Abstract
Mutations in the presenilin1 (PSEN1) and amyloid β-protein precursor (APP) genes account for the majority of cases of autosomal dominantly inherited Alzheimer’s disease (AD). We wished to assess and compare the patterns of cerebral loss produced by these two groups of mutations. Volumetric magnetic resonance imaging and neuropsychological assessments were performed in individuals with clinical AD carrying mutations in the APP (n = 10) and PSEN1 (n = 18) genes and in healthy controls (n = 18). Voxel-based morphometry (VBM), cortical thickness, and region of interest analyses were performed. Mini-Mental State Examination scores were similar in the two disease groups suggesting similar levels of disease severity. There was evidence that APP subjects have smaller hippocampal volume compared with PSEN1 subjects (p = 0.007), and weak evidence that they have larger whole-brain and grey matter volumes (both p = 0.07). Although there was no evidence of statistically significant differences between APP and PSEN1 in VBM or cortical thickness analyses, effect-maps were suggestive of APP subjects having more medial temporal lobe atrophy and conversely PSEN1 subjects showing more neocortical loss. Neuropsychological data were consistent with these regional differences and suggested greater memory deficits in the APP patients and greater impairment in non-memory domains in the PSEN1 group, although these differences were not statistically significant. We conclude that the mechanisms by which APP and PSEN1 mutations cause neuronal loss may differ which furthers our understanding of the neuropathology underlying AD and may inform future therapeutic strategies and trial designs.
Keywords: Alzheimer’s disease, APP, atrophy, autosomal dominant, magnetic resonance imaging, PSEN1
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia, affecting up to 20 million people worldwide [1]. Although AD is usually sporadic and of late onset, a minority of cases are autosomal dominantly inherited—familial AD (FAD). FAD can be caused by mutations in the amyloid beta (A4) precursor protein (APP) [2], presenilin 1 (PSEN1) [3], and PSEN2 genes [4] and by duplications of the APP gene [5]. Most recently, mutations in the sortilin-related receptor LR11 (SORL1) gene have been implicated in FAD [6]. Individuals with FAD tend to have a younger age at onset than those with the more typical sporadic AD (SAD) and the disease is inherited with virtually 100% penetrance. Studying subjects with such mutations has the potential to further our understanding of AD [7]; subjects can be diagnosed with greater certainty than those with SAD, comorbidity is rare, and at-risk individuals can be feasibly assessed many years prior to the onset of symptoms. These mutations have been incorporated into cellular models of AD, and transgenic mouse models have driven therapeutic developments. Recent initiatives have suggested a need to investigate treatments as early as possible in the disease process, ideally in healthy individuals who have a high risk of developing the disease, and FAD individuals form a promising cohort for such studies [7, 8]. Recently, the first ever therapeutic prevention trial in cognitively healthy individuals carrying familial AD mutations was announced (http://www.nia.nih.gov/newsroom/2012/05/obama-administrati-on-presents-national-plan-fight-alzheimers-disease).
According to the amyloid cascade hypothesis, the effects of mutations in the APP and PSEN genes are thought to feed into a final common pathway driven by the accumulation of toxic misfolded amyloid-β (Aβ) entities, resulting in neurodegeneration and cognitive decline. Previous work has suggested that individuals with FAD may have relative preservation of naming, spelling, and visuoperceptual function compared with SAD subjects [9]; conversely isolated slowly-progressive amnesia has been reported in an individual with a V717G mutation in the APP gene [10]. There is clearly heterogeneity in the clinical presentation of FAD [11] and it is likely that the location of the mutation within the gene can affect the phenotypic profile [12]. Additional neurological symptoms and signs appear to occur more commonly in FAD than SAD; myoclonus and seizures have been reported in both APP and PSEN1 families [5, 13, 14] and PSEN1 mutations producing spastic paraparesis [15] and cerebellar ataxia [16] have also been described. Extrapyramidal signs have been reported in both PSEN1 [17] and PSEN2 subjects [18].
Although previous pathological studies have shown some differences between the sporadic and familial forms of the disease (reviewed in [19]), there are few studies directly comparing pathology in the different mutations. Gregory and colleagues demonstrated more grey matter reduction in the frontotemporal regions in individuals with PSEN1 than those with an APP mutation [20], but Shepherd et al. failed to show a difference between these genotypes in terms of neuronal loss [21]. Amyloid PET studies have reported different patterns of Pittsburgh compound B (PiB) retention in FAD compared with SAD, most notably elevated PiB retention in the striatum of both PSEN1 and APP mutation carriers [22, 23]. The numbers of subjects in these studies are unsurprisingly small, and it appears that there can be considerable variability in pathology, even within the same family [24].
Understanding the neuropathological changes occurring early in the course of AD is important as novel disease-modifying therapies being developed may well be more effective at this stage. Patients with FAD in whom a definitive early diagnosis can be established in vivo can therefore provide valuable insights. Using this approach, we have previously demonstrated changes prior to symptom onset in FAD [25–27]. Examining variation in the patterns of atrophy in the different familial mutations at a group level (APP versus PSEN1) may be a means of understanding the extent to which genotype influences the distribution of neuropathology. We therefore aimed to investigate differences between individuals with FAD caused by mutations in the APP and PSEN1 genes. Using an automated whole-brain image analysis technique, voxel-based morphometry (VBM) [28], we examined differences in grey and white matter throughout the brain. We measured cortical thickness with an independent technique and also performed specific manual volumetric measures of the hippocampus and the cingulate gyrus in APP and PSEN1 cases. Correlates of these volumetric differences in terms of the cognitive profiles of each mutation group were examined.
Materials and Methods
Subjects
Twenty-eight FAD subjects with an autosomal dominant family history of AD were included; ten had a mutation in the APP gene and eighteen had a mutation in the PSEN1 gene. The majority of the mutations in this study have been reported elsewhere but three subjects with novel mutations were also included. The APP group comprised subjects with the following mutations: four V717I (of which three were from the same family), three V717G (all from the same family), one V717L, one A692G, and one with the novel T719N mutation. The PSEN1 group comprised subjects with the following mutations: six M139V (including three from one family and two from an unrelated family), two L235V (both from the same family), two intron 4 (from unrelated families), and one each of E280G, I143F, L250S, E184D, pL166del, and F237L, and the novel mutations Q222P and F283L. All novel sequence variants identified were absent from 100 healthy unrelated white control patients. The individuals with novel mutations all had a typical clinical presentation for FAD, with impairment of episodic memory progressing to global cognitive decline from their mid-late 40 s. Further information regarding these novel mutation cases is detailed in a separate paper undergoing preparation for publication. APOE status was available for nine of the APP group and 16 of the PSEN1 group and details are provided in Table 1.
Table 1. Demographics and neuropsychological assessments. Mean (SD).
| Controls(11M, 7F) | APP(5M, 5F) | PSEN1(10M, 8F) | |
|---|---|---|---|
| Age/years | 51.3 (12.8) | 55.0* (4.8) | 47.0*(7.8) |
| APOE status | |||
| 3/3 | 5 | 11 | |
| 3/4 | 4 | 5 | |
| MMSE/30 | 29.5† (0.8) | 18.6† (4.5) | 17.3† (6.0) |
| Dementia index | 17.6 (16.4) | 19.6 (14.7) | |
| NART – verbal IQ | |||
| Dementia Index | 25.6 (20.2) | 27.5 (18.8) | |
| NART – performance IQ | |||
| RMT – words/grade | 1.67 (1.12) | 1.88 (1.26) | |
| RMT – faces/grade | 2.00 (1.80) | 2.06 (1.48) |
NART, National Adult Reading Test; RMT, Recognition Memory Test.
PSEN1 < APP (p = 0.003).
Controls > AD groups (p ≤ 0.01).
All patients in this study fulfilled criteria for probable AD at the time of their MRI assessment [29]. Eighteen cognitively normal controls were also included, in order to improve the estimation of any age and gender effects. Subjects gave written informed consent according to the Declaration of Helsinki. Consent was taken by a clinician experienced in the assessment of patients with cognitive impairment and all subjects were considered to have capacity to consent according to the UK Mental Capacity Act of 2005. Ethical approval was received from the local ethics committee. Subject demographics are shown in Table 1.
MRI
All subjects underwent MR imaging on a 1.5 tesla Signa unit (General Electric, Milwaukee). T1-weighted volumes were acquired with 124 1.5 mm coronal partitions and a 256 × 256 matrix covering a 24 cm field of view. T2-weighted dual-echo images were acquired with 44 5 mm slices with 2.5 mm inter-slice spacing. All images were reviewed to identify artifacts or white matter hyperintensities or infarcts.
Voxel-based morphometry
VBM processing was performed using SPM5 (Statistical Parametric Mapping, Wellcome Trust Centre for Neuroimaging, London, UK; (http://www.fil.ion.ucl.ac.uk/spm/) running on Matlab 7 (release 14) platform (The Mathworks, MA, USA). Initial tissue classification was performed using the unified segmentation process with rigid alignment [30]. DARTEL (diffeomorphic anatomic registration through exponentiated lie algebra) was then used to spatially normalize (with volume-preserving modulation) the initial tissue segmentations to an iteratively estimated study-specific template [31]. We used a smoothing kernel of 4 mm full width at half maximum (FWHM) with the aim of allowing accurate anatomical localization of fine-scale group differences.
Cortical thickness
Cortical thickness measurements were performed using FreeSurfer version 4.0.3 (https://surfer.nmr.mgh.harvard.edu/). This technique has been described previously [32]. In summary, FreeSurfer generates a white-matter segmentation, from which the grey/white matter and pial surfaces are generated; cortical thickness measurements are computed using the average of the shortest distance from one surface to the other. Cortical thickness measures were smoothed on the average surface by 20 mm FWHM.
Volumetric analysis
The in-house MIDAS package was used to measure whole-brain volume semi-automatically [33]; hippocampal and cingulate volumes were measured manually using previously described protocols [34, 35]; left and right sides were summed together for hippocampal and cingulate volumes. Total intracranial volume (TIV) was measured according to the protocol of Whitwell et al. [36].
Neuropsychological assessment
All subjects underwent a neurological assessment including testing with the Mini-Mental State Examination (MMSE) [37]. In addition, FAD subjects were assessed on measures of verbal IQ and performance IQ, and estimated premorbid IQ (National Adult Reading Test (NART)) [38]. Two Dementia Index scores were calculated by subtracting the verbal IQ and performance IQ scores respectively from the NART IQ score. Assessment of visual and verbal memory was performed using the recognition memory tests (RMT) for words and faces [39]. Individuals completed the full, short [40] or easy [41] version of each test and all raw scores were converted to grades as follows: 1: <1st percentile; 2: 1st–5th percentile; 3: 6th–10th percentile; 4: 11th–25th percentile; 5: 26th–50th percentile and 6: >50th percentile. The neuropsychological test battery has been described in detail previously [13].
Statistical analysis
Regional volumetric data, VBM, and cortical thickness were all fitted with linear models comprising terms for group (Controls, APP, and PSEN1 as a three-level factor), gender, age, and TIV. Models were fitted which allowed the residual variance to differ between subject group. In addition to reporting (adjusted) means, we report the difference between the estimated (adjusted) means for APP and PSEN1 expressed as a percentage of the average of these two estimated means: 200 * (APP − PSEN1)/(APP + PSEN1)
The volumetric data were modeled using the PROC MIXED command in SAS 9.1 (SAS Institute Inc, Cary, NC, USA). The Kenward-Rogers degrees of freedom adjustment was used to allow for the small sample sizes [42].
VBM data were modeled in SPM5, with the analysis restricted to masks defined by binarising the mean tissue segments (for grey and white matter, separately) using correlation-maximizing thresholds [43]. Results were corrected for multiple comparisons using the false discovery rate (FDR) [44] at the 5% level. Cortical thickness data were modeled using Surf-Stat (http://www.nitrc.org/projects/surfstat/). FDR-corrected p-values are shown, again thresholded at the 5% level.
Because of non-normality in the neuropsychological data, unadjusted group differences were compared using the Kolmogorov-Smirnov test in Stata (StataCorp College Station, Texas).
Results
The PSEN1 group was, as expected, significantly younger than the APP group (p = 0.003) (Table 1).
Neuropsychological assessment
There was no evidence of a difference in mean MMSE between disease groups (p = 0.60). Although there was a suggestion that the PSEN1 subjects had greater impairment on the dementia indices and the APP group showed more impairment in memory as measured by the recognition memory tests for words and faces, these differences were not statistically significant (p > 0.6).
MRI
We found no evidence of significant artifacts or white matter lesions on visual inspection with the exception of one PSEN1 subject (F283L mutation) who had a small (clinically non-specific) localized lesion in the white matter of the superior frontal lobe. This lesion was not judged sufficient to exclude this individual.
VBM analysis
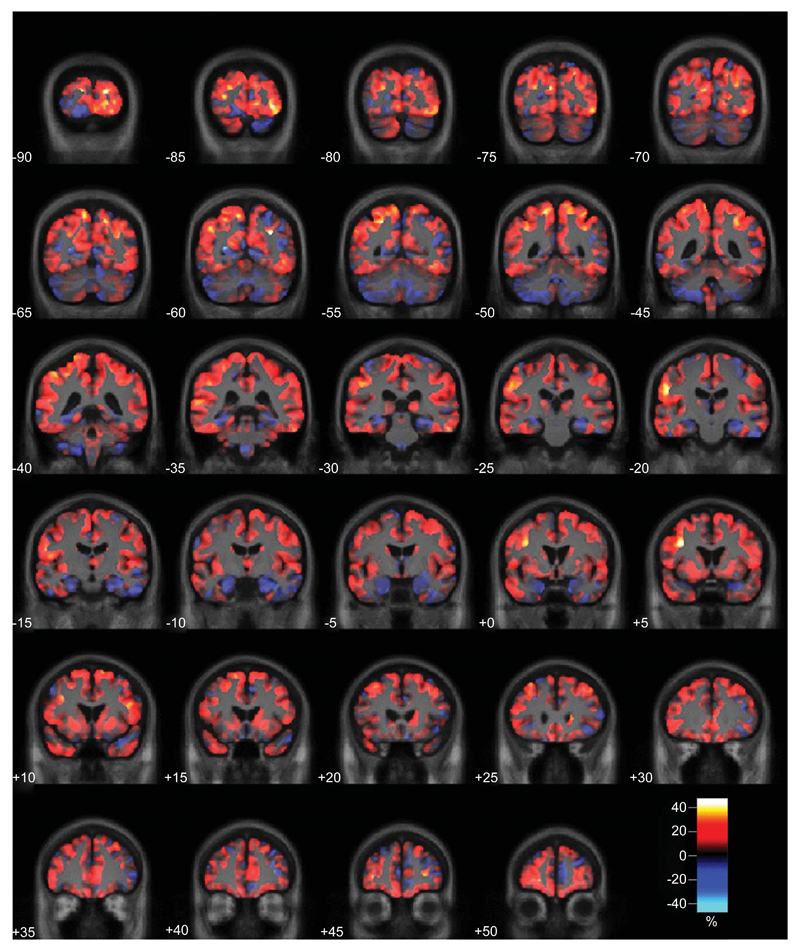
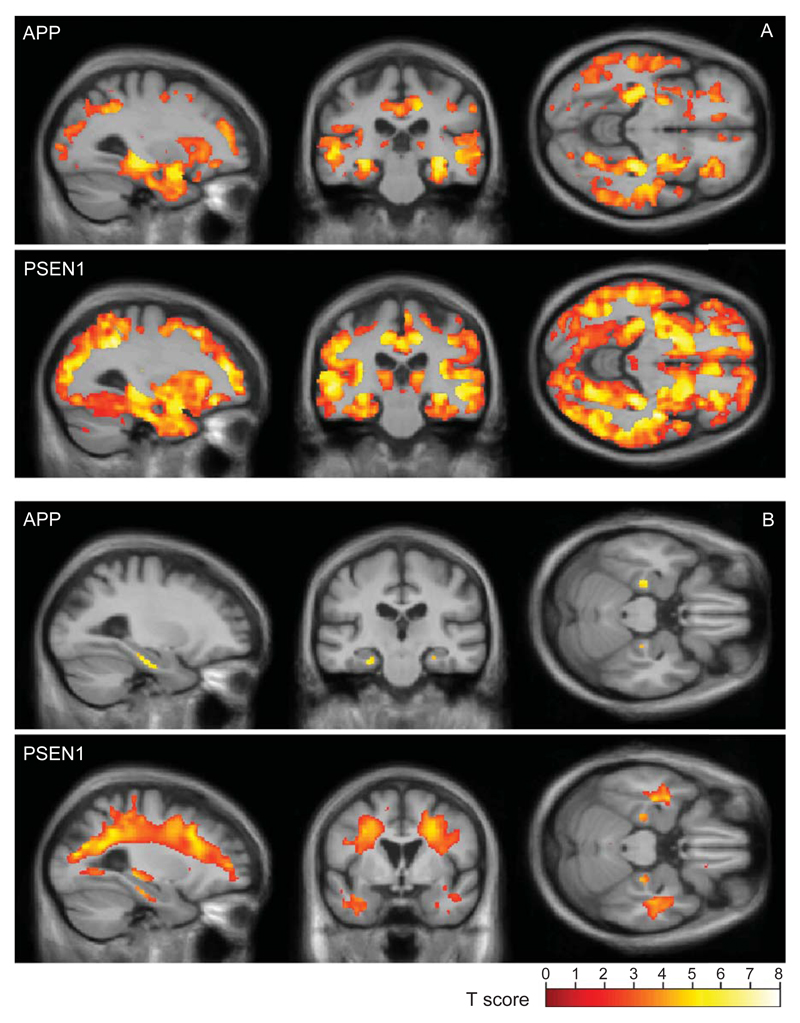
A direct comparison of the APP and PSEN1 groups did not reveal any significant differences in atrophy pattern in either grey or white matter at the FDR <5% level. However, effect-maps suggest that APP subjects had more atrophy in the medial temporal lobe regions and those with the PSEN1 mutation had more widespread cortical involvement (Fig. 1) and more widespread white matter atrophy (Fig. 2). Comparing each disease group with the controls (Fig. 3), we found evidence that the APP subjects have atrophy predominantly in the medial temporal lobes, with the white matter atrophy restricted to this region, whereas PSEN1 subjects showed widely distributed atrophy in both the grey and white matter.
Fig. 1.
Effect-map comparing grey matter volume in the APP and PSEN1 carriers. Regions showing reduced grey matter in the APP group are shown in blue and in PSEN1 in red. Differences between the two adjusted group-means are expressed as a percentage of their average and are overlaid on a mean study-specific template.
Fig. 2.
Effect-map comparing white matter volume in the APP and PSEN1 carriers. Regions showing reduced white matter in the APP group are shown in blue and in PSEN1 in red. Differences between the two adjusted group-means are expressed as a percentage of their average and are overlaid on a mean study-specific template.
Fig. 3.
Grey (A) and white (B) matter reductions compared with controls. SPM results for (top) APP and (bottom) PSEN1 mutation carriers are shown overlaid on a mean study-specific template and are corrected for multiple comparisons using the false discovery rate (FDR <5%).
Cortical thickness measurements
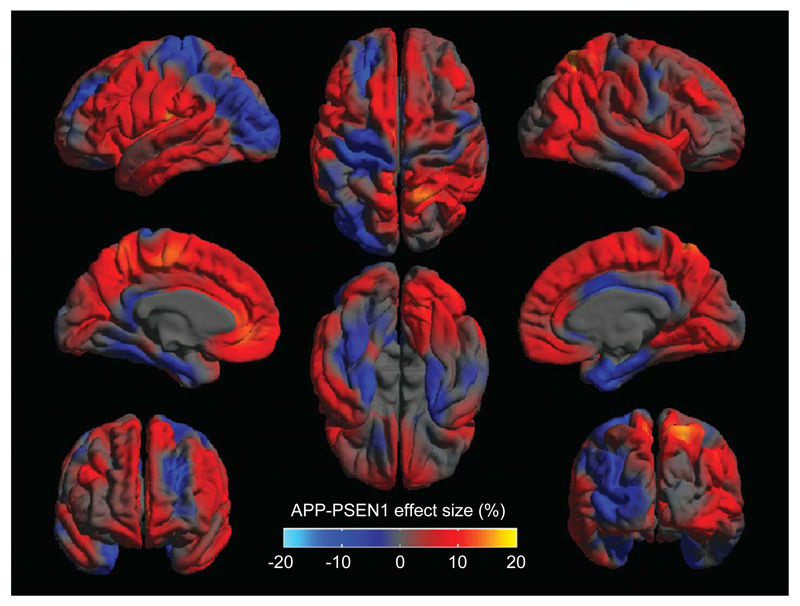
A direct comparison of the APP and PSEN1 groups did not reveal any significant differences in cortical thinning at the FDR <5% level. However, effect-maps suggest that there may be more focal medial temporal lobe thinning in the APP subjects and greater involvement of cortical areas in the PSEN1 group (Fig. 4). Comparisons of each mutation group with controls are shown in Fig. 5, which is consistent with a differential pattern of cortical thinning in these two groups.
Fig. 4.
Effect-map comparing cortical thickness in the APP and PSEN1 carriers. Regions showing thinner cortex in the APP group are shown in blue and in PSEN1 in red, differences between the two adjusted group-means are expressed as a percentage of their average.
Fig. 5.
Cortical thickness reductions for APP (A) and PSEN1 (B) compared with controls. Results are rendered on a mean study-specific cortical surface and are corrected for multiple comparisons using the false discovery rate (FDR <5%).
Volumetric analysis
There was weak evidence that after adjustment for age, gender, and TIV the APP group had larger (52 ml 95% CI (−3.6, 108), p = 0.065) whole-brain volumes and larger (44 ml 95% CI (−4, 91), p = 0.067) grey matter volumes (Table 2). There was no evidence of a difference in mean white matter volume (p = 0.15) or cingulate volume (p = 0.83). There was evidence that APP patients had significantly smaller hippocampi than the PSEN1 group (6.99 ml (95% CI 0.29, 13.69), p = 0.042). Supplementary Figures 1 to 5 show scatterplots of volumetric measures against age by group (available online: http://www.j-alz.com/issues/35/vol35-1.html#supplementarydata04). When adjusted for whole-brain volume (in addition to age, gender, and TIV), there was strong evidence that hippocampal volume was smaller in APP than PSEN1 (9.51 ml (95% CI 2.85, 16.17), p = 0.007).
Table 2.
Estimated mean volumes in mls, at overall mean age, gender, and total intracranial volume, with 95% CIs
| Control | APP | PSEN1 | APP-PSEN1a | % | |
|---|---|---|---|---|---|
| (n = 18) | (n = 10) | (n = 18) | |||
| Brain | 1204 (1160, 1247) | 1110†(1066, 1154) | 1058‡(1023, 1092) | 52 (−3.6, 108) p = 0.065 | 4.8 |
| GM | 669 (649, 690) | 597†(553, 640) | 553‡(531, 575) | 44 (−4, 91) p = 0.067 | 7.6 |
| WM | 480 (457, 503) | 460 (442, 479) | 438* (414, 462) | 22 (−8, 53) p = 0.15 | 4.9 |
| Cingulate | 16.95 (15.53, 18.37) | 13.19b‡(11.95, 14.42) | 13.39†(11.88, 14.90) | −0.20 (−2.15, 1.75) p = 0.83 | −1.5 |
| Hippocampus | 5.72 (5.45, 6.00) | 4.39‡(3.87, 4.91) | 5.09*(4.62, 5.55) | −0.70 (−13.7, −0.03) p = 0.042 | −14.8 |
| Hippocampus adjusted for brainc | 5.25 (4.84, 5.66) | 4.23†(3.72, 4.73) | 5.18 (4.72, 5.63) | −0.95 (−1.62, −0.29) p = 0.007 | −20.2 |
Differences between the APP and PSEN1 group-means as a percentage of their average.
Estimated hippocampal volumes adjusting for brain are for a brain volume of 1075 ml, which is approximately the average of the estimated means of the APP and PSEN1 groups.
Only n=9 APP subjects had data on cingulate volume.
Controls > AD groups (p < 0.05).
Controls > AD groups (p < 0.01).
Controls > AD groups (p < 0.001).
Discussion
This study suggests that there may be differing patterns of regional atrophy between APP and PSEN1 FAD mutation groups. We found statistically significant evidence that APP patients have smaller hippocampi compared with PSEN1 subjects, despite being similar in terms of disease severity. This difference became larger when adjustment was made for whole-brain volume, suggesting that the hippocampal volume difference cannot be ascribed to greater global atrophy in the APP group. Furthermore, there was weak evidence that the APP group had less global atrophy, with higher whole-brain and grey matter volumes than PSEN1 subjects.
Although direct group comparisons did not reveal statistically significantly different grey matter atrophy patterns, VBM and cortical thickness effect-maps suggest the PSEN1 subjects have at least as much, if not more cortical involvement compared with the APP patients, and while non-significant, the trends for reduced whole-brain, grey and white matter volumes in the PSEN1 group compared with the APP subjects further support this suggestion. Likewise, the effect-maps are suggestive of more focal temporal lobe grey matter atrophy in the APP subjects then the PSEN1 patients. There is some support for this finding from postmortem studies which have shown greater grey matter loss in PSEN1 subjects compared with those with APP mutations [20].
In parallel with the grey matter changes, white matter atrophy appears relatively more localized in the APP group, whereas the PSEN1 subjects had extensive white matter involvement of occipital, parietal, and frontal lobes. This marked white matter involvement might explain the greater occurrence of atypical clinical features including spastic paraparesis in some patients with PSEN1 mutations. Since VBM results may be affected by regional signal intensity differences as well as by atrophy we conducted a post hoc SPM analysis (not shown here) and were unable to detect any significant local intensity differences in white matter. It is likely that other imaging modalities such as diffusion-weighted MRI and quantitative mapping of parameters such as magnetization transfer, which reflect more specific properties of white matter, will allow for more thorough investigation of gene-specific white-matter pathology [45].
Amyloid β-protein precursor (AβPP) and presenilin (PSEN) have different biological functions [46]. AβPP is the substrate for the γ-secretase complex of which PSEN forms a part; differential abnormalities in function may therefore exert different regional neuropathological effects. PSEN has roles beyond the cleavage of AβPP, for example in NOTCH processing [47]. In AβPP, the precise location of the mutation has critical implications for its functional effects: mutations pathogenic for AD are particularly clustered around theβ- and γ-secretase cleavage sites that bracket the Aβ coding domain or within the Aβ domain around position 692/693 (http://molgen-www.uia.ac.be/ADMutations) [11, 48]. All but one of the APP subjects in our study had mutations just distal to the γ-secretase site at codon 717, which cause relative overproduction of Aβ peptides ending at residue 42/43. Irrespective of effects on other substrates, it is increasingly clear that different familial AD mutations may affect Aβ generation by different mechanisms which results in qualitative changes in the profiles of the Aβ peptides produced [49]. The proportion of Aβ fragments differ between the two mutation groups so the composition and kinetics of the toxic agent is also likely to differ [49]. Mechanistically, the apparent differences in regional atrophy between APP and PSEN1 subjects could reflect additional, non-AβPP related damage in PSEN1 mutation carriers. Alternatively, effects on AβPP metabolism of APP and PSEN1 mutations could be subtly different in vivo. It is noteworthy both that AβPP levels are highest in the entorhinal pathway [50] and that AβPP expression is upregulated by damage [51]. Under these circumstances, one could envisage that a mutant APP gene would initiate a more local and self-propagating hippocampal lesion in the early stages of disease, whereas the more widespread distribution of presenilin might predict a more global effect.
When we examined whether these regional atrophy patterns were reflected by differences in the neuropsychological profiles, we found greater memory impairment in APP, albeit not statistically significantly. This provides some clinical support for the finding of greater involvement of the medial temporal and limbic regions in these patients compared with the PSEN1 group. Conversely, compared with APP, PSEN1 patients showed greater impairment in the non-memory domains, although again non-significantly, which is also consistent with the imaging findings of greater cortical loss in PSEN1. The lack of significance between the groups may be due to a lack of power or possibly because many patients performed at floor on these tests. Hence, though the neuropsychological data are supportive of the double dissociation suggested by the imaging results, more sensitive tests in larger cohorts are required for confirmation.
It is possible that differences between these groups are due to factors other than the specific mutation, e.g., age, disease severity, or other genetic modifiers. However, the MMSE scores suggest that the patient groups were similarly affected, and the apparent double dissociation between the most atrophied brain areas could not be explained by a simple severity effect. Previous work has shown regional specificity in the decline in brain volume with age [52], and it can be hypothesized that the neurotoxic effects of the amyloid cascade increase with age, perhaps particularly so in the hippocampus which may have greater age-related vulnerability to Aβ oligomers [53]. The older APP subjects may be exhibiting this effect, but may not be as far advanced as the PSEN1 group in terms of cortical loss. Frisoni and colleagues report more medial temporal lobe atrophy in late-onset AD compared with non-familial early onset AD [54], supporting the idea that there may be additional age-related atrophy in this region. However, whole-brain volumes also decline with increasing age [52, 55]. By including age as a covariate in the models for VBM and cortical thickness, we have tried to minimize the effects of this potential confound. Nonetheless larger studies are needed to confirm this.
Our study has demonstrated differences in the extent of atrophy and its distribution between APP and PSEN1 mutations, suggesting that studies comparing FAD and SAD groups may differ depending on the proportions of APP and PSEN1 subjects included. The finding of greater atrophy in FAD compared with SAD reported previously may largely reflect the effect of PSEN1, the most common cause of FAD [19]. For example, a recent study suggested that visual assessment of hippocampal atrophy was insensitive in the identification of presymptomatic and affected subjects with FAD [56]. However, this sample was dominated by PSEN1 mutation carriers who may have relative hippocampal sparing. Our data suggest that it may therefore be important for individual mutation groups to be considered separately when drawing conclusions about the atrophy pattern and presentation of FAD. Phenotypic differences between APP and PSEN1 also have implications for generalization from animal models incorporating these mutations.
A limitation of the study is that while there are several lines of imaging and neuropsychological evidence to support a double dissociation in regional disease burden between the groups, most direct comparisons of the disease groups did not reveal statistically significant differences. Studying individuals with genetically confirmed AD allows definitive diagnosis during life; however, the numbers of subjects available for study are limited and despite the lack of co-morbidity a number of features may still lead to disease heterogeneity which reduces the power to detect group differences. First, different mutations within the same gene may exert different effects. Ideally, analysis of mutation subtypes would be performed; however insufficient data was available to make such comparisons meaningful. Second, other genetic factors may modify the disease, perhaps interacting with the different mutations. A recent report describes different clinical phenotypes in siblings carrying the same PSEN1 mutation [57]. Previous studies have also suggested greater medial temporal atrophy in AD subjects carrying the APOE ε4 allele compared with non-carriers [58, 59], while the APOE ε2 allele may have a protective effect regarding time of disease onset. Possession of an ε4 allele appears to hasten the onset of the disease in APP mutation carriers, but not so clearly in individuals with mutations in the PSEN1 gene [60]. Examination of other mutations, such as PSEN2 and modulating factors such as APOE status and newly identified risk factors such as clusterin and phosphatidylinositol binding clathrin assembly protein [61] may therefore provide further information about disease heterogeneity. Such analyses will require multi-center collaborations, such as the Dominantly Inherited Alzheimer Network (http://www.dian-info.org/) [7].
In this study, we aimed to match the mutation groups according to disease severity; we used the MMSE, a widely used clinical assessment tool for AD. Nevertheless, it is possible that the MMSE may not fully capture the severity of the disease when individuals have different phenotypic presentations. Other rating scales, such as the Clinical Dementia Rating, might provide additional information regarding disease severity but unfortunately were not available for this cohort. It would also be interesting to examine volumetric brain changes prior to the onset of disease in order to identify the earliest focus of neurodegeneration in each mutation type. Previous studies have highlighted volumetric change in these subjects years before clinical onset [25, 34], and a recent study from the DIAN consortium used a variety of imaging techniques to estimate that there were alterations in cerebrospinal fluid markers, brain glucose metabolism, and amyloid deposition decades before the expected age of onset [62] in a large FAD cohort. We await further studies using similar techniques to explore the differential effects of the specific mutations within such FAD cohorts.
In conclusion, we suggest that FAD subjects with the APP and PSEN1 mutations at a similar disease stage may have differing patterns of regional atrophy. The APP subjects had significantly smaller hippocampal volumes than the PSEN1 group and a trend for larger whole-brain and grey matter volumes. This pattern of more localized medial temporal lobe atrophy related to an APP mutation and more widespread cortical loss in PSEN1 patients was supported by VBM and cortical thickness analysis. APP and PSEN1 mutations may modulate the neurodegenerative process in subtly different ways resulting in distinct presentations and patterns of neurodegeneration, suggesting it may be important for individual mutation groups to be considered separately when drawing conclusions about the course of FAD and in considering future therapeutic strategies and clinical trial designs.
Supplementary Material
Supplementary data available online: http://dx.doi.org/10.3233/JAD-121255
Acknowledgments
This work was undertaken at UCLH/UCL which receives funding from the Department of Health’s NIHR for a Biomedical Research Centre and for a Biomedical Research Unit in Dementia. Recruitment was supported by NIHR Dementias and Neurodegenerative Disease Research Network (DeNDRoN). The Dementia Research Centre is an Alzheimer’s Research UK co-ordinating centre and has also received equipment funded by Alzheimer’s Research UK. The Wellcome Trust Centre for Neuroimaging is supported by core funding from the Wellcome Trust, grant number 091593/Z/10/Z and the MRC Prion Unit receives funding from the Medical Research Council UK. Additional support was provided by UK Medical Research Council Programme Grant (G9626876). RIS was supported by an MRC Bioinformatics Training Fellowship (G90/86). JB and SC are supported by Alzheimer’s Research UK Research Fellowships (UK). NSR holds an MRC Clinical Research Training Fellowship (G0900421). MJC is supported by TSB grant M1638A, and CBRC grant 168. SO is supported by TSB, CBRC and EPSRC. JMS is supported by Alzheimer Research UK and HEFCE. JDW is supported by a Wellcome Trust Senior Clinical Fellowship. MNR and NCF are NIHR senior investigators and NCF holds an MRC Senior Clinical Fellowship (G116/143). The authors wish to thank all the patients and their families who took part in this research project and Ray Young for his help with preparing the figures.
Footnotes
Authors’ disclosures available online (http://www.j-alz.com/disclosures/view.php?id=1641).
References
- [1].Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- [2].Goate A, Chartier Harlin M-C, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, et al. Segregation of a missence mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- [3].Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- [4].Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, Mar L, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- [5].Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- [6].Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, Legallic S, Paquet C, Bombois S, Pariente J, Thomas-Anterion C, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17:875–879. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- [7].Bateman RJ, Aisen PS, De Strooper B, Fox NC, Lemere CA, Ringman JM, Salloway S, Sperling RA, Windisch M, Xiong C. Autosomal-dominant Alzheimer’s disease: A review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res Ther. 2011;3:1. doi: 10.1186/alzrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Reiman EM, Langbaum JB, Fleisher AS, Caselli RJ, Chen K, Ayutyanont N, Quiroz YT, Kosik KS, Lopera F, Tariot PN. Alzheimer’s Prevention Initiative: A plan to accelerate the evaluation of presymptomatic treatments. J Alzheimers Dis. 2011;26(Suppl 3):321–329. doi: 10.3233/JAD-2011-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Warrington EK, Agnew SK, Kennedy AM, Rossor MN. Neuropsychological profiles of familial Alzheimer’s disease associated with mutations in the presenilin 1 and amyloid precursor protein genes. J Neurol. 2001;248:45–50. doi: 10.1007/s004150170268. [DOI] [PubMed] [Google Scholar]
- [10].Knight WD, Ahsan RL, Jackson J, Cipolotti L, Warrington EK, Fox NC, Rossor MN. Pure progressive amnesia and the APPV717G mutation. Alzheimer Dis Assoc Disord. 2009;23:410–414. doi: 10.1097/wad.0b013e31819cb7f3. [DOI] [PubMed] [Google Scholar]
- [11].Ryan NS, Rossor MN. Correlating familial Alzheimer’s disease gene mutations with clinical phenotype. Biomark Med. 2010;4:99–112. doi: 10.2217/bmm.09.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jayadev S, Leverenz JB, Steinbart E, Stahl J, Klunk W, Yu CE, Bird TD. Alzheimer’s disease phenotypes and genotypes associated with mutations in presenilin 2. Brain. 2010;133:1143–1154. doi: 10.1093/brain/awq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Godbolt AK, Cipolotti L, Watt H, Fox NC, Janssen JC, Rossor MN. The natural history of Alzheimer disease: A longitudinal presymptomatic and symptomatic study of a familial cohort. Arch Neurol. 2004;61:1743–1748. doi: 10.1001/archneur.61.11.1743. [DOI] [PubMed] [Google Scholar]
- [14].Shrimpton AE, Schelper RL, Linke RP, Hardy J, Crook R, Dickson DW, Ishizawa T, Davis RL. A presenilin 1 mutation (L420R) in a family with early onset Alzheimer disease, seizures and cotton wool plaques, but not spastic paraparesis. Neuropathology. 2007;27:228–232. doi: 10.1111/j.1440-1789.2007.00766.x. [DOI] [PubMed] [Google Scholar]
- [15].Crook R, Verkkoniemi A, Perez-Tur J, Mehta N, Baker M, Houlden H, Farrer M, Hutton M, Lincoln S, Hardy J, Gwinn K, et al. A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nat Med. 1998;4:452–455. doi: 10.1038/nm0498-452. [DOI] [PubMed] [Google Scholar]
- [16].Piccini A, Zanusso G, Borghi R, Noviello C, Monaco S, Russo R, Damonte G, Armirotti A, Gelati M, Giordano R, Zambenedetti P, et al. Association of a presenilin 1 S170F mutation with a novel Alzheimer disease molecular phenotype. Arch Neurol. 2007;64:738–745. doi: 10.1001/archneur.64.5.738. [DOI] [PubMed] [Google Scholar]
- [17].Dintchov Traykov L, Mehrabian S, Van den Broeck M, Radoslavova Raycheva M, Cruts M, Kirilova Jordanova A, Van Broeckhoven C. Novel PSEN1 mutation in a Bulgarian patient with very early-onset Alzheimer’s disease, spastic paraparesis, and extrapyramidal signs. Am J Alzheimers Dis Other Demen. 2009;24:404–407. doi: 10.1177/1533317509341464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Piscopo P, Marcon G, Piras MR, Crestini A, Campeggi LM, Deiana E, Cherchi R, Tanda F, Deplano A, Vanacore N, Tagliavini F, et al. A novel PSEN2 mutation associated with a peculiar phenotype. Neurology. 2008;70:1549–1554. doi: 10.1212/01.wnl.0000310643.53587.87. [DOI] [PubMed] [Google Scholar]
- [19].Shepherd C, McCann H, Halliday GM. Variations in the neuropathology of familial Alzheimer’s disease. Acta Neuropathol. 2009;118:37–52. doi: 10.1007/s00401-009-0521-4. [DOI] [PubMed] [Google Scholar]
- [20].Gregory GC, Macdonald V, Schofield PR, Kril JJ, Halliday GM. Differences in regional brain atrophy in genetic forms of Alzheimer’s disease. Neurobiol Aging. 2006;27:387–393. doi: 10.1016/j.neurobiolaging.2005.03.011. [DOI] [PubMed] [Google Scholar]
- [21].Shepherd CE, Grace EM, Mann DM, Halliday GM. Relationship between neuronal loss and ‘inflammatory plaques’ in early onset Alzheimer’s disease. Neuropathol Appl Neurobiol. 2007;33:328–333. doi: 10.1111/j.1365-2990.2006.00816.x. [DOI] [PubMed] [Google Scholar]
- [22].Villemagne VL, Ataka S, Mizuno T, Brooks WS, Wada Y, Kondo M, Jones G, Watanabe Y, Mulligan R, Nakagawa M, Miki T, et al. High striatal amyloid beta-peptide deposition across different autosomal Alzheimer disease mutation types. Arch Neurol. 2009;66:1537–1544. doi: 10.1001/archneurol.2009.285. [DOI] [PubMed] [Google Scholar]
- [23].Knight WD, Okello AA, Ryan NS, Turkheimer FE, Rodríguez Martinez de Llano S, Edison P, Douglas J, Fox NC, Brooks DJ, Rossor MN. Carbon-11-Pittsburgh compound B positron emission tomography imaging of amyloid deposition in presenilin 1 mutation carriers. Brain. 2011;134:293–300. doi: 10.1093/brain/awq310. [DOI] [PubMed] [Google Scholar]
- [24].Gómez-Isla T, Growdon WB, McNamara MJ, Nochlin D, Bird TD, Arango JC, Lopera F, Kosik KS, Lantos PL, Cairns NJ, Hyman BT. The impact of different presenilin 1 andpresenilin 2 mutations on amyloid deposition, neurofibrillary changes and neuronal loss in the familial Alzheimer’s disease brain: Evidence for other phenotype-modifying factors. Brain. 1999;122(Pt 9):1709–1719. doi: 10.1093/brain/122.9.1709. [DOI] [PubMed] [Google Scholar]
- [25].Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer’s disease: Unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci U S A. 2002;99:4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, Fox NC. Tracking atrophy progression in familial Alzheimer’s disease: A serial MRI study. Lancet Neurol. 2006;5:828–834. doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- [27].Knight WD, Kim LG, Douiri A, Frost C, Rossor MN, Fox NC. Acceleration of cortical thinning in familial Alzheimer’s disease. Neurobiol Aging. 2011;32:1765–1773. doi: 10.1016/j.neurobiolaging.2009.11.013. [DOI] [PubMed] [Google Scholar]
- [28].Ashburner J, Friston KJ. Voxel-based morphometry–the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- [29].McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s Disease: Report of the NINCDS- ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- [30].Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- [31].Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freeborough PA, Fox NC, Kitney RI. Interactive algorithms for the segmentation and quantitation of 3-DMRIbrain scans. Comput Methods Programs Biomed. 1997;53:15–25. doi: 10.1016/s0169-2607(97)01803-8. [DOI] [PubMed] [Google Scholar]
- 34.Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, Rossor MN. Presymptomatic hippocampal atrophy in Alzheimer's disease. A longitudinal MRI study. Brain. 1996;119(Pt 6):2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- 35.Jones BF, Barnes J, Uylings HB, Fox NC, Frost C, Witter MP, Scheltens P. Differential regional atrophy of the cingulate gyrus in Alzheimer disease: A volumetric MRI study. Cereb Cortex. 2006;16:1701–1708. doi: 10.1093/cercor/bhj105. [DOI] [PubMed] [Google Scholar]
- 36.Whitwell JL, Crum WR, Watt HC, Fox NC. Normalization of cerebral volumes by use of intracranial volume: Implications for longitudinal quantitative MR imaging. AJNR Am J Neuroradiol. 2001;22:1483–1489. [PMC free article] [PubMed] [Google Scholar]
- 37.Folstein M, Folstein S, McHughs P. The “Mini mental state”: A practical method for grading the cognitive state of patients for the clinician. J Psychiatric Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 38.Nelson HE, Willison J. The National Adult Reading Test (NART) manual. NFER-Nelson; Windsor, UK: 1991. [Google Scholar]
- 39.Warrington EK. Recognition Memory Test. NFERNelson; Windsor, UK: 1984. [Google Scholar]
- 40.Warrington EK. The Camden Memory Tests Manual. Psychology Press; Hove: 1996. [Google Scholar]
- 41.Clegg F, Warrington EK. Four easy memory tests for older adults. Memory. 1994;2:167–182. doi: 10.1080/09658219408258943. [DOI] [PubMed] [Google Scholar]
- 42.Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. [PubMed] [Google Scholar]
- 43.Ridgway GR, Omar R, Ourselin S, Hill DL, Warren JD, Fox NC. Issues with threshold masking in voxel-based morphometry of atrophied brains. Neuroimage. 2009;44:99–111. doi: 10.1016/j.neuroimage.2008.08.045. [DOI] [PubMed] [Google Scholar]
- 44.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 45.Acosta-Cabronero J, Williams GB, Pengas G, Nestor PJ. Absolute diffusivities define the landscape of white matter degeneration in Alzheimer’s disease. Brain. 2010;133:529–539. doi: 10.1093/brain/awp257. [DOI] [PubMed] [Google Scholar]
- 46.Wilquet V, De Strooper B. Amyloid-beta precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14:582–588. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Thinakaran G, Parent AT. Identification of the role of presenilins beyond Alzheimer’s disease. Pharmacol Res. 2004;50:411–418. doi: 10.1016/j.phrs.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 48.George-Hyslop PH, St, Petit A. Molecular biology and genetics of Alzheimer’s disease. C R Biol. 2005;328:119–130. doi: 10.1016/j.crvi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 49.Chávez-Gutiérrez L, Bammens L, Benilova I, Vandersteen A, Benurwar M, Borgers M, Lismont S, Zhou L, Van Cleynenbreugel S, Esselmann H, Wiltfang J, et al. The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts GW, Nash M, Ince PG, Royston MC, Gentleman SM. On the origin of Alzheimer’s disease: A hypothesis. Neuroreport. 1993;4:7–9. doi: 10.1097/00001756-199301000-00001. [DOI] [PubMed] [Google Scholar]
- 51.Van Den Heuvel C, Blumbergs P, Finnie J, Manavis J, Lewis S, Jones N, Reilly P, Pereira R. Upregulation of amyloid precursor protein and its mRNA in an experimental model of paediatric head injury. J Clin Neurosci. 2000;7:140–145. doi: 10.1054/jocn.1999.0168. [DOI] [PubMed] [Google Scholar]
- 52.Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch Neurol. 2003;60:989–994. doi: 10.1001/archneur.60.7.989. [DOI] [PubMed] [Google Scholar]
- 53.Balleza-Tapia H, Huanosta-Gutierrez A, Marquez-Ramos A, Arias N, Pena F. Amyloid beta oligomers decrease hippocampal spontaneous network activity in an age-dependent manner. Curr Alzheimer Res. 2010;7:453–462. doi: 10.2174/156720510791383859. [DOI] [PubMed] [Google Scholar]
- 54.Frisoni GB, Pievani M, Testa C, Sabattoli F, Bresciani L, Bonetti M, Beltramello A, Hayashi KM, Toga AW, Thompson PM. The topography of grey matter involvement in early and late onset Alzheimer’s disease. Brain. 2007;130:720–730. doi: 10.1093/brain/awl377. [DOI] [PubMed] [Google Scholar]
- 55.Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 56.Ringman JM, Pope W, Salamon N. Insensitivity of visual assessment of hippocampal atrophy in familial Alzheimer’s disease. J Neurol. 2010;257:839–842. doi: 10.1007/s00415-009-5436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishizuka T, Nakamura M, Ichiba M, Fujita S, Takeuchi K, Fujimoto T, Sano A. Different Clinical Phenotypes in Siblings with a Presenilin-1 P264L Mutation. Dement Geriatr Cogn Disord. 2012;33:132–140. doi: 10.1159/000338394. [DOI] [PubMed] [Google Scholar]
- 58.Schott JM, Ridha BH, Crutch SJ, Healy DG, Uphill JB, Warrington EK, Rossor MN, Fox NC. Apolipoprotein e genotype modifies the phenotype of Alzheimer disease. Arch Neurol. 2006;63:155–156. doi: 10.1001/archneur.63.1.155. [DOI] [PubMed] [Google Scholar]
- 59.Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, Boxer AL, Karydas A, Possin KL, Gorno-Tempini ML. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer’s disease and frontotemporal dementia. Proc Natl Acad Sci U S A. 2009;106:2018–2022. doi: 10.1073/pnas.0812697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorbi S, Nacmias B, Forleo P, Piacentini S, Latorraca S, Amaducci L. Epistatic effect of APP717 mutation and apolipoprotein E genotype in familial Alzheimer’s disease. Ann Neurol. 1995;38:124–127. doi: 10.1002/ana.410380120. [DOI] [PubMed] [Google Scholar]
- 61.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ, Xie X, Blazey TM, Holtzman DM, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.