Abstract
Background
This study was designed to explore the correlations of promoter methylation in Wnt inhibitory factor-1 (WIF-1), ras-association domain family member 1A (RASSF1A), and Cadherin 13 (CDH13) genes with the risk and prognosis of esophageal cancer (EC).
Material/Methods
A total of 71 EC tissues from resection and 35 adjacent normal tissues were collected. Methylation status in the promoter region was detected by methylation- and non-methylation-specific primers. Corresponding mRNA levels were detected by reverse transcriptase-polymerase chain reaction (RT-PCR). Correlations between the methylations of these 3 genes and clinicopathologic characteristics were analyzed. Kaplan-Meier method and Cox regression model were used to investigate the relationships between WIF-1, RASSF1A, and CDH13 promoter methylations and the prognosis of EC.
Results
Compared with adjacent normal tissues, the methylation frequencies of WIF-1, RASSF1A, and CDH13 genes were significantly higher but the mRNA levels of these 3 genes were significantly lower in EC tissues (all P<0.05). WIF-1 and CDH13 promoter methylations were associated with the degree of tumor differentiation and WIF-1 and RASSF1A promoter methylations were associated with age (all P<0.05). The survival rates of patients with WIF-1, RASSF1A, and CDH13 methylations were significantly lower than those of patients without methylation (all P<0.05). WIF-1, RASSF1A, and CDH13 promoter methylations were independent risk factors affecting the prognosis of EC (all P<0.05).
Conclusions
WIF-1, RASSF1A, and CDH13 promoter methylations are associated with EC. The methylation levels are negatively related with the prognosis in EC.
MeSH Keywords: Esophageal Neoplasms; Methylation; Promoter Regions, Genetic
Background
Esophageal cancer (EC) is the most aggressive malignant neoplasm of the alimentary canal and its mortality rate ranks sixth among all cancers worldwide [1]. In 2008, there were an estimated 482 300 new EC cases and 406 800 EC-caused deaths around the world [2]. EC risk factors include smoking, red meat consumption, hot tea drinking, low intake of vegetables and fresh fruits, and low socioeconomic status [3]. The 5-year survival rate for EC patients diagnosed at early stages is 90%, but it drops to 10~15% for those diagnosed at advanced stages, due to the lack of reliable early-stage diagnosis techniques [4]. Molecular techniques, especially the epigenetic changes in DNA methylation, have become a research focus in the early diagnosis and prognosis of EC [5,6].
Epigenetic silencing attributable to aberrant methylation of promoter regions was suggested as one of the main genetic alterations in the development and progression of cancers, including EC [7]. Wnt inhibitory factor-1 (WIF-1) is an antagonist that is down-regulated and methylated in many carcinomas, such as hepatocellular, esophageal, and gastrointestinal malignancies, and it triggers tumor formation by stimulating β-catenin [8]. The higher frequency of ras-association domain family member 1A (RASSF1A) gene methylation has been observed in many malignant tumor patients, indicating that RASSF1A inactivation is related to cancer pathogenesis [9]. Cadherin 13 (CDH13) is one of the atypical members of the cadherin family; it has been reported to have effects on cellular behavior, largely via its signaling properties, and it is down-regulated in various carcinomas with poorer prognosis [10,11]. Furthermore, promoter methylation of the WIF-1 gene is involved in the EC process, and the abnormal expression of WIF-1 mRNA might be related to EC oncogenesis [12]. RASSF1A was suggested to be one of the EC-related tumor suppressor genes, and hypermethylation of RASSF1A gene was associated with EC progression [13]. A previous study showed that CDH1 gene silencing contributed to by promoter hypermethylation might have a considerable impact on the development of EC, and CDH1 methylation predicted post-surgery survival status of EC patients [14]. However, no information has been published regarding interactions between promoter methylations of WIF-1, RASSF1A, and CDH13 genes in EC. Therefore, the present study aimed to explore the correlations of promoter methylations in the WIF-1, RASSF1A, and CDH13 genes with the risk and prognosis of EC.
Material and Methods
Ethical statement
The Ethics Committee of the Affiliated Hospital of Hebei University approved this study. Written informed consent was provided by all patients before study commencement. Study protocols complied with the ethics principles of medical research involving human subjects, which is based on the Helsinki Declaration [15].
Collection of EC tissues
A total of 71 EC tissues were collected from EC patients who underwent surgical resection at the Affiliated Hospital of Hebei University from January 2009 to September 2010. All patients were diagnosed pathologically with ESCC and received no radiotherapy or chemotherapy before the surgery. This study included 49 males and 22 females, with a mean age of 56.3±5.0 years (range, 43–74 years). The pathological grades of tumor tissues were observed as follows: 21 low differentiation cases, 36 moderate differentiation cases, and 14 high differentiation cases. There were 41 cases with tumor size <3 cm and 30 cases with tumor size ≥3 cm. There were 15 cases with lymph node metastasis (LNM) and 56 cases without LNM. According to the International Union Against Cancer (UICC) tumor node metastasis (TNM) staging system (2010) [16], 20 cases were diagnosed in stage I, 33 cases in stage II, 9 cases in stage III, and 9 cases in stage IV. Additionally, 35 samples of adjacent normal tissues, which were at least 5 cm away from the tumor margin, were collected. All tissues were flash-frozen in liquid nitrogen upon collection and stored at −80°C.
DNA extraction
DNA extraction kits were purchased from Sangon Biotech (Shanghai) Co., Ltd. Then, 30-mg tissue samples were crushed, placed in a sterile 1.5-ml centrifuge tube, and mixed with 200-μl Tris-ethylene diamine tetraacetic acid (TE) suspension. DNA extraction was carefully performed using the manufacturer’s instructions. The concentration and absorbance (A) of the extracted DNA were determined by use of a NanoDrop ND1000 (Thermo Fisher, CA, USA) ultraviolet (UV) spectrophotometer. The products of A260nm/A280nm in the ratio of 1.8~2.0 were used for downstream experiments.
Hydrosulfite treatment
DNA samples were treated with hydrosulfite to detect methylation. Sterile deionized water was added to 10 μl of DNA up to the total volume of 18 μl, shaken evenly, water-bathed at 95°C for 10 min, and placed in an ice bath for 5 min. At this stage, 2 μl of 3M sodium hydroxide (NaOH) was added. Samples were water-bathed at 42°C for 20 min, then a newly-configured 380 μl of 5M sodium hydrogen sulfite (NaHSO3) (containing 125 mM hydroquinol) was added and well mixed. Additionally, 200 μl of liquid paraffin was added, sealed by both Parafilm and silver paper. Tissues were kept away from light and water-bathed at 50°C for 16 min. After cleaning up the liquid paraffin, 1 ml of DNA purification liquid was added to the mixture. Progard was added to remove salt. Purification kits were purchased from Promega Company (USA). After purification, 1 1μl of 3M NaOH was added and mixed, then samples were water-bathed at 37°C for 15 min. Next, 80 μl of 10Mammonium acetate and 2.5 times volume of anhydrous ethanol were added and deposited for 1 h at 80°C. Finally, DNA samples were centrifuged at 12 000 rpm for 20 min with 200 μl of 70% ethanol added for depositing after removing the supernatant. After air drying and addition of 30 μl of TE solution, samples were stored at -−20°C.
Methylation-specific polymerase chain reaction (PCR)
The modified DNA was amplified using specific methylation sequences and non-specific methylation sequences in methylated and non-methylated alleles of WIF-1, RASSF1A, and CDH13 gene promoter regions. The primer sequences are shown in Table 1. PCR reaction was performed with the Biometra Thermocycler thermal cycling device (Germany). PCR reaction used a system with a volume of 25 μl, established according to the instructions of the PCR amplification kit (Sangon Biotech Co., Ltd., Shanghai). Specific PCR setting conditions were: pre-denatured at 95°C for 10 min, denatured at 95°C for 1 min, annealed at 57°C for 45 s, and extended at 72°C for 45 s with a total of 35 cycles, and finally extended at 72°C for 10 min.
Table 1.
Primers for methylation-specific polymerase chain reaction (PCR).
| Sequence | Length of product (bp) | |
|---|---|---|
| WIF-1 | ||
| MSP | F: 5′-GGTTTTATTGGGCGTATCGT-3′ | 145 |
| R: 5′-ACTAACGCGAACGAAATACGA-3′ | ||
| USP | F: 5′-GGGTGTTTTATTGGGTGTATTGT-3′ | 154 |
| R: 5′-AAAAAAACTAACACAAACAAAATACAAAC-3′ | ||
| RASSF1A | ||
| MSP | F: 5′-GTGTTAACGCGTTGCGTATC-3′ | 93 |
| R: 5′-AACCCCGCGAACTAAAAACGA-3′ | ||
| USP | F: 5′-TTTGGTTGGAGTGTGTTAATGTG-3′ | 105 |
| R: 5′-CAAACCCCACAAACTAAAAACAA-3′ | ||
| CDH13 | ||
| MSP | F: 5′-TGTATGAATGAAAACGTCGTC-3′ | 136 |
| R: 5′-GAATACAAAAACGAAACGCA-3′ | ||
| USP | F: 5′-GTGTATGAATGAAAATGTTGTT-3′ | 136 |
| R: 5′-CAAATACAAAAACAAAACACA-3′ | ||
MSP – methylation specific primer; USP – un-methylation specific primer; WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13.
Reverse transcription-PCR (RT-PCR)
Total RNA was isolated using the Trizol method. Tissues were broken into homogenate and 1000 μl of Trizol was added. Next, 200 μl of chloroform was added, followed by violent shaking for 5 min. Tissues were then centrifuged at 12 000 rpm for 15 min. The upper layer was extracted into new EP tubes and 2 volumes of isopropanol (ISO) were added. At 20 min after being placed in the tubes, tissues were centrifuged at 12 000 rpm for 15 min and the supernatant was removed. After the addition of 700 μl of 75% ethanol, tissues were centrifuged at 12 000 rpm for 5 min. After the removal of the supernatant, 20 μl of diethyl pyrocarbonate (DEPC) was added to the tissues, which were then evenly air dried. The products were stored at −80°C for later usage. RNA was measured with a UV spectrophotometer (ThermoFisher, CA, USA) and the products of A260 nm/A280 nm in the ratio of 1.8~2.0 were selected. The integrity of RNA bands was assessed with agarose gel electrophoresis and the clear bands of 28 s, 18 s, and 5 s were used to carry out the downstream experiments. RT was conducted using the RT kit (Sangon Biotech (Shanghai) Co., Ltd.). We added 3 μl of template, 1 μl of Oligo DT primer, and 6 μl of DEPC to 200-μl EP tubes. These tubes were water-bathed at 70°C for 10 min and placed on ice. We then added 5 μl of 5×reaction buffer, 1 μl of RNase, 1 μl of dNTP, 1 μl of RT enzyme, and 7 μl of DEPC to the tubes. After water-bathing at 42°C for 3 h, these tubes were stored at −20°C until further use. The implementation of RT-PCR complied with the instructions of the TaKaRa One Step RNA PCR Kit (TaKaRa Biomedicals, Otsu, Japan), with cDNA obtained by RT as a template, and glyceraldehyde phosphate dehydrogenase (GAPDH) as an internal reference. Reaction-related primers are shown in Table 2. The reaction conditions were: 50°C for 30 min; 94°C for 2 min; 94°C for 30 s, 60°C for 30 s, and 72°C for 6 min, with a total of 30 cycles; extended at 72°C for 5 min, and the reaction was ended at 4°C.
Table 2.
Reverse transcription-polymerase chain reaction (RT-PCR) primers.
| Sequence | Length of product (bp) | |
|---|---|---|
| WIF-1 | F: 5′-CCGAAATGGAGGCTTTTGTA-3′ | 188 |
| R: 5′-TGGTTGAGCAGTTTGA-3′ | ||
| RASSF1A | F: 5′-GGCGTCGTGCGCAAAGGCC-3′ | 280 |
| R: 5′-GAACCTTGATGAAGCCTGTG-3′ | ||
| CDH13 | F: 5′-CATGGTTCCCCCAGCAAGAA-3′ | 572 |
| R: 5′-CTTTCCAGTGAGCCGGAACT-3′ | ||
| GAPDH | F: 5′-GCCTCGCTGTCCACCTTCCA-3′ | 253 |
| R: 5′-CACCTTCACCGTTCCAGTTT-3′ |
WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13; GAPDH – glyceraldehyde phosphate dehydrogenase; F – forward; R – reverse.
Follow-up
The patients were followed up by telephone and outpatient records with a follow-up period of 60 months. Patients were examined once every 3 months for tumor recurrence and metastasis using chest-computed tomography (CT), abdominal ultrasound, and tumor markers. Additionally, survival status of these patients was recorded. The study endpoint was the total survival time, defined as the observed survival time from the start of the follow-up period until death, loss to follow-up, or the end of follow-up. Patients who were still alive at the follow-up deadline and those who were lost during the follow-up period were processed as censored data. Survival time of patients was measured in months.
Statistical methods
SPSS 21.0 statistical software was used for statistical analysis. The enumeration data are presented as rate or percentage, whereas measurement data are presented as mean ± standard deviation (χ̄±s). Comparison of 2 samples was analyzed using the t test. Differences in enumeration data between 2 groups were calculated using the χ2 test and Fisher exact probability test. P<0.05 provided evidence of statistical significance. Kaplan-Meier method and log-rank test were used to determine potential prognostic factors. Clinical pathological factors were analyzed using the Cox proportional hazards regression model.
Results
WIF-1, RASSF1A, and CDH13 gene promoter methylations and mRNA expression levels
After hydrosulfite treatment, cytosine containing no methylation modification in the sequence was converted to uracil; there was no transformation in the methylation-modified cytosine. Methylation levels of WIF-1, RASSF1A, and CDH13 gene promoter were detected using this method. When only methylation amplified fragments were identified, samples were designated as “complete methylation”; when only non-methylation amplified fragments were detected, samples were designated as “non-methylation”; and when both non-methylation and methylation amplified fragments were observed, samples were designated as “partial methylation”. Both complete methylation and partial methylation cases were regarded as methylated cases. The detection of these 3 genes is exhibited in Figure 1.
Figure 1.
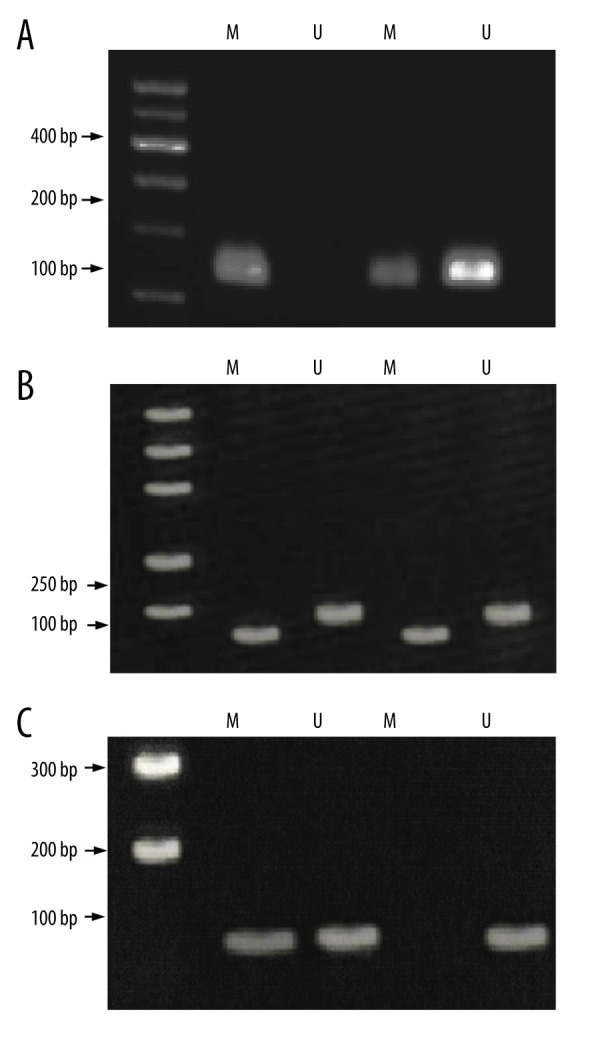
PCR detection for WIF-1, RASSF1A, and CDH13 gene promoter methylations. (A) Samples for WIF-1 methylation and un-methylation. (B) Samples for RASSF1A methylation and un-methylation. (C) Samples for CDH13 methylation and un-methylation. M – methylation; U – un-methylation; PCR – polymerase chain reaction; WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13.
All promoters of WIF-1, RASSF1A, and CDH13 in the adjacent normal tissues and EC tissues exhibited the methylation phenomenon (Table 3). Additionally, methylation frequencies of these genes in the EC tissues were significantly higher than those in adjacent normal tissues (all P<0.05). We also discovered that expressions of WIF-1, RASSF1A, and CDH13 genes in EC tissues were significantly decreased compared with adjacent normal tissues (all P<0.05) (Figure 2). There were significant differences in the mRNA expression levels of WIF-1, RASSF1A, and CDH13 genes between tissues with methylation and tissues without methylation, and expression levels in the methylation group were significantly lower than those in the non-methylation group (all P<0.05) (Figure 3).
Table 3.
Methylation status of promoters of WIF-1, RASSF1A and CDH13 in the adjacent normal tissues and EC tissues.
| Adjacent normal tissues | EC tissues | P | |||||
|---|---|---|---|---|---|---|---|
| M | U | Positive rate (%) | M | U | Positive rate (%) | ||
| WIF-1 | 4 | 31 | 11.43 | 41 | 30 | 57.75 | <0.001 |
| RASSF1A | 1 | 34 | 2.86 | 35 | 36 | 46.48 | <0.001 |
| CDH13 | 3 | 32 | 8.57 | 28 | 43 | 39.44 | 0.001 |
M – methylation; U – un-methylation; WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13; EC – esophageal cancer.
Figure 2.
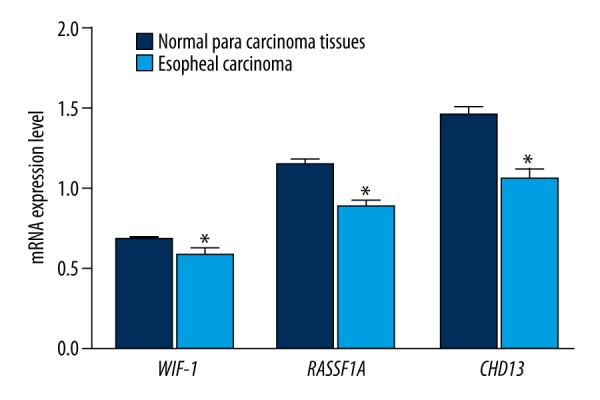
The mRNA expression levels of WIF-1, RASSF1A, and CDH13 genes in esophageal cancer tissues and adjacent normal tissues (* refers to P<0.05 when compared with adjacent normal tissues). WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13.
Figure 3.
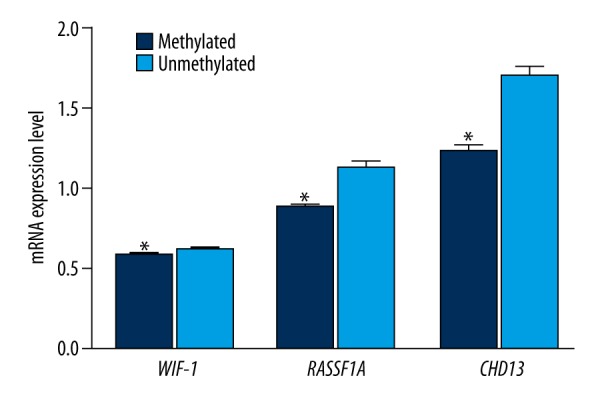
The mRNA expression levels of WIF-1, RASSF1A, and CDH13 genes in promoter region methylation and un-methylation (* refers to P<0.05 when compared with adjacent normal tissues un-methylated). WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13.
Correlations of WIF-1, RASSF1A, and CDH13 gene promoter methylations with clinicopathologic characteristics of EC
As shown in Table 4, the correlations of WIF-1, RASSF1A, and CDH13 gene promoter methylations with clinicopathologic characteristics of EC were further analyzed. Results indicated that methylation levels of WIF-1 and CDH13 gene promoters were associated with degree of tumor differentiation (all P<0.05) and methylation levels of WIF-1 and RASSF1A gene promoters were associated with age (both P<0.05). Other clinicopathologic characteristics of EC were independent from the 3 gene promoter methylations (all P>0.05).
Table 4.
Correlations of WIF-1, RASSF1A and CDH13 gene promoter methylations with clinicopathologic characteristics of EC.
| WIF-1 | P | RASSF1A | P | CDH13 | P | ||||
|---|---|---|---|---|---|---|---|---|---|
| M | U | M | U | M | U | ||||
| Gender | 0.160 | 0.553 | 0.222 | ||||||
| Male | 31 | 18 | 23 | 26 | 17 | 32 | |||
| Female | 10 | 12 | 12 | 10 | 11 | 11 | |||
| Age | 0.017 | 0.012 | 0.099 | ||||||
| <60 | 17 | 21 | 24 | 14 | 20 | 18 | |||
| >60 | 24 | 9 | 11 | 22 | 8 | 25 | |||
| Lymph node metastasis | 0.169 | 0.130 | 0.215 | ||||||
| With | 11 | 4 | 10 | 5 | 8 | 7 | |||
| Without | 30 | 26 | 25 | 31 | 20 | 36 | |||
| Tumor size | 0.520 | 0.069 | 0.119 | ||||||
| <3 cm | 25 | 16 | 24 | 17 | 13 | 28 | |||
| ≥3 cm | 16 | 14 | 11 | 19 | 15 | 15 | |||
| Differentiation degree | 0.018 | 0.210 | 0.032 | ||||||
| Low and moderate | 29 | 28 | 26 | 31 | 26 | 31 | |||
| High | 12 | 2 | 9 | 5 | 2 | 12 | |||
| TNM staging | 0.340 | 0.666 | 0.487 | ||||||
| Stage I | 12 | 8 | 9 | 11 | 5 | 15 | |||
| Stage II | 16 | 17 | 15 | 18 | 15 | 18 | |||
| Stage III | 6 | 3 | 5 | 4 | 4 | 5 | |||
| Stage IV | 7 | 2 | 6 | 3 | 4 | 5 | |||
M – methylation; U – un-methylation; WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13; EC – esophageal cancer; TNM – tumor node metastasis.
Correlations of WIF-1, RASSF1A, and CDH13 gene promoter methylations with the prognosis of EC
Follow-up data was eventually collected up to the 5th year. A total of 39 out of the 71 patients died during the follow-up period and the overall survival rate was 45.1%. Results from the Kaplan-Meier analysis demonstrated that methylation levels of WIF-1, RASSF1A, and CDH13 gene promoters significantly affected the prognosis of EC patients (Figure 4). Prognostic outcomes and the overall survival (OS) status were significantly better in EC patients without WIF-1, RASSF1A, or CDH13 methylation compared to those in patients with methylation (all P<0.05).
Figure 4.
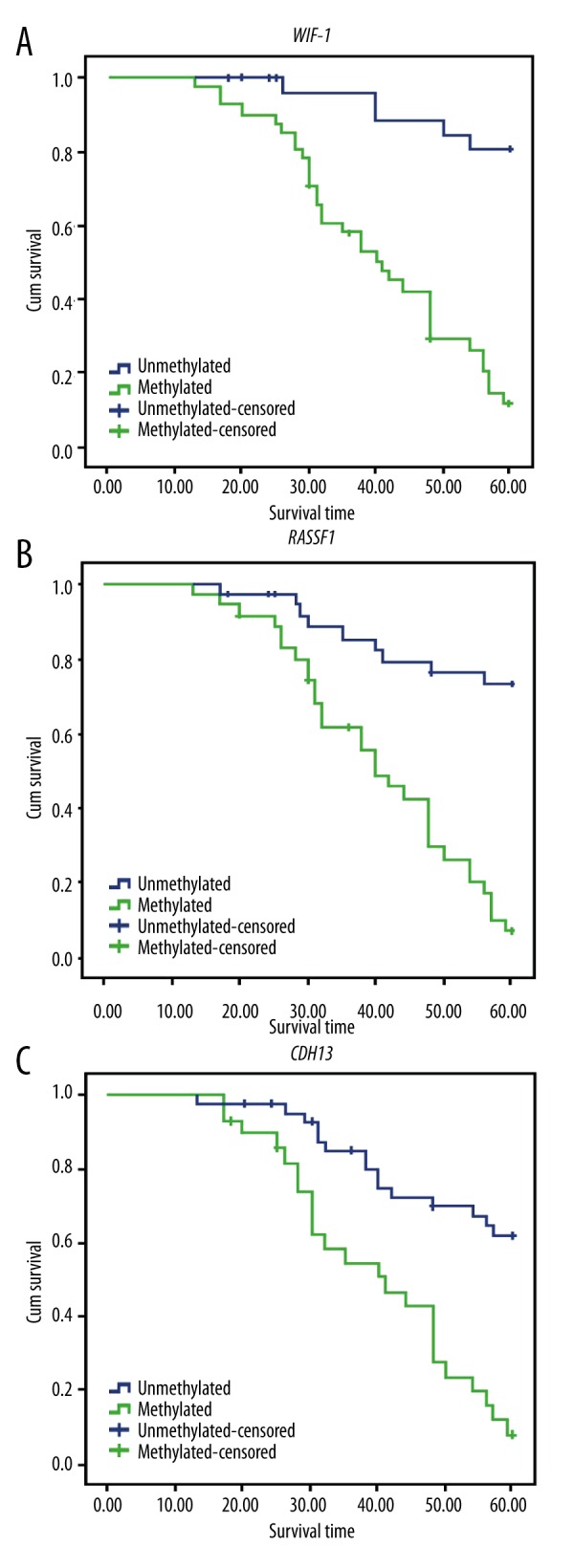
Survival analysis of WIF-1 (A), RASSF1A (B), and CDH13 (C) gene methylation. WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13.
The Cox proportional hazards regression model was used to analyze the clinicopathologic factors and relevant results are presented in Table 5. WIF-1, RASSF1A, and CDH13 gene promoter methylations were significant risk factors that independently affected the prognosis of EC patients (all P<0.05). No significant association was found between other clinicopathologic factors and the prognosis of EC patients (all P>0.05).
Table 5.
Cox regression analysis of prognostic factors in patients with EC.
| B | SE | Wald | df | Sig. | Exp (B) | 95.0% CI for Exp (B) | ||
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| Age | 0.566 | 0.356 | 2.522 | 1 | 0.112 | 1.761 | 0.876 | 3.541 |
| Differentiation degree | 0.442 | 0.584 | 0.573 | 1 | 0.449 | 1.556 | 0.495 | 4.893 |
| Tumor size | 0.136 | 0.415 | 0.108 | 1 | 0.743 | 1.146 | 0.508 | 2.585 |
| Lymph node metastasis | 0.691 | 0.453 | 2.324 | 1 | 0.127 | 1.996 | 0.821 | 4.852 |
| TNM staging | 0.700 | 0.420 | 2.783 | 1 | 0.095 | 2.014 | 0.885 | 4.587 |
| WIF-1 | 1.490 | 0.538 | 7.664 | 1 | 0.006 | 4.435 | 1.545 | 12.731 |
| RASS | 1.210 | 0.462 | 6.851 | 1 | 0.009 | 3.353 | 1.355 | 8.295 |
| CDH13 | 0.875 | 0.427 | 4.198 | 1 | 0.040 | 2.398 | 1.039 | 5.536 |
B – regression coefficient; S.E. – standard error; Wald – the statistical interference of partial regression coefficients; df – free degree; Sig. – significant level; Exp (B) – index of regression coefficient; WIF-1 – Wnt inhibitory factor-1; RASSF1A – ras-association domain family member 1A; CDH13 – Cadherin 13; EC – esophageal cancer; CI – confidence interval.
Discussion
DNA methylation is one of the most common epigenetic modifications [17]. One of the main changes resulting from DNA methylation is transcriptional silencing of tumor suppressor genes, which is caused by hypermethylation of CpG islands in the promoter regions. This change often occurs during the pathogenesis and progression of tumors, which has been described as the mechanism of gene inactivation in neoplasms [18,19]. Our study demonstrates that WIF-1, RASSF1A, and CDH13 promoter region methylations are associated with EC. In addition, methylation levels were found to be negatively related to the prognosis of EC.
One of the most significant findings of the present study was that the methylation frequencies of WIF-1, RASSF1A, and CDH13 genes in EC tissues were significantly higher than those in adjacent normal tissues; mRNA expression levels of these 3 genes in EC tissues were significantly decreased when compared with adjacent normal tissues. The obtained results indicate that WIF-1, RASSF1A, and CDH13 gene methylations are associated with the progression of EC. WIF-1 is a key restrainer of the Wnt/b-catenin signaling pathway and it directly binds to the extracellular Wnt ligands to inhibit their interaction with receptors, thus contributing to the degradation of cytosolic b-catenin through the APC Axin1 destruction complex [20]. Studies have determined that the epigenetic silencing of WIF-1 is a common mechanism of promoter hypermethylation and it causes aberrant activation of the Wnt/b-catenin pathway in EC [21,22]. RASSF1A is a tumor suppressor gene that plays significant roles in cell functions of apoptosis, microtubule stabilization, cell cycle arrest, and metaphase arrest [23]. As suggested by Dammann et al., the expression of RASSF1A in tumor cell lines can decrease the colony formation in vitro and tumorigenicity in vivo. In addition, DNA methylation of RASSF1A is expected to trigger the loss of function along with an increase in both spontaneous and induced tumor formation [24]. As a specific molecule of cadherin cell adhesion, CDH13 is important in establishing cell polarity via inhibiting tumor amplification and invasion, and inducing cell cycle arrest [25]. However, when CDH13 gene promoter region exhibits the hypermethylation status, the CDH13 gene silencing can elevate the risk of cancer [26]. Hibi et al. proposed that the aberrant methylation of CDH13 gene is common in oesophageal and gastric cancers. Additionally, the abnormal methylation could be found in patients with gastric cancers at all clinical stages, which means that both oesophageal and gastric cancers can be methylated at an early stage. Therefore, CDH13 methylation could act as a tumor marker for early detection of digestive tract cancers [27]. These studies confirmed that WIF-1, RASSF1A, and CDH13 gene methylations exert functions in the formation and development of EC. Our study shows that methylation levels of WIF-1 and CDH13 gene promoters are significantly associated with the degree of tumor differentiation. It has been verified that the aberrant DNA methylations of some particular genes were related to the clinicopathologic features and clinical outcomes of cancer cancers [28].
Another result in our study was that the overall survival rates of patients with WIF-1, RASSF1A, and CDH13 methylations were significantly lower than those of the patients with non-methylations. WIF-1, RASSF1A, and CDH13 gene promoter methylations appear to be independent risk factors that influence the prognosis of EC patients. Methylation profile can predict responses to radiotherapy and chemotherapy agents, and thus affects the prognosis of cancer [29]. This study indicates that the OS time is substantially reduced with the methylation of WIF-1; therefore, WIF-1 gene methylation could be used as a biomarker for predicting the prognosis of EC patients [30]. Inactivation of RASSF1A is closely associated with poor outcomes of some cancers, including advanced-stage tumors [24]. A study by Jia Xu et al. suggested that hyper-methylated RASSF1A is related to less favorable OS [31]. It has been reported that patients with CDH13 methylation had worse progression-free survival time and shorter recurrence-free survival when compared to patients without CDH13 methylation. Therefore, CDH13 methylation might be used to assess the severity of disease in order to tailor appropriate therapeutic approaches [32].
Conclusions
Our study provides strong evidence that WIF-1, RASSF1A, and CDH13 promoter region methylations are associated with EC. Higher methylation levels were associated with decreased survival in patients with EC. Therefore, WIF-1, RASSF1A, and CDH13 gene methylations may be considered as biomarkers for predicting the prognosis of EC patients.
Footnotes
Competing interests
We declare that no conflicts of interest exist in this article.
Source of support: Departmental sources
References
- 1.Groblewska M, Siewko M, Mroczko B, Szmitkowski M. The role of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in the development of esophageal cancer. Folia Histochem Cytobiol. 2012;50:12–19. doi: 10.2478/18691. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin RJ, Xiao DW, Liao LD, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–82. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 5.Guo W, Wang G, Dong Y, et al. Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog. 2013;52:265–74. doi: 10.1002/mc.21853. [DOI] [PubMed] [Google Scholar]
- 6.Guo W, Zhou RM, Wan LL, et al. Polymorphisms of the DNA repair gene xeroderma pigmentosum groups A and C and risk of esophageal squamous cell carcinoma in a population of high incidence region of North China. J Cancer Res Clin Oncol. 2008;134:263–70. doi: 10.1007/s00432-007-0283-0. [DOI] [PubMed] [Google Scholar]
- 7.Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008;9:215–34. doi: 10.2217/14622416.9.2.215. [DOI] [PubMed] [Google Scholar]
- 8.Zhou S, Chen L, Mashrah M, et al. Expression and promoter methylation of Wnt inhibitory factor-1 in the development of oral submucous fibrosis. Oncol Rep. 2015;34:2636–42. doi: 10.3892/or.2015.4264. [DOI] [PubMed] [Google Scholar]
- 9.Grawenda AM, O’Neill E. Clinical utility of RASSF1A methylation in human malignancies. Br J Cancer. 2015;113:372–81. doi: 10.1038/bjc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T, Wada S, Eguchi H, et al. Cadherin 13 overexpression as an important factor related to the absence of tumor fluorescence in 5-aminolevulinic acid-guided resection of glioma. J Neurosurg. 2013;119:1331–39. doi: 10.3171/2013.7.JNS122340. [DOI] [PubMed] [Google Scholar]
- 11.Hefti M, von Campe G, Moschopulos M, et al. 5-aminolevulinic acid induced protoporphyrin IX fluorescence in high-grade glioma surgery: A one-year experience at a single institutuion. Swiss Med Wkly. 2008;138:180–85. doi: 10.4414/smw.2008.12077. [DOI] [PubMed] [Google Scholar]
- 12.Clement G, Guilleret I, He B, et al. Epigenetic alteration of the Wnt inhibitory factor-1 promoter occurs early in the carcinogenesis of Barrett’s esophagus. Cancer Sci. 2008;99:46–53. doi: 10.1111/j.1349-7006.2007.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao WM, Li P, Zheng QQ, et al. Hypermethylation-modulated downregulation of RASSF1A expression is associated with the progression of esophageal cancer. Arch Med Res. 2011;42:182–88. doi: 10.1016/j.arcmed.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Ling ZQ, Li P, Ge MH, et al. Hypermethylation-modulated down-regulation of CDH1 expression contributes to the progression of esophageal cancer. Int J Mol Med. 2011;27:625–35. doi: 10.3892/ijmm.2011.640. [DOI] [PubMed] [Google Scholar]
- 15.Glas J, Seiderer J, Bues S, et al. IRGM variants and susceptibility to inflammatory bowel disease in the German population. PLoS One. 2013;8:e54338. doi: 10.1371/journal.pone.0054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobin LH, Gospodarowicz MK, Wittekind CH. TNM Classification of malignant tumours. New York: Wiley-Blackwell; 2009. International union against cancer (UICC) p. 7. [Google Scholar]
- 17.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–42. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 18.Zhang XM, Guo MZ. The value of epigenetic markers in esophageal cancer. Front Med China. 2010;4:378–84. doi: 10.1007/s11684-010-0230-3. [DOI] [PubMed] [Google Scholar]
- 19.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 20.Ge XS, Ma HJ, Zheng XH, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–82. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran I, Thavathiru E, Ramalingam S, et al. Wnt inhibitory factor 1 induces apoptosis and inhibits cervical cancer growth, invasion and angiogenesis in vivo. Oncogene. 2012;31:2725–37. doi: 10.1038/onc.2011.455. [DOI] [PubMed] [Google Scholar]
- 22.Chan SL, Cui Y, van Hasselt A, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest. 2007;87:644–50. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 23.Agathanggelou A, Cooper WN, Latif F. Role of the ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 24.Dammann R, Schagdarsurengin U, Seidel C, et al. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645–63. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, Geng J, Ma K, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135:1675–84. doi: 10.1007/s00432-009-0614-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhai X, Li SJ. Methylation of RASSF1A and CDH13 genes in individualized chemotherapy for patients with non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15:4925–28. doi: 10.7314/apjcp.2014.15.12.4925. [DOI] [PubMed] [Google Scholar]
- 27.Hibi K, Kodera Y, Ito K, et al. Methylation pattern of CDH13 gene in digestive tract cancers. Br J Cancer. 2004;91:1139–42. doi: 10.1038/sj.bjc.6602095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng W, Shen L, Wen S, et al. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res. 2007;9:R57. doi: 10.1186/bcr1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton JP, Sato F, Greenwald BD, et al. Promoter methylation and response to chemotherapy and radiation in esophageal cancer. Clin Gastroenterol Hepatol. 2006;4:701–8. doi: 10.1016/j.cgh.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Abdelmaksoud-Dammak R, Miladi-Abdennadher I, Saadallah-Kallel A, et al. Downregulation of WIF-1 and Wnt5a in patients with colorectal carcinoma: clinical significance. Tumour Biol. 2014;35:7975–82. doi: 10.1007/s13277-014-2015-9. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Shetty PB, Feng W, et al. Methylation of HIN-1, RASSF1A, RIL and CDH13 in breast cancer is associated with clinical characteristics, but only RASSF1A methylation is associated with outcome. BMC Cancer. 2012;12:243. doi: 10.1186/1471-2407-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YL, Xie PG, Ma JG. Aberrant methylation of CDH13 is a potential biomarker for predicting the recurrence and progression of non muscle invasive bladder cancer. Med Sci Monit. 2014;20:1572–57. doi: 10.12659/MSM.892130. [DOI] [PMC free article] [PubMed] [Google Scholar]


