 by SFN has been reported to protect the cells against oxidative stress,13 and Wasabi-derived ITCs were also found to upregulate their expressions. In addition, 6-MSITC and 6-MTITC also upregulated the expressions of other oxidative stress-related genes, such as AKR1C1, AKR1C2, NQO1, FTH1, FTL, and SQSTM1. These genes are well documented to be involved in detoxification and anti oxidant defense, neuronal proliferation and differentiation, and signal transduction.35 Thus, this suggests that SFN, 6-MSITC, and 6-MTITC might also exert neuroprotective activity via pathways governing the regulation of antioxidant defense genes. In order to confirm the regulation of these genes in signal network level, we performed signal pathway analyses by IPA software. Nrf2-mediated oxidative stress response pathway came up to be the most significant pathway modulated by SFN, 6-MSITC, and 6-MTITC. Moreover, detailed evaluation showed that 6-MSITC enhanced higher number of genes associated with Nrf2-mediated stress response pathway than SFN and 6-MTITC, suggesting that 6-MSITC is a stronger inducer of Nrf2-mediated oxidative stress response pathway than SFN and 6-MTITC (Fig. 2, Table 3). 6-MSITC stimulation in animal study exhibited a stronger HO-1 protein expression than the SFN treatment.11 Recent cancer cell model study demonstrated that lengthening the carbon chain between the sulfinyl sulfur and the ITC group from 4 to 6 carbon atoms has a beneficial effect on Nrf2 activation, whereas the increasing stearic size of the substituent on the sulfur atom contributes a negative effect on the biological activity.36 On the other hand, we observed that SFN did not affect the gene regulation of other upstream transcription factors similar to what 6-MSITC and 6-MTITC did. We also noted that the TR/RXR activation pathway ranked the second significantly regulated pathway for ITC-induced gene expression, and the expressions of AKR1C1, AKR1C2, and AKR1C3 linked to this pathway were highly upregulated by the SFN, 6-MSITC, and 6-MTITC. However, an understanding of how these ITC interactions with TR/RXR activation is linked to neuroprotection will require further study. Other significantly regulated pathways include metabolism of xenobiotics by cytochrome P450, GSH metabolism, and C21-steroid hormone metabolism. These denote that Wasabi ITCs also act as xenobiotics, causing induction of metabolizing enzymes and intracellular GSH.
Finally, we confirmed the products of these genes at the protein level using 6-MSITC, a representative of these Wasabi-derived ITCs. 6-MSITC treatment induced a higher level of Nrf2 protein, but no significant effect on Keap1 protein (Fig. 4). These data are in agreement with the results in primary cortical neurons, in which SFN enhanced Nrf2 protein level in a time-dependent manner, but no effect on Keap1 protein.37 On the other hand, Wasabi ITCs could covalently modify Keap1, preventing Nrf2 ubiquitination and promoting Nrf2 stability to mediate the ARE-driven activation in human hepatoblastoma cells, HepG2.27,29 These data suggest that Wasabi ITCs may have different actions on Keap1 protein in different cell types. As indicated in Figure 4, the activation of Nrf2 followed increase in the levels of NQO1, TXNRD1, AKR1C1, and AKR1C3 protein expressions. These data further demonstrated that Wasabi ITCs exert the neuroprotective effects in IMR-32 cells via activating Nrf2-mediated oxidative stress response pathway until the protein level. Furthermore, in vivo experiment showed that ITC could penetrate the blood brain barrier and deliver its neuroprotective function in the central nervous system.8 In addition, ITCs such as 6-phenethyl isothiocyanate and SFN are also rapidly accumulated in various types of cell lines with intracellular concentration within millimolar level.38 In rats and humans, pharmacoki-netic data revealed that SFN can be absorbed in the body and reach micromolar concentration in the blood. Specifically in rats, detectable amount of SFN was evident after an hour and peaked at ∼20 μM after four hours, following 50 μM gavages of SFN.39 On the other hand, single doses of 200 μM broccoli sprouts ITC preparation given to human subjects showed that ITC plasma concentrations peaked between 0.943 and 2.27 μM/L one hour after intake.40 Thus, the dose of 10 μM ITCs used in this study maybe achievable in vivo.
In summary, our DNA microarray data revealed for the first time the gene expression profiles of Wasabi-derived ITCs in a neuronal cell model, IMR−32. 6-MSITC had the strongest regulation on gene expression among the three ITCs, showing a positive relationship in a carbon chain length of ITCs. Specifically, 6-MSITC could stimulate Nrf2-mediated gene expressions through the stabilization of Nrf2 protein at post-transcription. Taken our data together with previous findings, 6-MSITC can exert the neuroprotective effect by activating the Nrf2-mediated oxidative stress response pathway (Fig. 6).
Figure 6.
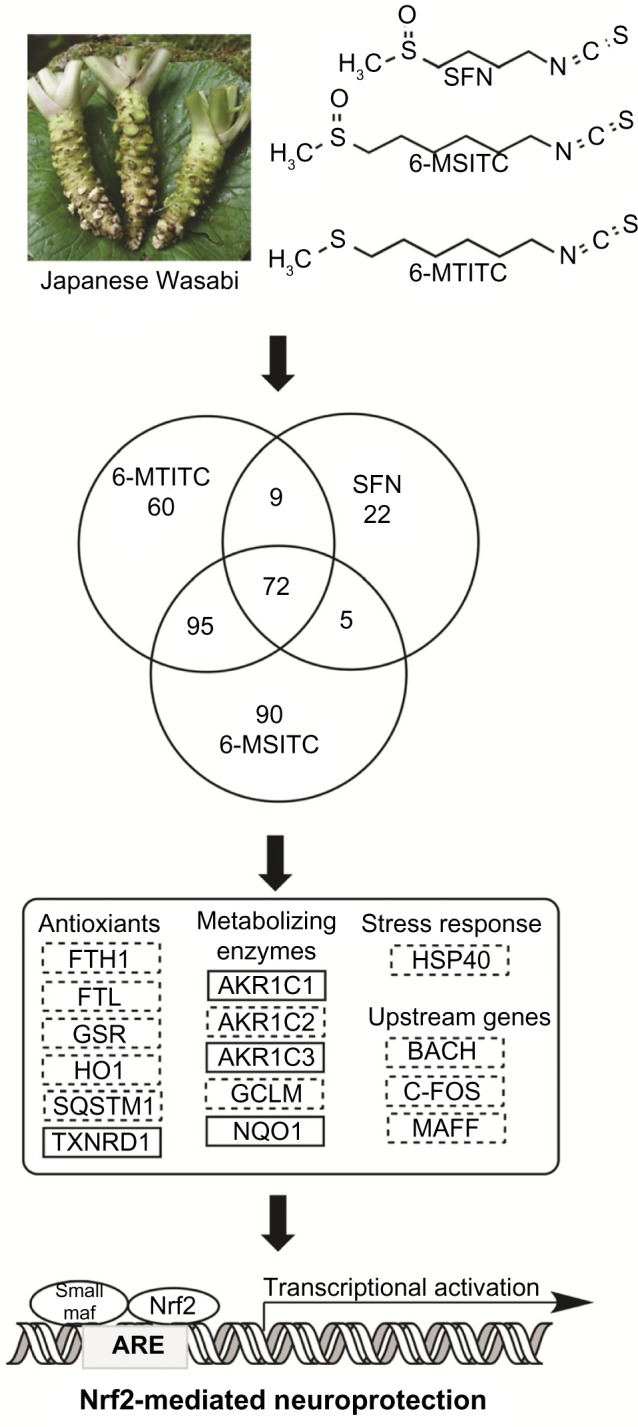
Proposed mechanisms for the neuroprotective effects by Wasabi-derived 6-MSITC in IMR-32 cells. Wasabi-derived ITCs activate Nrf2-mediated oxidative stress pathway and subsequently induce the expression of antioxidant proteins/enzymes to exert the neuroprotective effects. The mRNA and protein of marker in solid box were confirmed by RT-PCR and Western blotting.
Supplementary Material
Supplementary Figure 1. Cytotoxicity assay results showing no significant decrease in the number of viable cells after treatment with 0–20 μM of SFN, 6-MSITC, or 6-MTITC for 12 hours.
Footnotes
ACADEMIC EDITOR: James Willey, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,141 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by the Scholar Research of Kagoshima University (DXH). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the experiments and wrote the manuscript as corresponding author: DXH. Primary investigator including DNA array analysis, Western blotting and wrote the first draft of the manuscript: PZT. Contributed to partial cell culture and DNA array operation: ST. Contributed to partial cell culture and Western blotting: SF, AH, KS. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Isshiki K, Tokuoka K. Allyl isothiocyanate and wholesomeness of food. Jpn J Food Microbiol. 1993;12:1–6. [Google Scholar]
- 2.Kumagai H, Kashima N, Seki T, Sakurai H, Ishii K, Ariga T. Analysis of components in essential oil of upland Wasabi and their inhibitory effects on platelet aggregation. Biosci Biotech Biochem. 1994;58:2131–5. [Google Scholar]
- 3.Nagai M, Okunishi I. The effect of Wasabi rhizome extract on atopic dermatitis-like symptoms in HR-1 hairless mice. J Nutr Sci Vitaminol. 2009;55:195–200. doi: 10.3177/jnsv.55.195. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki M, Ogawa T, Wang L, et al. Anti-obesity effects of hot water extract from Wasabi (Wasabi japonica Matsum.) leaves in mice fed high-fat diets. Nutr Res Pract. 2013;7:267–72. doi: 10.4162/nrp.2013.7.4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsuan SW, Chyau CC, Hung HY, Chen JH, Chou FP. The induction of apop-tosis and autophagy by Wasabi japonica extract in colon cancer. Eur J Nutr. 2016;55(2):491–503. doi: 10.1007/s00394-015-0866-5. [DOI] [PubMed] [Google Scholar]
- 6.Ono H, Adach K, Fuke Y, Shinohara K. Purification and structural analysis of substances in Wasabi (Eutrema Wasabi Maxim.) that suppress the growth of MKN-28 human stomach cancer cells. J Jap Food Sci Technol. 1996;43:1092–7. [Google Scholar]
- 7.Uto T, Fuji M, Hou DX. Inhibition of lipopolysaccharide-induced cyclooxy-genase-2 transcription by 6-(methylsulfinyl) hexyl isothiocyanate, a chemo-preventive compound from Wasabi japonca (Miq.) Matsumura, in mouse macrophages. Biochem Pharmacol. 2005;70:1772–84. doi: 10.1016/j.bcp.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Tarozzi A, Angeloni C, Malaguti M, Morroni F, Hrelia S, Hrelia P. Sul-foraphane as a potential protective phytochemical against neurodegenerative diseases. Oxid Med Cell Longev. 2013;2013:415078. doi: 10.1155/2013/415078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang J, Alesi GN, Zhou N, Keep RF. Protective effects of isothiocyanates on blood-CSF barrier disruption induced by oxidative stress. Am J Physiol Regul Integr Comp Physiol. 2012;303:R1–7. doi: 10.1152/ajpregu.00518.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danilov C, Chandrasekaran K, Racz J, Soane L, Zielke C, Fiskum G. Sul-foraphane protects astrocytes against oxidative stress and delayed death caused by oxygen and glucose deprivation. Glia. 2009;57:645–56. doi: 10.1002/glia.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno K, Kume T, Muto C, et al. Glutathione biosynthesis via activation of the nuclear factor E2–related factor 2 (Nrf2) – antioxidant-response element (ARE) pathway is essential for neuroprotective effects of sulforaphane and 6-(methyl-sulfinyl) hexyl isothiocyanate. J Pharmacol Sci. 2011;115:320–8. doi: 10.1254/jphs.10257fp. [DOI] [PubMed] [Google Scholar]
- 12.Han J, Lee Y, Lee S, et al. Protective effect of sulforaphane against dopaminergic cell death. J Pharmacol Exp Ther. 2007;321:249–56. doi: 10.1124/jpet.106.110866. [DOI] [PubMed] [Google Scholar]
- 13.Ye L, Yu T, Li Y, et al. Sulforaphane enhances the ability of human retinal pigment epithelial cell against oxidative stress, and its effect on gene expression profile evaluated by microarray analysis. Oxid Med Cell Longev. 2013;2013:413024. doi: 10.1155/2013/413024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morroni F, Sita G, Tarozzi A, Cantelli-Forti G, Hrelia P. Neuroprotection by 6-(methylsulfinyl)hexyl isothiocyanate in a 6-hydroxydopamine mouse model of Parkinson’s disease. Brain Res. 2014;1589:93–104. doi: 10.1016/j.brainres.2014.09.033. [DOI] [PubMed] [Google Scholar]
- 15.Petri S, Korner S, Kiaei M. Nrf2/ARE signaling pathway mediator in oxidative stress and potential therapeutic target in ALS. Neurol Res Int. 2012;2012:878030. doi: 10.1155/2012/878030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsey CP, Glass CA, Montgomery MB, et al. Expression of Nrf2 in neurode-generative diseases. J Neuropathol Exp Neurol. 2007;66:75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related fator-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–38. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 18.Shih AY, Imbeault S, Barakauskas V, et al. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J Biol Chem. 2005;280:22925–36. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- 19.Calkins MJ, Johnson DA, Townsend JA, et al. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid Redox Signal. 2009;11:497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Y, Yan W, Chen S, Sun CR, Zhang JM. The role of Nrf2 signaling in the regulation of antioxidants and detoxifying enzymes after traumatic brain injury in rats and mice. Acta Pharmacol Sin. 2010;31:1421–30. doi: 10.1038/aps.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otterbein LE, Soares MP, Yamashita K, Bach FH. Heme oxygenase-1: unleashing the protective properties of heme. Trends Immunol. 2003;24:449–55. doi: 10.1016/s1471-4906(03)00181-9. [DOI] [PubMed] [Google Scholar]
- 22.Siegel D, Gustafson DL, Dehn DL, et al. NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol. 2004;65:1238–47. doi: 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Kawasaki ES. Microarrays: the use of oligonucleotides and cDNA for the analysis of gene expression. Drug Discov Today. 2003;8:134–41. doi: 10.1016/s1359-6446(02)02578-3. [DOI] [PubMed] [Google Scholar]
- 24.Rushmore TH, Kong ANT. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Curr Drug Metab. 2002;3:481–90. doi: 10.2174/1389200023337171. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Uto T, Tanigawa S, Yamada-Kato T, Fujii M, Hou DX. Microarray-based determination of anti-inflammatory genes targeted by 6-(methylsulfinyl) hexyl isothiocyanate in macrophages. Exp Ther Med. 2010;1:33–40. doi: 10.3892/etm_00000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou DX, Fukuda M, Fujii M, Fuke Y. Transcriptional regulation of nicoti-namide adenine dinucleotide phosphate: quinone oxidoreductase in murine hepatoma cells by 6-(methylsulfinyl)hexyl isothiocyanate, an active principle of Wasabi (Eutrema Wasabi Maxim.) Cancer Lett. 2000;20:195–200. doi: 10.1016/s0304-3835(00)00611-x. [DOI] [PubMed] [Google Scholar]
- 27.Korenori Y, Tanigawa S, Kumamoto T, et al. Modulation of Nrf2/Keap1 system by Wasabi 6-methylthiohexyl isothiocyanate in ARE-mediated NQO1 expression. Mol Nutr Food Res. 2013;57:854–64. doi: 10.1002/mnfr.201200689. [DOI] [PubMed] [Google Scholar]
- 28.Thimmulappa RK, Mai KH, Srisuma S, et al. Identification of Nrf2-regulated genes induced by chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- 29.Hou DX, Korenori Y, Tanigawa S, et al. Dynamics of Nrf2 and Keap1 in ARE-mediated NQO1 expression by Wasabi-6-(methylsulfinyl)hexyl isothiocyanate. J Agric Food Chem. 2011;59:11975–82. doi: 10.1021/jf2032439. [DOI] [PubMed] [Google Scholar]
- 30.Dash PK, Zhao J, Orsi SA, Zhang M, Moore AN. Sulforaphane improves cognitive function administered following traumatic brain injury. Neurosci Lett. 2009;460:103–7. doi: 10.1016/j.neulet.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moehlenkamp J, Johnson JA. Activation of antioxidant/electrophile-responsive elements in IMR-32 human neuroblastoma cells. Arch Biochem Biophys. 1999;363:98–106. doi: 10.1006/abbi.1998.1046. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci U S A. 1992;89:2399–403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ina K, Ina H, Ueda M, Yagi A, Kishima I. ω-Methylthioalkyl isothiocyanates in Wasabi. Agric Biol Chem Tokyo. 1989;53:537–8. [Google Scholar]
- 34.Etoh H, Nishimura A, Takasawa R, et al. ω-Methylsulfinylalkyl isothiocyanates in Wasabi, Wasabia japonica Matsum. Agric Biol Chem Tokyo. 1990;54:1587–9. [Google Scholar]
- 35.Li J, Johnson JA. Time-dependent changes in ARE-driven gene expression by use of a noise-filtering process for microarray. Physiol Genomics. 2002;9:137–44. doi: 10.1152/physiolgenomics.00003.2002. [DOI] [PubMed] [Google Scholar]
- 36.Elhalem E, Recio R, Werner S, et al. Sulforaphane homologues: enantiodiver-gent synthesis of both enantiomers, activation of the Nrf2 transcription factor and selective cytotoxic activity. Eur J Med Chem. 2014;87:552–63. doi: 10.1016/j.ejmech.2014.09.052. [DOI] [PubMed] [Google Scholar]
- 37.Vauzour D, Buonfiglio M, Corona G, et al. Sulforaphane protects cortical neurons against 5-S-cysteinyl-dopamine-induced toxicity through the activation of ERK1/2, Nrf-2 and the upregulation of detoxification enzymes. Mol Nutr Food Res. 2010;54:532–42. doi: 10.1002/mnfr.200900197. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y. Molecular mechanism of rapid cellular accumulation of anticarcino-genic isothiocyanates. Carcinogenesis. 2001;22:425–31. doi: 10.1093/carcin/22.3.425. [DOI] [PubMed] [Google Scholar]
- 39.Hu R, Hebbar V, Kim BR, et al. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther. 2004;310:263–71. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- 40.Ye L, Dinkova-Kostova AT, Wade KL, Zhang Y, Shapiro TA, Talalay P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin Chim Acta. 2002;316:43–53. doi: 10.1016/s0009-8981(01)00727-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cytotoxicity assay results showing no significant decrease in the number of viable cells after treatment with 0–20 μM of SFN, 6-MSITC, or 6-MTITC for 12 hours.