Abstract
The propensity of protein molecules to self-assemble into highly ordered, fibrillar aggregates lies at the heart of the understanding of many disorders such as Alzheimer’s disease or lysozyme systemic amyloidosis. In this article we use highly accurate kinetic measurements of amyloid fibril growth in combination with spectroscopic tools to quantify the effect of modifications in the solution conditions and the amino acid sequence of human lysozyme on its propensity to form amyloid fibrils under acidic conditions. We elucidate and quantify the correlation between the rate of amyloid growth and the population of non-native states, and show that changes in amyloidogenicity are almost entirely due to alterations in the stability of the native state, while other regions of the global free energy surface remain largely unmodified. These results provide insight into the complex dynamics of a macromolecule on a multidimensional energy landscape, and point the way for a better understanding of misfolding diseases.
Introduction
Lysozyme systemic amyloidosis is a fatal hereditary disease associated with the deposition of amyloid fibrils in the spleen, liver and kidney.1 These fibrils are composed of mutational variants of the 130-residue anti-bacterial protein lysozyme that possess a decreased native state stability and folding cooperativity, and predispose the protein to populate partially folded states under conditions that can readily lead to aggregation.2–6 The general link between mutations, partial unfolding and amyloid formation is robust and has been well-documented for a variety of systems.7–18 However, a quantitative relationship between the population of partially folded states and aggregation rates would provide direct and definitive evidence of the way in which the free energy landscape is modified by mutations. To achieve this objective requires the ability to make measurements of both parameters with high accuracy; in this paper, we describe the means by which such data can be obtained.
A crucial point in establishing the connection between changes in the free energy surface and aggregation rates in a quantitative way is the availability of precise kinetic data for protein aggregation. Recent developments, based on quartz crystal microbalance (QCM) measurements now make it possible to determine highly accurate values for the rates of elongation of amyloid fibrils.19,20 Biosensor-based methods such as QCM have the advantage over conventional solution state assays in that small differences in elongation rates due to small variations in the reaction conditions can be resolved quantitatively.21,22 In this approach, a constant ensemble of pre-formed seed fibrils, covalently attached to the microbalance sensor,23 is exposed to a solution of soluble amyloidogenic precursor molecules. The elongation reaction is then monitored in real time through the resulting increase in the surface-bound hydrodynamic mass that can be detected through changes in the mechanical resonant frequency of the piezoelectric sensor. Under conditions where successful cross-seeding between mutants of a protein can be achieved, this technique can be used in a particularly straightforward manner to determine accurately the influence of changes in the amino acid sequence on the free energy surface governing misfolding and aggregation.
We have recently characterized the energy landscape of monomeric human lysozyme in great detail under acidic conditions using temperature as a means of unfolding the protein.24 We have demonstrated that, rather than the classical three-state model (native, intermediate and unfolded states), an extended two-state-model is needed to describe the thermal unfolding process: the native state is in equilibrium with a structurally heterogeneous ‘denatured ensemble’ whose constituent non-native species gradually unfold further from an initially compact ’molten globular’25 state as the temperature is increased. Monitoring the locally cooperative unfolding of this denatured ensemble has revealed that the regions that unfold most readily (the β-domain and helix C) are also those that form the core of amyloid fibrils grown under similarly acidic conditions, suggesting that the unfolding process is closely linked to the aggregation of human lysozyme into amyloid fibrils.26
In the present paper, we describe the enhancement of our knowledge of the energy landscape of human lysozyme by performing at high accuracy measurements of elongation rates of amyloid fibrils formed from human lysozyme under conditions where we have detailed spectroscopic and structural information about the populations of different species present in solution.24 We focus our study on the WT protein and two single-point mutational variants: the I56T variant, associated with systemic lysozyme amyloidosis, and the non-natural I59T variant, which exhibits an unfolding behavior that is intermediate between that of the WT protein and the I56T variant.3,24,27 The energy landscape of a protein can be examined by probing the effect of small perturbations, such as single point mutations and changes in solution pH, on the dynamics of the system.28 Here the elongation rates for amyloid fibrils were measured using a quartz crystal microbalance (QCM), a highly accurate method for kinetic measurements of amyloid growth.19 We chose to use low pH conditions, where the population of partially folded states can be determined at temperatures close to physiological ones using near-UV circular dichroism (CD) spectroscopy, and readily varied, therefore allowing us to quantify the link between the nature of the species populated under given conditions24 and the propensity of lysozyme to aggregate.
Results and discussion
QCM measurements of the elongation of amyloid fibrils from human lysozyme variants
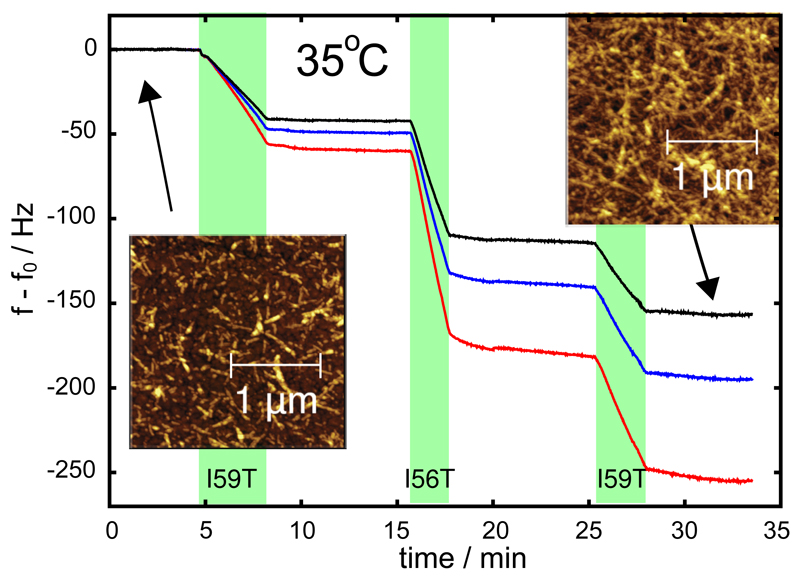
After attaching WT fibril seeds to the gold surface of the sensor (see Supporting Information for experimental procedures and Buell et al23), we exposed the surface to a 70 µM solution of the I59T variant, at pH 1.2, 100 mM of NaCl and 35°C (see Figure 1). This led to a decrease in the resonant frequency, proportional to the increase in mass of protein attached to the sensor surface, due to the elongation of seed fibrils by addition of the amyloidogenic precursor.19 After flushing the sensor surface with buffer solution, a solution of the I56T variant at identical concentration, pH and ionic strength was then introduced into the flow cell containing the QCM sensor. A further decrease in resonant frequency was then measured for this variant; the results shown in Figure 1 demonstrate that the growth rate was significantly higher than that obtained in the presence of the I59T solution. After washing with buffer and upon a second exposure of the sensor to a solution of I59T, a further decrease in resonant frequency was observed, yielding a slope similar to that obtained in the first exposure to a solution of the same variant. The frequencies of the three overtones with N = 3, 5, 7 are plotted in Figure 1; in the remaining figures, for clarity, we instead only show the overtone with N = 3. In the analysis carried out in this work, we always used the average of the three overtones.19 The origin of the difference in the mass sensitivity coefficients between the three overtones, which minimally affects our analysis of the relative changes in rate, lies in the complex material and hydrodynamic properties of the surface-bound fibrils (see Supporting Information). Similarly to the experiment described above, we have observed that the WT, I59T and I56T variants can all elongate both WT or I59T fibrils with rates that depend only on the identity of the soluble precursor and not on the seed (see Figure 1, Figure 2 and Fig. S2 in the Supporting Information). These experiments show that the different forms of the proteins can all access regions of the free energy surface corresponding to fibril growth at pH 1.2 and elevated temperatures, and that the QCM measurements allow us to monitor the fibril elongation process. Atomic force microscopy images of the sensor surface before and after the experiment confirm that the amyloid seed-fibrils have indeed elongated over the course of the experiment (insets in Figure 1). The elongation rates measured by QCM are in agreement with the more qualitative aggregation rates obtained using bulk solution experiments monitored by thioflavin T (ThT) fluorescence29 (see Figure 2 and Fig. S1 of the Supporting Information). The QCM experiments described above have the unique advantage over the conventional solution based assays, such as ThT fluorescence, in that the growth rates of a constant ensemble of fibrils are monitored; this feature allows the rates under different conditions to be readily compared. Moreover, this experimental strategy allows a single process on the aggregation pathway to be monitored, namely the elongation of mature amyloid fibrils, rather than the multiple processes that contribute to the overall mechanism of fibril formation.30 In addition, our approach makes complex cross-seeding experiments like the one shown in Figure 2 straightforward, rapid and highly accurate.
Figure 1.
Growth rates of WT lysozyme fibril seeds in contact with solutions of the I59T or I56T variants (at pH 1.2, 100 mM NaCl and 35°C), measured through the changes in resonant frequency (f-f0) of the sensor system. The green bands indicate the times when the cell contained a solution of the protein under investigation. We used the average of the rates of frequency change of the three overtones with N = 3 (black line), 5 (blue) and 7 (red) as a measure for the aggregation rate. The differences between the mass sensitivities of the frequency overtones is discussed in the main text. The numerical values of the average for the three growth periods shown in this figure are: -12.8, -35.9 and -17.5 Hz/min. Within experimental error, the rate of mass addition depends only on the nature of the monomer, and not on the history of the seed fibrils. The insets show AFM images of the gold coated quartz crystal before and after a growth experiment, yielding an independent confirmation of the elongation of the surface-bound fibrils as the origin of the decrease in resonant frequency. Experiments such as these ones were used to compute the ratios of rates reported in Table S1 and a summary of which can be found in Table 1.
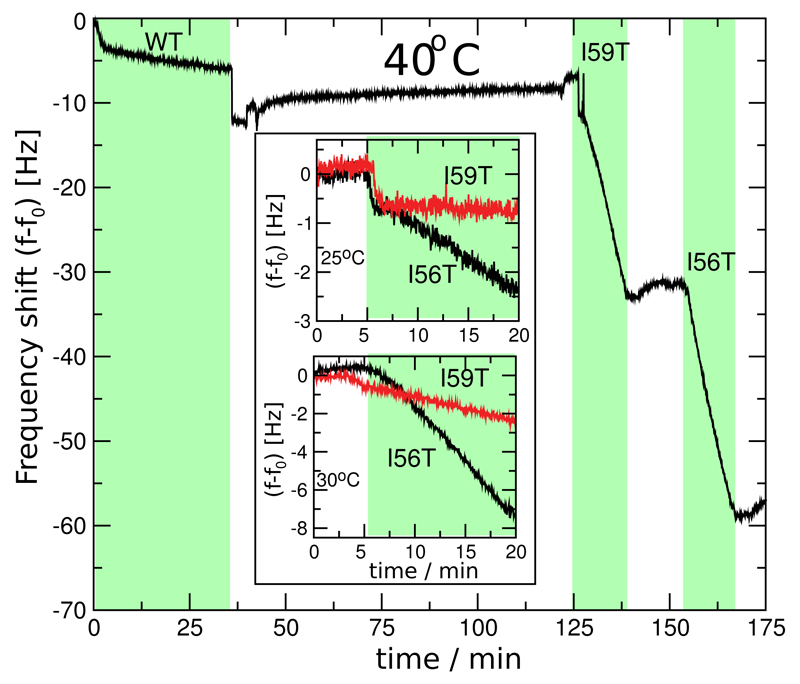
Figure 2.
Comparison of the elongation rates of amyloid fibrils from lysozyme variants at different temperatures. The main part of the figure shows a direct comparison of the rates of monomer addition of all three proteins (WT, I59T and I56T) to WT seeds at 40°C (a similar experiment with I59T seeds is shown in Fig. S2 in the Supporting Information), demonstrating a large difference in absolute rates between the WT protein and the two variants. The two inserts show the fibril growth rates of the two variants at 25°C and 30°C and reveal that the relative kinetics are temperature dependent; at higher temperatures the aggregation rates are substantially more similar than at lower temperature. Equivalent experiments were used to compute the ratios of rates reported in Table S1, a summary of which can be found in Table 1.
Cross-seeding of WT human lysozyme and its destabilized I59T and I56T variants
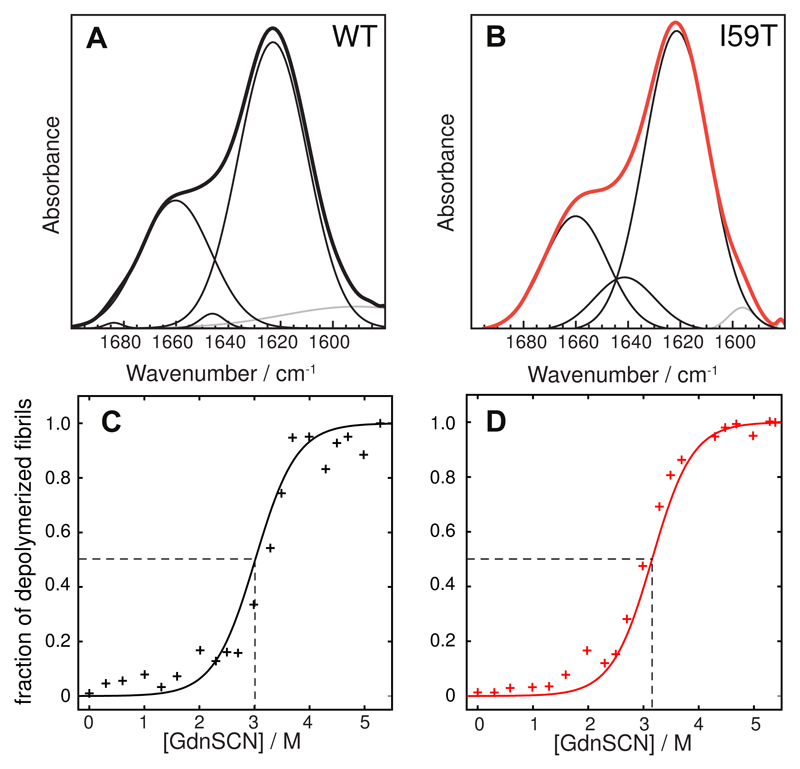
The experiments described in the previous paragraph (see Figure 1, Figure 2 and Fig. S2 in the Supporting Information) show that lysozyme amyloid fibrils elongate with a rate that is independent of the nature of the seeds, within experimental error, and so depends only on the nature of the soluble protein in the aggregation reaction. At a given pH and temperature, the I59T monomer, for example, elongates WT and I59T seeds with a very similar rate, but at a different rate from that of the monomer of the I56T and WT variants. The observed cross-seeding between WT human lysozyme and its associated single point mutational variants I59T and I56T is in agreement with previous results for lysozyme and other proteins.21,31–35 Cross-seeding between the conformational states of the different variants in the vicinity of the midpoints of their thermal denaturation can be rationalized by considering the structural and physico-chemical similarity of the resulting fibrils formed at pH 1.2 and at high temperature. Indeed, an analysis by Fourier Transform Infra Red (FTIR) spectroscopy36 of the conformation of the protein molecules embedded in the fibrils formed from the WT and I59T variants shows that these adopt very similar conformations (see Figure 3A-B). In addition, the fibrils have very similar resistance to de-polymerization by denaturants37 such as guanidinium thiocyanate (GdnSCN), indicating that the nature of the core structures is very similar in both cases (see Figure 3C-D).
Figure 3.
(A) and (B): Amide I region of the FTIR spectra of WT (black) and I59T (red) fibrils grown at pH 1.2. The measured spectra are shown in thick black lines, whereas the deconvoluted Gaussian contributions are shown in thin lines (black for backbone and grey for side chains). The relative intensities are given in Table S3 in the Supporting Information. (C) and (D): Conformational stability of WT (black) and I59T fibrils (red) formed in the presence of WT seeds, measured by de-polymerization experiments performed using GdnSCN. Continuous lines represent fits to a sigmoidal function (see Supporting Information).
Differential effects of small changes in solution conditions on the fibril elongation rates of the I59T, I56T and WT variants
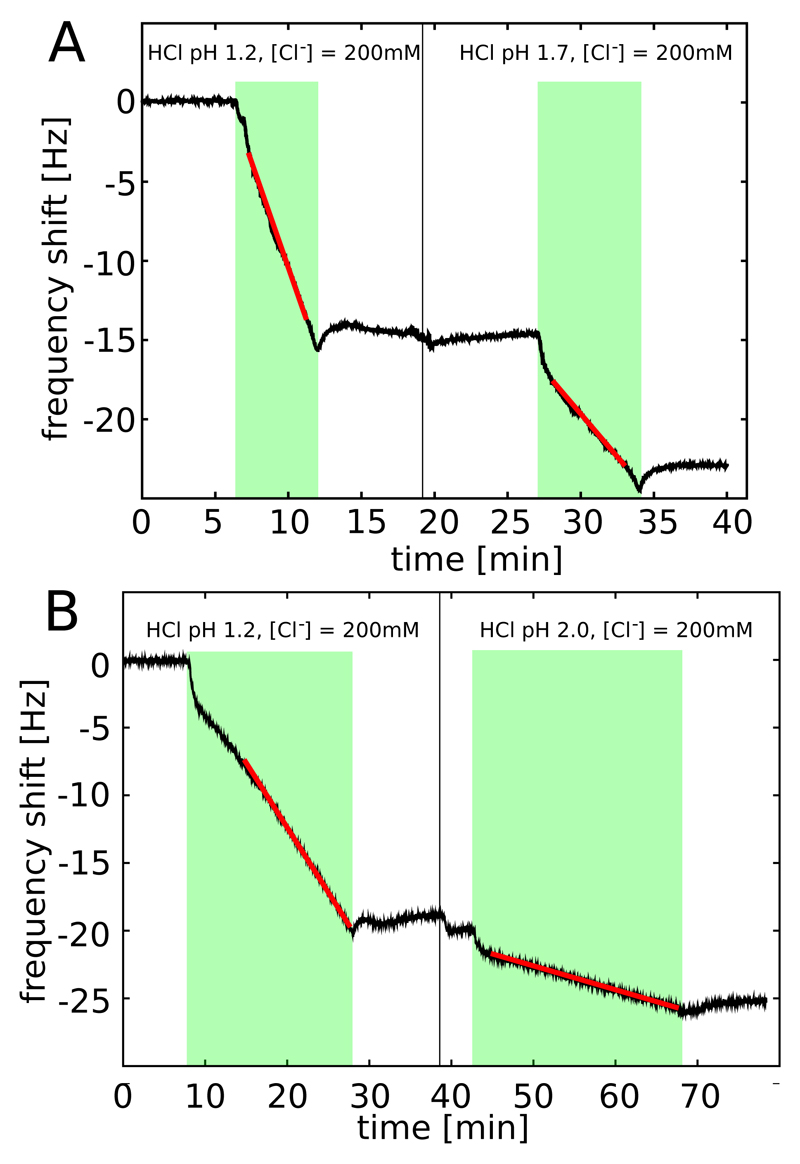
We next examined the elongation rates of the protein fibrils in greater detail by exposing a given QCM sensor to different solution conditions. To investigate the link between the differences in native state stability of the lysozyme variants and the kinetics of aggregation, we studied the impact of temperature (from 25°C to 40°C) and pH (1.2 to 2.0), two variables that strongly influence the relative populations of native and non-native states of the three lysozyme variants under low pH conditions.24 We first compared the rates of incorporation of the monomeric WT protein and its I59T and I56T variants into WT fibril seeds at 25°C, 30°C and 40°C (see Figure 2). The rate for the WT protein is so slow under these conditions that it could only be measured at 40°C, although the rates of the two variants I56T and I59T could readily be measured at all temperatures. Interestingly, the difference in aggregation rates between the I56T and the I59T variants decreased substantially as the temperature was increased: fibrils of the I56T variant grew ca. 10 times faster than those of the I59T variant at 25°C but at approximately the same rate at 40°C. We then investigated the effect of small increases in pH (from pH 1.2 to 1.7 and 2.0) at constant ionic strength (I = 0.2 M; adjusted by addition of sodium chloride) and found that lowering the acidity of the solution induced a significant decrease in the aggregation rate of the I59T variant at 35°C (see Figure 4). These results show that temperature, pH and the location of single point mutations have dramatic and differential effects on the aggregation rates of human lysozyme.
Figure 4.
Growth of surface-bound I59T lysozyme fibrils in contact with solutions of I59T at different pH values and a constant ionic strength of I = 0.2 M, measured through the changes in resonant frequency of the sensor (T = 35°C). The green bands indicate the times when the cell was filled with a solution of the relevant protein, and the black vertical lines indicate a change in solution pH. The red lines show linear fits to the frequency shifts and provide a direct probe of the filament growth rate. A) pH 1.2 and 1.7; B) pH 1.2 and 2.0; the measurements in A and B were acquired using two different QCM sensors. Experiments such as these ones were used to compute the ratios of rates reported in Table S1, a summary of which can be found in Table 1.
Link between elongation rates and populations of non-native states
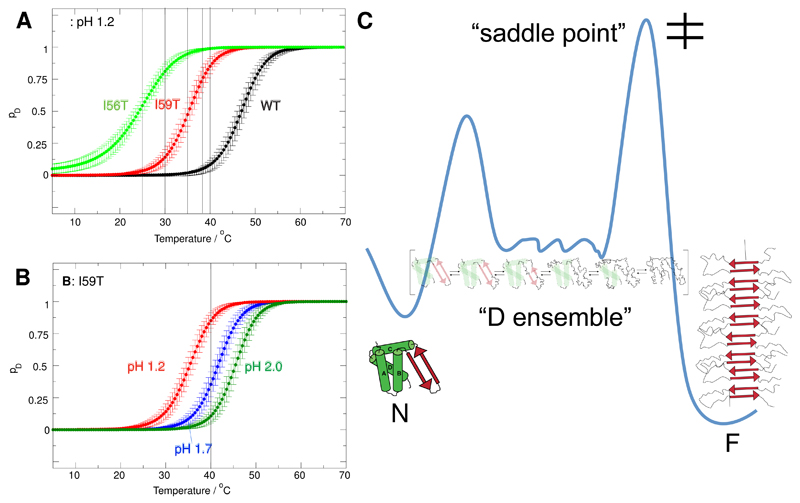
In order to determine the specific origins of the observed differences in aggregation rates, we determined the relative populations of the native and non-native states under the same sets of conditions that we had investigated during the kinetic experiments. As shown in our recent work,24 the populations of the native and non-native states of each protein can be determined by fitting the near-UV circular dichroism (CD) thermal unfolding curves to an apparent two-state model. The unfolding of the I56T, I59T and WT variants at pH 1.2 was monitored by near-UV CD at 270 nm (data taken from Dhulesia et al.24), as well as the unfolding of I59T at pH 1.7 and 2.0 (see Supporting Information Fig. S3). The unfolding traces were fitted globally to an apparent two-state model by imposing common slopes for the signals arising from the native and non-native states: the resulting populations are shown in Figure 6A-B (see Supporting Information for more details on the fitting procedures).
Figure 6.
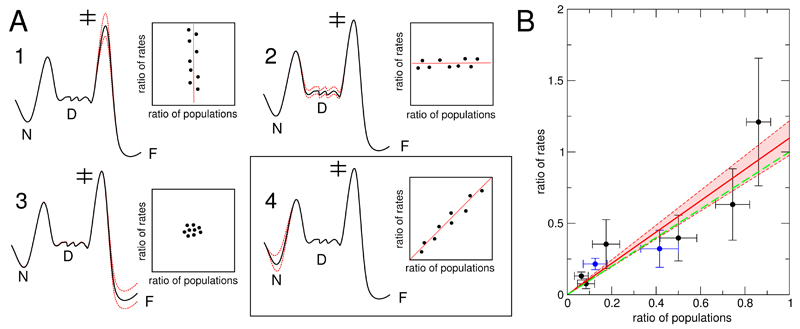
A: Four scenarios of the influence of modifications in amino acid sequence and solution conditions on the free energy landscape are presented. Variations in energy are indicated with red dashed lines. Only modifications that change the energy difference between the lowest state on the energy landscape (“N”) and the saddle point for aggregation (“‡”) will change the aggregation rate (scenarios 1 and 4). Of those two cases, only the latter will lead to a direct relationship between ratios of rates and ratios of populations, with a limiting case of slope 1.0. Changes in the stability of the denatured ensemble (“D”) and the fibrillar state (“F”) do not influence the aggregation rate. B: Plot of the ratios of elongation rates and fractions of populations of non-native states (data from Table 1). The data shown in black correspond to pH 1.2, whereas the data shown in blue correspond to changes in pH for the I59T mutant (to pH 1.7 or 2.0). The linear fit (red) is given by the equation: y = a * x, with a = 1.10 ± 0.12 (correlation coefficient 0.90). The zone between the red dashed lines corresponds to the error in the fitting, as estimated by the variation in slope. As a guide to the eye, y = x is shown (green dashed line), the limiting case 4 and most direct link between the fraction of denatured states and aggregation rates. Our finding that the experimental data very closely follow this prediction indicates that our landscape model of lysozyme can be used to quantitatively understand the origin of amyloidogenicity.
Since both the temperature and the number of fibril ends attached to the QCM sensor will influence the observed rate of frequency change, we directly compared only QCM experiments carried out on identical sensors (where the number of fibril ends is fixed) and at the same temperature. For each different variant (WT, I59T and I56T) at pH 1.2 we calculated the ratios between the amyloid growth rates, as well as for I59T at different pH values (pH 1.2, 1.7 and 2.0). Most of these measurements were repeated several times, therefore allowing estimation of the errors associated with the ratios of the growth rates of two variants under a given set of conditions or of the I59T variant at different pH values. When only one measurement was performed, we used the reproducibility of our initial experiments, where we tested for successful cross-seeding, as an estimate of the error which was determined to be approximately 40% (see e.g. the difference between the first and third growth periods in Figure 1). The ratios of all independent experiments are reported in Table S1, where we indicate from which variant the initial seed fibrils were made, and a summary of the data is shown in Table 1. The link between the ratios of rates and those of the population will be established in the next section.
Table 1.
Summary of the experimental results of this study that are reported in full in Table S1. The averages of the ratios of amyloid fibril growth rates from independent experiments (see text for details) and the populations of the denatured state for different variants, under different conditions (from Figure 5 A and B) are listed. Data from experiments using seed fibrils from different variants are taken together to compute a single average. The data presented here are used to plot Figure 5 C.
| Monomeric proteins and conditions | Temperature | Ratio of amyloid fibril growth rates | Ratio of populations of denatured state |
|---|---|---|---|
| I59T/I56T pH 1.2 | 25°C | 0.13 ± 0.03 | 0.06 ± 0.03 |
| 30°C | 0.35 ± 0.17 | 0.17 ± 0.06 | |
| 35°C | 0.40 ± 0.16 | 0.50 ± 0.08 | |
| 38°C | 0.63 ± 0.25 | 0.74 ± 0.08 | |
| 40°C | 1.21 ± 0.44 | 0.86 ± 0.05 | |
| WT/I59T pH 1.2 | 40°C | 0.08 ± 0.04 | 0.08 ± 0.04 |
| I59T pH 2.0/1.2 | 35°C | 0.22 ± 0.04 | 0.13 ± 0.05 |
| I59T pH 1.7/1.2 | 35°C | 0.32 ± 0.13 | 0.42 ± 0.08 |
Energy landscape model for human lysozyme at low pH
We have recently discussed a framework for describing protein aggregation on the basis of diffusive dynamics on an energy landscape38 in a manner analogous to the behavior encountered during protein folding.39,40 The key determinants of protein aggregation kinetics are the barriers on this energy surface separating the soluble state from the fibrillar configuration. It can be shown that under many conditions the principal factor affecting the overall conversion rate is the height of the highest saddle point that the reactive trajectories cross relative to the energy of the soluble state. Many other barriers can be present, and for proteins such as human lysozyme, that show a cooperative unfolding transition, a second barrier exists under most conditions that separates the native state from a denatured ensemble.24 We have illustrated schematically a one-dimensional projection of a free energy landscape with these features (Figure 5C). This illustration does not imply that all of the denatured ensemble is "on-pathway" for aggregation in a mechanistic sense.
Figure 5.
Fractional populations pD of non-native denatured (D) states, derived from fitting near UV-CD data to a pseudo two-state model (native state, non-native ’denatured ensemble’). A) the I56T, I59T variants and WT lysozyme at pH 1.2. B) I59T at pH 1.2, pH 1.7 and 2.0. C) Free energy landscape of human lysozyme. The native state N is in equilibrium with the ’denatured ensemble D’, a collection of non-native states populated according to the Boltzmann distribution. In addition, the presence of fibril seeds makes the fibrillar state F accessible; the monomeric state is separated from the aggregated state by a free energy barrier. The relative stabilities of the various states can be modulated by changes in solution conditions and amino acid sequence. We use this free energy landscape model to rationalize the origin of increased amyloidogenicity of lysozyme variants.
The landscape picture allows us to generate predictions for the effect of changes in different parts of the energy landscape and thus by comparison with experimental data pinpoint the origin of the amyloidogenicity of human lysozyme. We have analysed the limiting cases of several scenarios where we consider the influence that variations in amino acid sequence can have on the global free energy landscape (Figure 6A), and on measurable quantities such as the ratios of rates and populations listed in Table 1. Since the energy difference between the lowest energy state of the landscape and the saddle point of the aggregation reaction is the principal determinant of aggregation kinetics, energy variations in regions that do not change this overall energy difference will not influence the kinetics (Figure 6A, cases 2 and 3). On the other hand, if the region of the free energy landscape in proximity of the saddle point is modified, we expect changes in the rate, but the relative populations of the native and denatured states should remain unaffected (case 1). We also consider a scenario where the modifications solely affect the native state and where they lead to changes in both the relative populations and the aggregation rate (case 4). The limiting case of such a scenario yields a slope of 1.0, demonstrating a direct relationship between the destabilization of the native state and the increased aggregation rate.
In order to gain insight into the origin of the increased amyloidogenicity of the lysozyme mutants considered in this study, we therefore compare our experimental data (summarized in Table 1) with the predictions of the landscape picture in Figure 6A. This comparison reveals a situation very close to the limiting case 4 (Figure 6B). A linear fit of the data to y = a · x yields a slope of 1.10± 0.12. We therefore conclude that the mutations I59T and I56T increase the amyloidogenicity of lysozyme principally by affecting the stability of the native structure.
Also remarkable is the fact that variations in pH between 1.2 and 2.0 for the I59T variant have a comparable effect on the overall energy landscape as do the single point mutations I59T or I56T (Figure 6B). Several experimental studies have shown that hen lysozyme, which has a sequence identity of ca. 60% to human lysozyme and a closely similar fold, possesses aspartic acid residues with pKa values lower than 2.0;41–43 this is the case, for example, for aspartic acid at position 66, that is also present in the sequence of human lysozyme, with a pKa value of ca. 1.5. A decrease in pH to values below 2.0 therefore modifies the charge state of the protein and affects its thermal stability. Such a change in charge can in principle influence the stability of the native state as well as the height of the barrier separating the denatured ensemble and the fibrillar state, as a result of a change in the electrostatic repulsion between the fibril ends and the monomers. Our finding that the correlation between aggregation rates and populations of denatured states holds under different pH conditions shows, however, that this effect must be negligible in comparison with the change in the overall kinetic barrier resulting from the shift in energy of the native state. This can be rationalized through efficient electrostatic screening at the ionic strength of 0.2 M, where the Debye length is shorter than 1 nm;44 such screening is likely to be more efficient between a partly unfolded monomer and a fibril end than within a folded monomer.
Concluding remarks
In the present study, we have combined CD spectroscopy and QCM, an emerging method in the field of protein aggregation, to study the link between the effects of mutations and solution conditions on the amyloidogenicity of a globular protein. We have shown that variations in the population of non-native conformations of human lysozyme at low pH quantitatively account for changes in the rates of formation of amyloid fibrils under a variety of conditions. The fraction of non-native states populated in solution, as determined by CD spectroscopy, correlates remarkably well with the amyloid fibril elongation rates measured by QCM, with a closely similar proportionality factor for all conditions studied in the work discussed in this article. This finding can be easily rationalized, as changes in charge state or amino acid composition modulate almost exclusively the energy landscape of the protein in the proximity of the native state, and the accuracy of our data allows us to go beyond a purely qualitative understanding of this effect. We therefore corroborate the well-known correlation between native state stability and amyloidogenicity in a uniquely quantitative and systematic way.
At pH 1.2 and the temperatures studied here (up to 40°C), we have recently shown that the non-native states of the amyloidogenic I56T variant comprise both ‘molten globular’ and highly unfolded regions, and that the unfolding of the molten globular region occurs gradually with temperature, with the first unfolded region being part of the β-domain and helix C, while the last residues to unfold are part of the rest of the α-domain (helices A, B and D).24 Our results show that the population of all non-native sub-states that constitute the denatured ensemble correlates very well with the aggregation rate, therefore suggesting that aggregation is generally driven by the presence of non-native states (that are locally molten globular or unfolded, but non-native like) rather than by the formation of non-native species with specific and well-defined properties. Given that residues of the β-domain are the first ones to fully unfold in non-native states, and that residues that form the β-domain and helix C in the native state are found at the core of the fibrils, we hypothesize that the unfolding of those residues allows the formation of first intermolecular contacts in the self-assembly process.
In conclusion, the ability to make highly quantitative measurements of amyloid fibril elongation kinetics, and to compare these data with detailed descriptions of the conformational states populated under different conditions, have enabled us to obtain mechanistic insights into the process of fibril formation and to characterize the free energy landscape governing this process. Such information and its interpretation represent the first and crucial step towards rational intervention to inhibit the misfolding and aggregation of proteins that can give rise to disease.
Supplementary Material
Supporting Information Available
Experimental procedures for bulk solution aggregation, QCM, chemical de-polymerization and FTIR. Figures showing the kinetics of formation of amyloid fibrils in bulk solution and the integrity of the protein inside the fibrils, QCM experiments on I59T seeds and normalized CD signal for the unfolding of human lysozyme. Tables presenting the ratios of populations and growth rates, the thermodynamic parameters obtained by fitting the near-UV CD data and the secondary structure deconvolution of the FTIR spectra recorded on WT and I59T fibrils. This material is available free of charge via the Internet at http://pubs.acs.org/.
Acknowledgement
CMD thanks the Wellcome and the Leverhulme Trusts for their support of this work. AKB acknowledges support from the EPSRC and thanks Magdalene College, Cambridge for support. AD is grateful for support from Boehringer Ingelheim Fonds through a PhD scholarship and from Murray Edwards College, Cambridge, through a Junior Research Fellowship. MFM acknowledges support from IRB Barcelona. CMD and JRK acknowledge support through a BBSRC (BB/E019927/1). NC is a recipient of a Human Frontier Science Program Long-term Fellowship (LT000795/2009). MD is a research associate of the Belgian FRS-FNRS. This work was also supported by the Belgian Government (IAP P6/19 to CMD and MD). TPJK acknowledges support from St John’s College, Cambridge, and XS is grateful to ICREA, IRB and MICINN for their support.
References
- (1).Pepys MB, Hawkins PN, Booth DR, Vigushin DM, Tennent GA, Soutar AK, Totty N, Nguyen O, Blake CC, Terry CJ. Nature. 1993;362:553–557. doi: 10.1038/362553a0. [DOI] [PubMed] [Google Scholar]
- (2).Takano K, Funahashi J, Yutani K. Eur J Biochem. 2001;268:155–159. doi: 10.1046/j.1432-1327.2001.01863.x. [DOI] [PubMed] [Google Scholar]
- (3).Booth DR, Sunde M, Bellotti V, Robinson CV, Hutchinson WL, Fraser PE, Hawkins PN, Dobson CM, Radford SE, Blake CC, Pepys MB. Nature. 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- (4).Canet D, Last AM, Tito P, Sunde M, Spencer A, Archer DB, Redfield C, Robinson CV, Dobson CM. Nat Struct Biol. 2002;9:308–315. doi: 10.1038/nsb768. [DOI] [PubMed] [Google Scholar]
- (5).Dumoulin M, Canet D, Last AM, Pardon E, Archer DB, Muyldermans S, Wyns L, Matagne A, Robinson CV, Redfield C, Dobson CM. J Mol Biol. 2005;346:773–788. doi: 10.1016/j.jmb.2004.11.020. [DOI] [PubMed] [Google Scholar]
- (6).Mossuto MF, Dhulesia A, Devlin G, Frare E, Kumita JR, de Laureto PP, Dumoulin M, Fontana A, Dobson CM, Salvatella X. J Mol Biol. 2010;402:783–796. doi: 10.1016/j.jmb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chiti F, Taddei N, Bucciantini M, White P, Ramponi G, Dobson CM. EMBO J. 2000;19:1441–1449. doi: 10.1093/emboj/19.7.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hammarstrom P, Jiang X, Hurshman AR, Powers ET, Kelly JW. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16427–16432. doi: 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhu L, Zhang X-J, Wang L-Y, Zhou J-M, Perrett S. J Mol Biol. 2003;328:235–254. doi: 10.1016/s0022-2836(03)00249-3. [DOI] [PubMed] [Google Scholar]
- (10).Smith DP, Jones S, Serpell LC, Sunde M, Radford SE. J Mol Biol. 2003;330:943–954. doi: 10.1016/s0022-2836(03)00687-9. [DOI] [PubMed] [Google Scholar]
- (11).Foss TR, Kelker MS, Wiseman RL, Wilson IA, Kelly JW. J Mol Biol. 2005;347:841–854. doi: 10.1016/j.jmb.2005.01.050. [DOI] [PubMed] [Google Scholar]
- (12).Jahn TR, Parker MJ, Homans SW, Radford SE. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- (13).Campioni S, Mossuto MF, Torrassa S, Calloni G, de Laureto PP, Relini A, Fontana A, Chiti F. J Mol Biol. 2008;379:554–567. doi: 10.1016/j.jmb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- (14).Calloni G, Lendel C, Campioni S, Giannini S, Gliozzi A, Relini A, Vendruscolo M, Dobson CM, Salvatella X, Chiti F. Journal of the American Chemical Society. 2008;130:13040–13050. doi: 10.1021/ja8029224. [DOI] [PubMed] [Google Scholar]
- (15).Platt GW, Radford SE. FEBS Letters. 2009;583 doi: 10.1016/j.febslet.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Platt GW, Routledge KE, Homans SW, Radford SE. Journal of Molecular Biology. 2008;378 doi: 10.1016/j.jmb.2008.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gerum C, Silvers R, Wirmer-Bartoschek J, Schwalbe H. Angew Chem Int Ed Engl. 2009;48:9452–9456. doi: 10.1002/anie.200903771. [DOI] [PubMed] [Google Scholar]
- (18).Gerum C, Schlepckow K, Schwalbe H. Journal of Molecular Biology. 2010;401:7–12. doi: 10.1016/j.jmb.2010.06.008. [DOI] [PubMed] [Google Scholar]
- (19).Knowles TPJ, Shu W, Devlin GL, Meehan S, Auer S, Dobson CM, Welland ME. Proc Natl Acad Sci U S A. 2007;104:10016–10021. doi: 10.1073/pnas.0610659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hovgaard MB, Dong M, Otzen DE, Besenbacher F. Biophys J. 2007;93:2162–2169. doi: 10.1529/biophysj.107.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Buell AK, Tartaglia GG, Birkett NR, Waudby CA, Vendruscolo M, Salvatella X, Welland ME, Dobson CM, Knowles TPJ. Chembiochem. 2009;10:1309–1312. doi: 10.1002/cbic.200900144. [DOI] [PubMed] [Google Scholar]
- (22).White DA, Buell AK, Knowles TPJ, Welland ME, Dobson CM. J Am Chem Soc. 2010;132:5170–5175. doi: 10.1021/ja909997e. [DOI] [PubMed] [Google Scholar]
- (23).Buell AK, White DA, Meier C, Welland ME, Knowles TPJ, Dobson CM. J Phys Chem B. 2010;114:10925–10938. doi: 10.1021/jp101579n. [DOI] [PubMed] [Google Scholar]
- (24).Dhulesia A, Cremades N, Kumita JR, Hsu S-TD, Mossuto MF, Dumoulin M, Nietlispach D, Akke M, Salvatella X, Dobson CM. J Am Chem Soc. 2010 doi: 10.1021/ja103524m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ohgushi M, Wada A. FEBS Lett. 1983;164:21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- (26).Frare E, Mossuto MF, de Laureto PP, Dumoulin M, Dobson CM, Fontana A. J Mol Biol. 2006;361:551–561. doi: 10.1016/j.jmb.2006.06.055. [DOI] [PubMed] [Google Scholar]
- (27).Hagan CL, Johnson RJK, Dhulesia A, Dumoulin M, Dumont J, De Genst E, Christodoulou J, Robinson CV, Dobson CM, Kumita JR. Protein Engineering Design and Selection. 2010;23:499–506. doi: 10.1093/protein/gzq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nguyen H, Jager M, Moretto A, Gruebele M, Kelly JW. Proc Natl Acad Sci U S A. 2003;100:3948–3953. doi: 10.1073/pnas.0538054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Nilsson MR. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- (30).Knowles TPJ, Waudby CA, Devlin GL, Cohen SIA, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- (31).Wood SJ, Wypych J, Steavenson S, Louis JC, Citron M, Biere AL. J Biol Chem. 1999;274:19509–19512. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- (32).Krebs MRH, Wilkins DK, Chung EW, Pitkeathly MC, Chamberlain AK, Zurdo J, Robinson CV, Dobson CM. Journal of Molecular Biology. 2000;300:541–549. doi: 10.1006/jmbi.2000.3862. [DOI] [PubMed] [Google Scholar]
- (33).Krebs MRH, Morozova-Roche LA, Daniel K, Robinson CV, Dobson CM. Protein Science. 2004;13:933–1938. doi: 10.1110/ps.04707004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Morozova-Roche LA, Zurdo J, Spencer A, Noppe W, Receveur V, Archer DB, Joniau M, Dobson CM. J Struct Biol. 2000;130:339–351. doi: 10.1006/jsbi.2000.4264. [DOI] [PubMed] [Google Scholar]
- (35).Wright CF, Teichmann SA, Clarke J, Dobson CM. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- (36).Hiramatsu H, Kitagawa T. Biochim Biophys Acta. 2005;1753:100–107. doi: 10.1016/j.bbapap.2005.07.008. [DOI] [PubMed] [Google Scholar]
- (37).Narimoto T, Sakurai K, Okamoto A, Chatani E, Hoshino M, Hasegawa K, Naiki H, Goto Y. FEBS Lett. 2004;576:313–319. doi: 10.1016/j.febslet.2004.09.024. [DOI] [PubMed] [Google Scholar]
- (38).Buell AK, Blundell JR, Dobson CM, Welland ME, Terentjev EM, Knowles TPJ. Phys Rev Lett. 2010;104:228101. doi: 10.1103/PhysRevLett.104.228101. [DOI] [PubMed] [Google Scholar]
- (39).Wolynes PG, Onuchic JN, Thirumalai D. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- (40).Lazaridis T, Karplus M. Science. 1997;278:1928–1931. doi: 10.1126/science.278.5345.1928. [DOI] [PubMed] [Google Scholar]
- (41).Kuramitsu S, Hamaguchi K. J Biochem. 1980;87:1215–1219. [PubMed] [Google Scholar]
- (42).Bartik K, Redfield C, Dobson CM. Biophys J. 1994;66:1180–1184. doi: 10.1016/S0006-3495(94)80900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Abe Y, Ueda T, Iwashita H, Hashimoto Y, Motoshima H, Tanaka Y, Imoto T. J Biochem. 1995;118:946–952. doi: 10.1093/jb/118.5.946. [DOI] [PubMed] [Google Scholar]
- (44).Girault HH. Analytical and Physical Electrochemistry. EPFL Press; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.