Abstract
We present an acute promyelocytic leukemia (APL) patient with two subtypes of IRF2BP2–RARA, in which the IRF2BP2 gene showed completely new breakpoints. Bone marrow examination revealed morphologic features indicative of APL. However, promyelocytic leukemia–RARA fusion was not detected. A paired‐end mRNA sequencing followed by RT‐PCR and direct sequencing revealed two types of fusion transcripts between exon 1B of IRF2BP2 and exon 3 of RARA. The patient received all‐trans retinoic acid and conventional chemotherapy, but showed resistance. This is the second report of IRF2BP2 involvement in APL, and we describe various breakpoints for the IRF2BP2–RARA fusion gene.
Keywords: Acute promyelocytic leukemia, all‐trans retinoic acid, fluorescence in situ hybridization, gemtuzumab ozogamicin, interferon regulatory protein 2 binding protein 2
Acute promyelocytic leukemia (APL) is an acute myeloid leukemia subtype characterized by promyelocytes with abnormal morphology, coagulopathy, specific chromosomal abnormalities, and unique response to retinoic acid treatment. Most APL patients carry t(15;17)(q22;q12), which results from the fusion of the promyelocytic leukemia (PML) and retinoic acid receptor α (RARA) genes.1 Consequently, the retinoid sensitivity of RARA is reduced, blocking myeloid differentiation. Other fusion partners of RARA have been found.2, 3, 4, 5, 6, 7, 8, 9 Recently, interferon regulatory protein 2 binding protein 2 (IRF2BP2) emerged as a new RARA partner; this IRF2BP2–RARA fusion gene includes exon 2 of IRF2BP2 and intron 2 of RARA.10
Herein, we describe the second case of APL with the IRF2BP2–RARA fusion gene, in which novel breakpoints in IRF2BP2 and unique clinical characteristics were noted.
Case
A 68‐year‐old Japanese woman was admitted to Otemae Hospital (Osaka City, Japan) with pancytopenia in May 2012. Initial complete blood counts showed: hemoglobin, 8.2 g/dL; platelets, 48 × 109/L; and white blood cells, 1.65 × 109/L (including 1% promyelocytes). No disseminated intravascular coagulation (DIC) was detected. A bone marrow sample showed marked hypercellularity; 36.8% were promyelocytes with azurophilic granules and Auer rods (Fig. 1a). Flow cytometry revealed APL cells positive for cluster of differentiation (CD) 33, CD34, CD64, CD117, and human leukocyte antigen (HLA)‐DR, and negative for CD2, CD7, CD13, and CD19. Fluorescence in situ hybridization (Fig. 1b) and RT‐PCR failed to detect PML–RARA fusion, and chromosomal examination of leukemia cells showed –X. Fluorescence in situ hybridization with dual fusion translocation probes detected no typical RARA split signals, but rather a small RARA signal and two large signals (46/100) (Fig. 1c). Accordingly, the patient was diagnosed with APL, and remission induction therapy was initiated with all‐trans retinoic acid (ATRA; 45 mg/m2/day). On day 12, promyelocytes in the peripheral blood increased to >10 × 109/L; therefore, we added idarubicin (9 mg/m2/day from days 12 to 14) and cytosine arabinoside (75 mg/m2/day from days 12 to 18). However, complete remission (CR) was not achieved. Re‐induction therapy was initiated with gemtuzumab ozogamicin (GO) (9 mg/m2) because of high CD33 expression. One month later, morphological examination and FISH of bone marrow aspirate showed hematological remission. In April 2013, the patient experienced molecular relapse as determined by an increase of Wilms tumor‐1 mRNA in the peripheral blood. She was re‐administered with GO (9 mg/m2), and remained in hematological remission until August 2013 when she developed syncope. Magnetic resonance images revealed several enhanced regions in the right brain, and her cerebrospinal fluid (CSF) clearly contained APL cells. Because her level of consciousness was depressed despite high‐dose methotrexate and cytarabine infusion, whole brain irradiation was undertaken. One month later, her neurological symptoms disappeared, and her CSF cytology revealed no APL cells. In February 2014, she developed bilateral leg weakness and walking difficulty. Lumbar magnetic resonance imaging showed enhancement in the conus and nerve roots, and APL cells were once again detected in her CSF. Craniospinal irradiation was undertaken, and her symptoms disappeared temporarily. However, she relapsed in August 2014 and died of APL.
Figure 1.
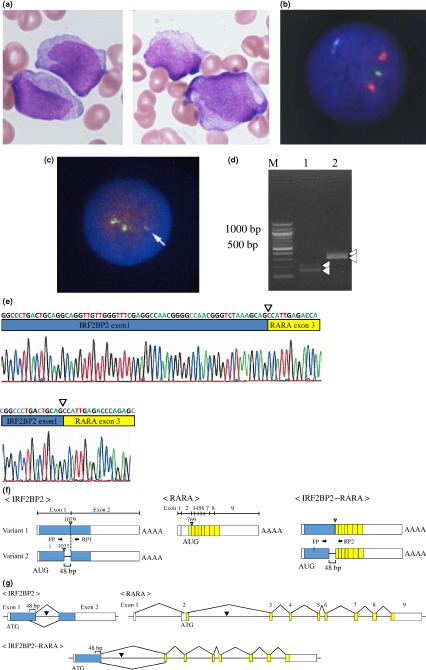
Morphological, cytogenetic, and molecular features of a bone marrow sample from a 68‐year‐old Japanese woman with acute promyelocytic leukemia. (a) May–Giemsa staining. Promyelocytes with azurophilic granules and Auer rods are observed. (b) Interphase FISH using promyelocytic leukemia (PML) (15q22; green) and retinoic acid receptor α (RARA) (17q21; red) probes did not reveal a PML–RARA fusion signal (yellow). (c) Interphase FISH assay to detect RARA split signal (using 3′‐RARA [green] and 5′‐RARA [red] probes) showed no split signals but rather a small yellow signal (46/100; arrow). (d) mRNA expression of interferon regulatory factor 2 binding protein 2 (IRF2BP2) and IRF2BP2–RARA. Lane 1 shows the result of using two complementary oligonucleotide primers, FP1 and RP1. The upper detected band was predicted to be 206 bp and the lower band was predicted to be 158 bp; these bands were supposed to correspond to the expression of normal splice variants of IRF2BP2 in the patient's bone marrow. Lane 2 shows the result of FP1 and RP2. The upper detected band was predicted to be 398 bp and the lower band was predicted to be 350 bp; these bands were supposed to correspond to the expression of the fusion variants of IRF2BP2–RARA. (e) Direct sequencing of the PCR products. The sequences of each PCR product obtained in (d) were determined. The results confirmed that this patient expressed two variants of IRF2BP2 (data not shown), as well as the predicted fusion mRNAs of IRF2BP2–RARA; the upper and lower panels show the sequences of the fusion genes from IRF2BP2 variants 1 and 2, respectively. (f) Schematic illustrations of the transcripts. The blue and yellow solid bars represent the coding regions of the IRF2BP2 and RARA genes, respectively. IRF2BP2 generates the transcript variants through alternative splicing. Variant 1 uses an alternate in‐frame splice junction in the 3′‐ coding region of exon 1, resulting in an mRNA transcript 48 bp longer than variant 2. The inverted triangles represent each predicted fusion point on the transcript. The IRF2BP2–RARA fusion is expected to generate two types of fusion mRNA depending on the IRF2BP2 variants. In the longer type, base 1079 of the IRF2BP2 mRNA is likely to be fused to base 769 of the RARA mRNA. In the shorter type, the base 1031 of the IRF2BP2 mRNA is likely to be fused to base 769 of the RARA mRNA. (g) Overviews of genomic DNA of the IRF2BP2,RARA, and predicted IRF2BP2–RARA fusion genes.
After obtaining written informed consent and institutional review board approval, paired‐end mRNA sequencing was carried out to identify the fusion gene partner of RARA. Consequently, we identified a chromosomal translocation between chr.1q42.3 and 17q21.2. This translocation gives rise to a putative fusion protein comprising exon 1 of IRF2BP2 and exons 3–9 of RARA. IRF2BP2 comprises two exons with an intron, and there are two splicing sites within IRF2BP2 exon 1. Accordingly, we generated primer pairs to amplify the cDNAs of IRF2BP2 (FP and RP1) and IRF2BP2–RARA (FP and RP2). This primer information is shown in Appendix S1. As predicted, two fragments were amplified by RT‐PCR for both IRF2BP2 and IRF2BP2–RARA (Fig. 1d). The PCR product sequences corresponded well to the splicing sites (at the 1031 and 1079 bp positions of IRF2BP2 cDNA) (Fig. 1e). The RARA transcript started at the beginning of exon 3 (at 769 bp of RARA cDNA). Thus, our additional mRNA analysis clearly indicated that the IRF2BP2–RARA fusion proteins showed variant forms that were dependent on IRF2BP2 splicing. The transcript structures for IRF2BP2 and RARA, as well as IRF2BP2–RARA, are shown in Figure 1f. Taken together, the IRF2BP2–RARA gene is likely to be positioned between intron 1 of IRF2BP2 and intron 2 of RARA (Fig. 1g).
Discussion
To our knowledge, this is the second report of a novel APL variant with IRF2BP2‐RARA; this case uniquely showed atypical morphological and clinical features with no DIC detected at admission.
Flow cytometry is a powerful ancillary tool for diagnosing APL. Classic APL cells with PML–RARA typically express mature myeloid markers, including CD13 and CD33, and are negative for CD34 and HLA‐DR. Moreover, several reports have shown expression of CD34 and/or HLA‐DR in a significant proportion of newly diagnosed APL patients.11, 12, 13 In our case, the APL cells were positive for CD33, CD34, CD64, CD117, and HLA‐DR, and negative for CD2, CD7, CD13, and CD19.
Fluorescence in situ hybridization with dual fusion translocation probes is useful for detecting unknown rearrangements of the RARA gene.14 Herein, we failed to detect PML–RARA fusion; however, use of a RARA dual fusion translocation probe allowed us to detect a novel rearrangement of the RARA gene. Cases of APL with cryptic PML–RARA rearrangement that were negative by FISH and conventional karyotyping but positive by RT‐PCR have been reported;15 these findings were caused by small cryptic insertions. Abnormal FISH and normal karyotyping also suggested small cryptic insertions in our case. A normal yellow signal indicated normal 17q while another indicated a small deletion in 17q; furthermore, an abnormal small yellow signal indicated a small insertion of RARA (Fig. 1).
The IRF2BP2–CDX‐1 fusion gene has been reported in mesenchymal chondrosarcoma,16 and there is one previous report of IRF2BP2–RARA in APL.10 Yin et al.10 reported a case of APL with IRF2BP2–RARA that involved exon 2 of IRF2BP2 and intron 2 of RARA, with breakpoints at positions 1687 bp in IRF2BP2 and 41620 bp in RARA. The patient achieved CR with ATRA, arsenic trioxide, and GO; however, relapse occurred 2 months after consolidation therapy. Herein, IRF2BP2 showed distinct breakpoints, and two subtypes of IRF2BP2–RARA were noted. Our case differed in its atypical morphology and clinical course, as well as the lack of DIC on admission. Tissue factors and cancer procoagulant expressed in APL cells are associated with DIC; the lack of DIC in our case may be explained by the lower number of APL cells compared to that in the previously reported patient. Most patients with APL can achieve CR with ATRA with or without anthracycline, resulting in favorable clinical outcomes.17, 18, 19 In the previous IRF2BP2–RARA case, clinical sensitivity to ATRA was not properly evaluated because of combination therapy with arsenic trioxide and GO. Our patient did not respond to ATRA monotherapy. In the presence of pharmacological concentrations of ATRA, transcriptional activation can be induced by dissociation of the co‐repressor complex from PML–RARA as well as proteasome‐dependent PML–RARA degradation. In cases with ligand binding domain mutations in the RARA gene, ATRA fails to recognize RARA, and co‐repressor dissociation does not occur, resulting in ATRA resistance.14 Other fusion genes such as PLZF–RARA 5 and STAT5b–RARA 20 also show ATRA resistance. The Nuclear receptor corepressor (N‐CoR)/Silencing mediator for retinoid and thyroid hormone receptors (SMRT) co‐repressor complex interaction with PLZF and STAT5b translocation is thought to be involved in this resistance. IRF2BP2 also interacts with N‐CoR co‐repressor proteins;21 we speculated that IRF2BP2–RARA is involved in ATRA resistance because of this.
In conclusion, we described a case of APL with IRF2BP2–RARA involving exon 1B of IRF2BP2. The patient had some peculiar clinical characteristics, such as no coagulopathy, a unique cell surface phenotype, and ATRA resistance. Although APL with IRF2BP2–RARA is rarely observed, its unique characteristics have clinical implications for the diagnosis and treatment of APL patients.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Appendix S1 Primer information for amplification of cDNAs of IRF2BP2 (FP and RP1) and IRF2BP2–RARA (FP and RP2).
Acknowledgments
The authors thank the medical and nursing staff at Otemae Hospital.
Cancer Sci 107 (2016) 1165–1168
Funding Information
Takeda Pharmaceutical Co., Ltd.; Sumitomo Dainippon Pharma; Asahi Kasei Pharma; Shionogi Co., Ltd.; Yakult Pharmaceutical Industry Co., Ltd.
References
- 1. de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature 1990; 347: 558–61. [DOI] [PubMed] [Google Scholar]
- 2. Chen Z, Brand NJ, Chen A et al Fusion between a novel Kruppel‐like zinc finger gene and the retinoic acid receptor‐alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J 1993; 12: 1161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin‐retinoic acid receptor fusion. Blood 1996; 87: 882–6. [PubMed] [Google Scholar]
- 4. Wells RA, Catzavelos C, Kamel‐Reid S. Fusion of retinoic acid receptor alpha to NuMA, the nuclear mitotic apparatus protein, by a variant translocation in acute promyelocytic leukaemia. Nat Genet 1997; 17: 109–13. [DOI] [PubMed] [Google Scholar]
- 5. Arnould C, Philippe C, Bourdon V, Grégoire MJ, Berger R, Jonveaux P. The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic‐like leukaemia. Hum Mol Genet 1999; 8: 1741–9. [DOI] [PubMed] [Google Scholar]
- 6. Catalano A, Dawson MA, Somana K et al The PRKAR1A gene is fused to RARA in a new variant acute promyelocytic leukemia. Blood 2007; 110: 4073–6. [DOI] [PubMed] [Google Scholar]
- 7. Kondo T, Mori A, Darmanin S, Hashino S, Tanaka J, Asaka M. The seventh pathogenic fusion gene FIP1L1‐RARA was isolated from a t(4;17)‐positive acute promyelocytic leukemia. Haematologica 2008; 93: 1414–6. [DOI] [PubMed] [Google Scholar]
- 8. Yamamoto Y, Tsuzuki S, Tsuzuki M, Handa K, Inaguma Y, Emi N. BCOR as a novel fusion partner of retinoic acid receptor alpha in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood 2010; 116: 4274–83. [DOI] [PubMed] [Google Scholar]
- 9. Won D, Shin SY, Park CJ et al OBFC2A/RARA: a novel fusion gene in variant acute promyelocytic leukemia. Blood 2013; 121: 1432–5. [DOI] [PubMed] [Google Scholar]
- 10. Yin CC, Jain N, Mehrotra M et al Identification of a novel fusion gene, IRF2BP2‐RARA, in acute promyelocytic leukemia. J Natl Compr Canc Netw 2015; 13: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stasi R, Bruno A, Venditti A et al A microgranular variant of acute promyelocytic leukemia with atypical morpho‐cytochemical features and an early myeloid immunophenotype. Leuk Res 1997; 21: 575–80. [DOI] [PubMed] [Google Scholar]
- 12. Thalhammer‐Scherrer R, Mitterbauer G, Simonitsch I et al The immunophenotype of 325 adult acute leukemias: relationship to morphologic and molecular classification and proposal for a minimal screening program highly predictive for lineage discrimination. Am J Clin Pathol 2002; 117: 380–9. [DOI] [PubMed] [Google Scholar]
- 13. Lin P, Hao S, Medeiros LJ et al Expression of CD2 in acute promyelocytic leukemia correlates with short form of PML‐RARalpha transcripts and poorer prognosis. Am J Clin Pathol 2004; 121: 402–7. [DOI] [PubMed] [Google Scholar]
- 14. Goto E, Tomita A, Hayakawa F, Atsumi A, Kiyoi H, Naoe T. Missense mutations in PML‐RARA are critical for the lack of responsiveness to arsenic trioxide treatment. Blood 2011; 118: 1600–9. [DOI] [PubMed] [Google Scholar]
- 15. Kim MJ, Cho SY, Kim MH et al FISH‐negative cryptic PML‐RARA rearrangement detected by long‐distance polymerase chain reaction and sequencing analyses: a case study and review of the literature. Cancer Genet Cytogenet 2010; 203: 278–83. [DOI] [PubMed] [Google Scholar]
- 16. Nyquist KB, Panagopoulos I, Thorsen J et al Whole‐transcriptome sequencing identifies novel IRF2BP2‐CDX1 fusion gene brought about by translocation t(1;5)(q42;q32) in mesenchymal chondrosarcoma. PLoS ONE 2012; 7: e49705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frankel SR, Eardley A, Lauwers G, Weiss M, Warrell RP Jr. The “retinoic acid syndrome” in acute promyelocytic leukemia. Ann Intern Med 1992; 117: 292–6. [DOI] [PubMed] [Google Scholar]
- 18. Lo CF, Nervi C, Avvisati G, Mandelli F. Acute promyelocytic leukemia: a curable disease. Leukemia 1998; 12: 1866–80. [DOI] [PubMed] [Google Scholar]
- 19. Tallman MS, Nabhan C, Feusner JH, Rowe JM. Acute promyelocytic leukemia: evolving therapeutic strategies. Blood 2002; 99: 759–67. [DOI] [PubMed] [Google Scholar]
- 20. Chen Z, Guidez F, Rousselot P et al PLZF‐RAR alpha fusion proteins generated from the variant t(11;17)(q23;q21) translocation in acute promyelocytic leukemia inhibit ligand‐dependent transactivation of wild‐type retinoic acid receptors. Proc Natl Acad Sci USA 1994; 91: 1178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stadhouders R, Cico A, Stephen T et al Control of developmentally primed erythroid genes by combinatorial co‐repressor actions. Nat Commun 2015; 6: 8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Primer information for amplification of cDNAs of IRF2BP2 (FP and RP1) and IRF2BP2–RARA (FP and RP2).


