Abstract
There is a critical need for monitoring physiologically relevant, sustainable, human adipose tissues in vitro to gain new insights into metabolic diseases. To support long term culture, a 3D silk scaffold assisted culture system was developed that maintained mature unilocular adipocytes ex vivo in co-culture with pre-adipocytes, endothelial cells, and smooth muscle cells obtained from small volumes of liquefied adipose samples. Without the silk scaffold, adipose tissue explants could not be sustained in long term culture (3 months) due to their fragility. Adjustments to media components were used to tune lipid metabolism and proliferation, in addition to responsiveness to an inflammatory stimulus. Interestingly, patient specific responses to TNFα stimulation were observed, providing a proof of concept translational technique for patient specific disease modeling in the future. In summary, our novel 3D scaffold assisted approach is required for establishing physiologically relevant, sustainable, human adipose tissue systems from small volumes of lipoaspirate, making this methodology of great value to studies of metabolism, adipokine-driven diseases, and other diseases where the roles of adipocytes are only now becoming uncovered.
Keywords: Adipose, tissue engineering, explant, unilocular morphology, patient specific approach
2. Introduction
White adipose tissues are endocrine organs that secrete adipokines[1] and store excess energy in unilocular lipid-filled vacuoles of adipocytes.[2] Lipid metabolism includes storing triglycerides and lipolysis, where the triglycerides are broken down into glycerol and free fatty acids to meet energy needs.[3] In healthy individuals there is homeostasis between the release of fatty acids from adipose tissues into the circulation and the uptake and oxidation in peripheral tissues. However, in obese individuals energy intake exceeds the storage capacity of adipose tissue, triggering an inflammatory response (as the tissue expands), additional lipolysis, and altered concentrations of fatty acids and glucose in the circulation.[4] Obesity predisposes individuals to complex metabolic disorders including dyslipidemia and type 2 diabetes.[5] As the prevalence of obesity reaches epidemic proportions in many countries, there is a critical need for physiologically relevant, sustainable, human adipose tissue engineered models to generate new insights into disease initiation and progression and to test potential treatment options.
Current in vitro models of white adipose depots fail to recapitulate the complex human tissue. The models utilize animal cells,[6-8] cell suspension cultures,[9] ceiling cultures,[2, 10-12] or differentiation of human adipose derived stem cells.[13, 14, 15] Animal cell lines (including murine 3T3-L1 and 3T3-F442A) undergo adipogenesis during in vitro culture, however they lack the single large lipid droplet characteristic of mature in vivo adipocytes.[6] Additionally murine differentiated cells secrete only 1-2% of the leptin secreted by primary mature adipocytes[7] and often do not translate well to human adipocyte function.[16] Primary cell suspension cultures consist of adipocytes floating in medium, and hence the cells are not equally exposed to treatments of interest. Moreover, this culture method is only a short term solution, as the floating cells are not exposed to adequate nutrition and lyse within 72 hours of incubation.[10] Ceiling cultures also take advantage of the buoyancy of adipocytes: a culture flask is filled completely with medium to allow the adipocytes to adhere to the top surface of the flask. Although the adipocytes proliferate and exhibit some adipocyte functions such as lipogenesis (accumulation of multilocular lipid droplets) and lipolysis, they display a fibroblast-like phenotype rather than the round unilocular phenotype typical of adipocytes.[2, 12] Finally, the most common method of generating white adipose tissue involves differentiating human adipose-derived stem cells in two dimensional or three dimensional (3D) systems. To enhance physiological relevance, endothelial cells have been incorporated with differentiated stem cells,[15, 17, 18] demonstrating improved adipogenic outcomes. However, these methods require lengthy culture times to differentiate the stem cells into adipocytes, and in the absence of perfusion, contain multilocular lipid droplets.[19] Therefore, better in vitro models are required that incorporate the physiologically relevant mature unilocular adipocytes.
Ex vivo culture of unilocular adipocytes is challenging, as lipid laden mature cells are fragile, highly buoyant, and prone to dedifferentiation in culture. Maintenance of cells in their native tissue matrix (explanted tissue) would be ideal to maintain their 3D morphology in vitro; however, explants are delicate and lack the structural integrity required for extended culture periods (>14 days). Culturing tissue fragments in collagen gels has been proposed as a solution to the fragile and buoyant nature of adipocytes/explants[20] for establishing human ex vivo adipose tissue models. However, in collagen gels, preadipocytes only develop actively at the periphery of the adipose tissue fragments. It was hypothesized that a silk scaffold would provide a 3D framework that would capture and maintain mature unilocular adipocytes ex vivo while having the structural integrity required for long term culture. Silk is a naturally occurring and clinically accepted biocompatible protein material that has tunable mechanical strength, low inflammatory and immunogenic responses, an absence of cell-specific signaling domains, and can be tailored to degrade slowly for long term culture.[21] Furthermore, silk has demonstrated compatibility with adipose tissue engineering applications.[17, 22, 23] Liquefied adipose tissue was used in this study in combination with silk scaffolds as this heterogeneous mixture contained the relevant cell types required to create a physiologically relevant adipose system (Figure 1) and could penetrate into the scaffold farther than larger tissue fragments used in studies with collagen.[20] In addition, for implanting in vivo, collagen has been shown to rapidly degrade while silk provides longer-term structural integrity to promote the maintenance of soft tissue.[24] Moreover, an important aspect of this method is the applicability to patient-specific in vitro systems. To create a patient specific model of an adipose depot, a small volume of liquefied adipose tissue would be obtained by non-invasive lipoaspirate procedures and cultured within the silk scaffolding material. The adipose depot could then be used to explore treatment options (including high throughput screens of multiple drug targets as well as optimizing the doses), adverse effects to drug treatments (including side effects and toxicity), disease mechanisms (including type II diabetes and cancers that affect the adipose tissue) and other metabolic parameters that vary between patients.
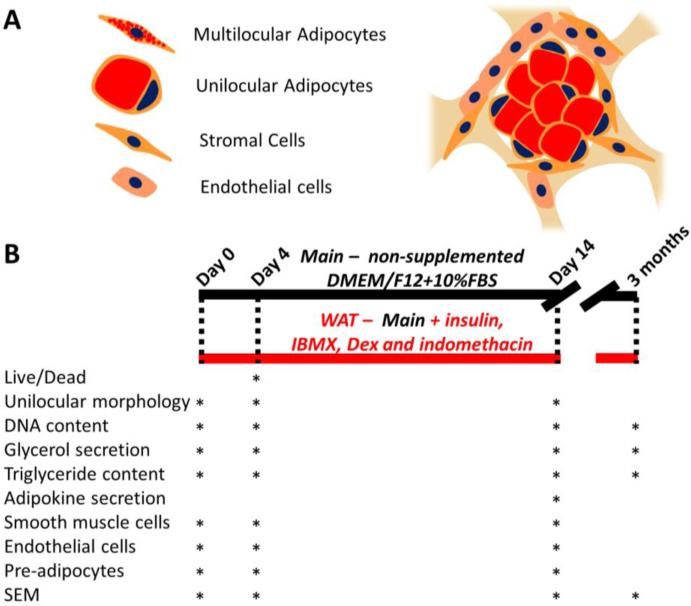
Figure 1. Schematic of cells in the adipose tissue model and culture timeline.
Adipose tissue containing unilocular adipocytes (as opposed to multilocular immature adipocytes), stromal cells (pre-adipocytes, fibroblasts, smooth muscle pericytes), and endothelial cells was liquefied and seeded into porous silk scaffolds (A) in different media conditions (B). The maintenance media group, referred throughout as Main, was a minimally supplemented media (DMEM/F12 and 10% FBS), while the white adipose tissue stimulation media group, referred throughout as WAT, was created by supplementing the maintenance media with insulin, IBMX, dexamethasone and indomethacin. The different assays that were performed at each time point are shown.
3. Results and Discussion
3.1 Viability and distribution of cells in the scaffolds
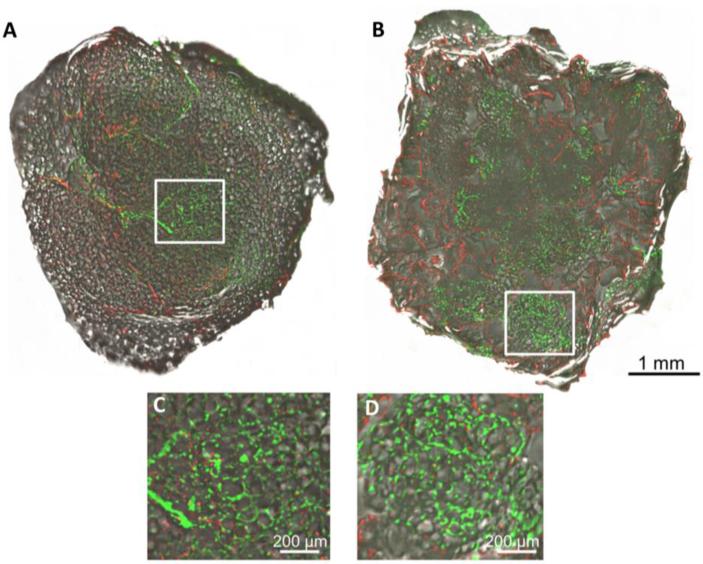
Since adipose tissue explants rapidly deteriorate in culture, the goal of this work was to determine whether a white adipose tissue model could be maintained ex vivo in silk scaffold cultures. Liquified adipose tissue was chosen to improve penetration in the scaffolds. In addition, lipoaspirates are an ideal source of obtaining adipose tissue minimally invasively. To validate that cells remained viable through the seeding process, calcein and ethidium staining was used to visualize the distribution of cells throughout the cultures. Live cells were dispersed throughout the explants and scaffolds 4 days after seeding (Figure 2) demonstrating a safe and effective isolation, seeding, and culturing process for both culture techniques. Pockets of lipid laden adipocyte cell clusters were also evident in the pores of the scaffolds, demonstrating that the cells were viable, distributed throughout the constructs, and may have maintained their in vivo unilocular morphology. Since current models contain cells with a multilocular morphology and not the characteristic unilocular phenotype observed in vivo,[2, 12] maintenance of a unilocular morphology would indicate the model maintained physiologically relevant characteristics of an adipose depot.
Figure 2. Distribution of cells in explant and scaffold cultures.
Calcein and ethidium staining of an explant (A, C) and scaffold (B,D) demonstrating the distribution of live (green) and dead (red) cells, respectively, throughout the explants and scaffolds after 4 days of culture (silk is also evident in the red channel, but to a lesser degree). Magnified insets (C,D) are expanded from the square white region in the lower magnification images (A,B).
3.2 Determination of cell types present
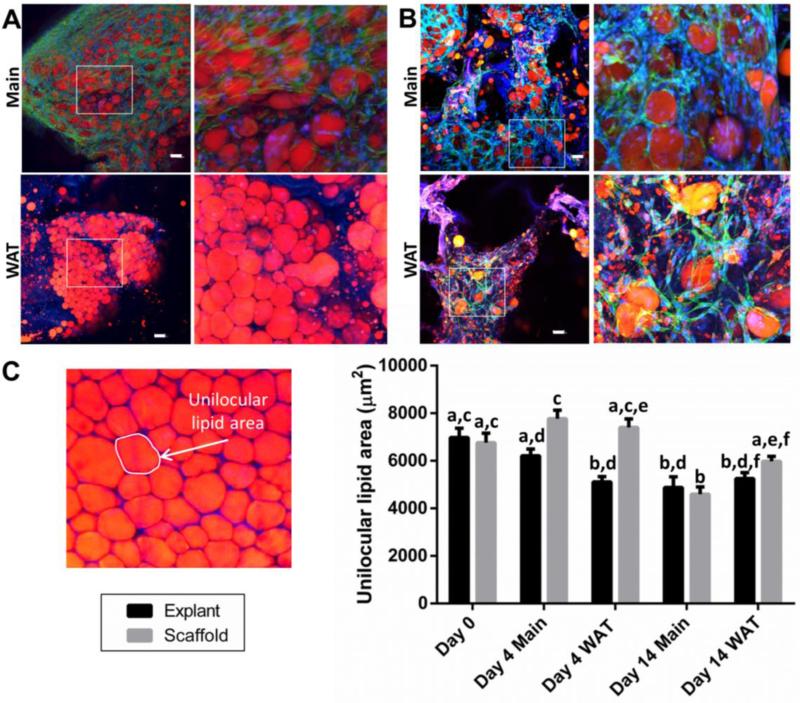
To further test if the unilocular lipid morphology was maintained, lipophilic AdipoRed staining was performed. AdipoRed staining demonstrated that mature adipocytes were maintained in both explant and scaffold cultures under both media conditions tested (Figure 3). The two media conditions were maintenance (Main) media (DMEM/F12, 10% fetal bovine serum) and white adipose tissue (WAT) stimulation media (DMEM/F12, 10% fetal bovine serum, 1X antibiotic-antimycotic, 1 μM insulin, 0.5 mM IBMX, 1 μM dexamethasone, 0.05 mM indomethacin). Non-supplemented maintenance media (Main) demonstrated greater numbers of cells while stimulatory supplemented media (WAT) showed smaller lipid droplets accumulating in both explant and scaffold cultures. To quantify whether the larger unilocular lipid droplet size was maintained ex vivo, lipid areas were traced and are shown for one patient (Figure 3C). After 2 weeks, there were no differences between scaffold and explant unilocular lipid droplet areas. While there was a significant decrease in unilocular lipid droplet area from day 0 for the explants, scaffold cultures had similar areas as the values measured directly at the time of seeding (day 0). Therefore, this experiment confirmed that the scaffold pores were filled with mature unilocular adipocytes, as well as other populations of cells indicated by phalloidin staining in the absence of AdipoRed staining.
Figure 3. The unilocular morphology is maintained in explant and scaffold cultures.
Adipose tissue was either cultured directly as explants (A) or liquefied and seeded into scaffolds (B) and stained with a lipophilic dye (AdipoRed, red = lipids), DAPI (blue = nuclei) and phalloidin (green = actin cytoskeleton) at 14 days (silk fluorescence is visible in the blue and red wavelengths and therefore is purple). Non-supplemented media (Main) demonstrated greater numbers of cells in both explant and scaffold cultures while white adipose tissue stimulation media (WAT = Main + insulin, IBMX, dexamethasone and indomethacin) showed smaller lipid droplets accumulating in both culture conditions. Quantification of AdipoRed unilocular morphology (C) demonstrated no significant differences in sizes of cells at 14 days of culture for explants (black) and scaffolds (grey). An example unilocular lipid area from an explant culture is shown for reference. Scale bars are 100 μm in length. Groups with different letters are significantly different (two way ANOVA significant effects of: culture condition, media supplementation and the two factors interacting, p<0.001). Error bars represent standard error of the mean. 3 different patient samples were stained, imaged, and measured with at least 50 cells measured for each condition, 1 patient is shown for clarity as each patient had different lipid sizes. See Figure S1 for positive staining for smooth muscle cells, endothelial cells, and preadipocytes.
To identify what other cell types remained in the cultures (the phalloidin positive cells), further imaging was performed. Multiple cell types are residents of adipose tissue, including adipocytes, preadipocytes, endothelial cells, fibroblasts, vascular smooth muscle cells and immune cells[25] therefore markers of preadipocytes, smooth muscle cell lineages, and endothelial cells were immunostained with preadipocyte factor 1 (PREF1),[26] alpha smooth muscle actin (αSMA),[27] and cluster of differentiation 31 (CD31),[28] respectively. It was thought that different media conditions might not support all three cell types; however, positive staining was present under all of the conditions for all three cell types (Figure S1). Since culturing multiple cell types improves the accuracy of tissue engineered systems,[29] this model system will likely have improved physiological relevance over other adipose tissue engineered systems that incorporate only one[13, 15] or two[15, 17, 23] cell types.
3.3 Proliferation and lipid metabolism
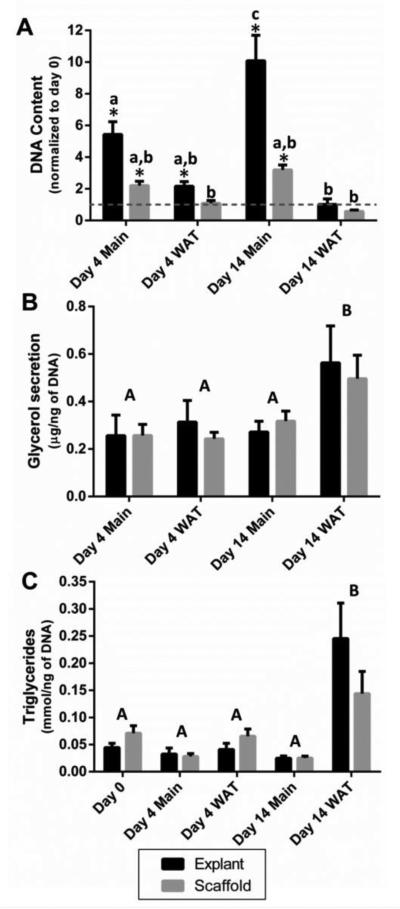
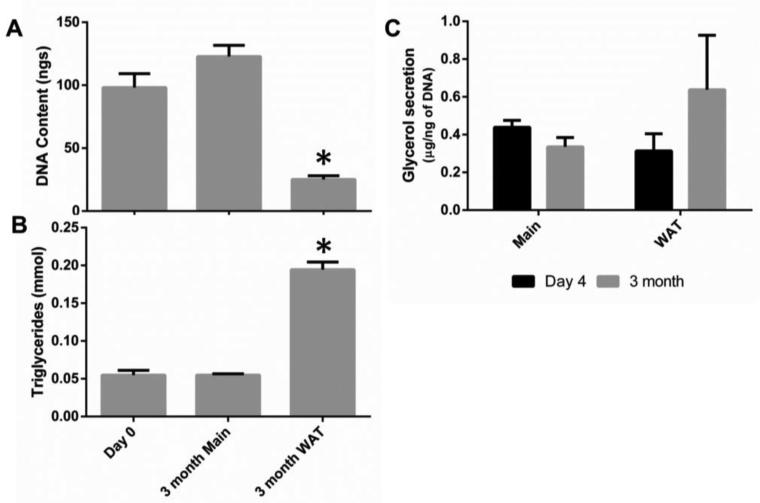
Adipocytes store excess energy in unilocular lipid-filled vacuoles of adipocytes.[2] Lipid metabolism includes not only storage of triglycerides, but also lipolysis, where the triglycerides are broken down into glycerol and free fatty acids to meet energy needs.[3] Therefore a hallmark of adipocyte functionality in vitro is quantification of lipid accumulation (triglyceride content) and glycerol secretion.[30] Media composition was an important factor in determining the number of cells, lipogenesis and secretion of glycerol and other proteins. The media without supplements; the Main group, had increased levels of DNA compared to the initial seeding values, suggesting it encouraged proliferation (Figure 4A, consistent with more cells seen in Figure 3). The WAT group on the other hand was stimulated by IBMX, indomethacin, dexamethasone and insulin and stimulated preadipocytes to accumulate lipids, rather than proliferate. Cells cultured in this media demonstrated an increase in triglyceride accumulation (Figure 4C consistent with small lipid droplets observed in Figure 3) and glycerol secretion (Figure 4B).
Figure 4. Media supplementation can be used to enhance triglyceride accumulation and glycerol secretion or withheld to enhance proliferation.
Media conditions significantly (p<0.001) affected the DNA content (A) and the triglyceride content (B). Non-supplemented media (Main) enhanced DNA content, while media supplementation (WAT) enhanced triglyceride accumulation and glycerol secretion (C). DNA content was also dependent on culture condition and was higher in explant cultures compared to scaffold cultures (p<0.001, significant interaction between media and culture condition p<0.001). Triglyceride content, however, was not significantly different between explant and scaffold cultures (p=0.445, interaction between media and culture condition p=0.046). Likewise, after two weeks of culture, glycerol secretion was enhanced with media supplementation (p=0.002) without any differences between explant and scaffold culture conditions (p=0.667, no significant interaction between media and culture condition p=0.846). Groups with different letters are significantly different. A two way ANOVA was used, where lower case letters indicate significant differences between groups when there a significant interaction between the two factors and upper case letters indicate significant differences when there was a factor effect. * indicates significant differences from day 0 (one sample t test) for DNA content. Error bars represent standard error of the mean. Assays were performed with two or more patients with 5 samples from each patient (run in duplicate) for each of the experiments.
3.4 Evaluation of the adipose secretome
White adipose tissue is actively involved in many physiologic and pathologic processes, including immunity, inflammation, and communication with other tissues.[1] Since adipose tissues are endocrine organs that function by secreting cytokines,[1] an adipose specific cytokine array (for a full list of proteins and their abbreviations see Table S1) was used to screen the in vitro cultures for proteins that are known to be secreted in vivo. For a comparison of scaffold secretion levels to explant secretion levels see Figure S2 (data repeated from Figure 5). In general, explant cultures secreted decreased numbers of adipokines compared to scaffold cultures. This suggests that the scaffold-assisted cultures are a more valid model of endocrine function than explant cultures.
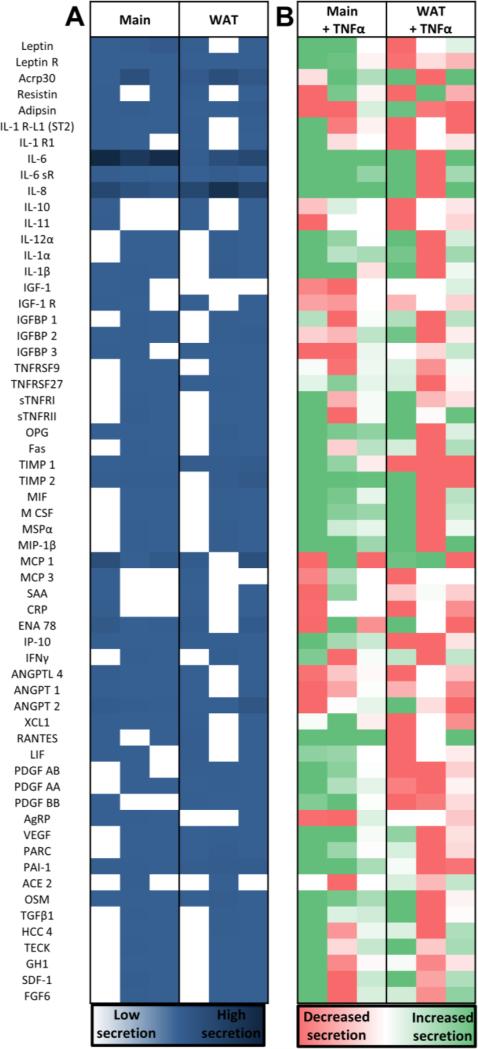
Figure 5. Cells cultured in scaffolds secreted a variety of proteins.
Common adipokines were evaluated for the different culture conditions and media supplementations at 14 days of culture (for full protein names see Table S1). Each column in these heat maps represents the secretion of constructs from 3 different patients normalized by media only values and negative control intensity values (run in duplicate and averaged). The left columns are secretion levels without stimulation (A, navy blue represents high secretion levels) while the right columns represent the change when stimulated with TNFα for 24 hours (B, decreases in secretion levels are red while increases in secretion levels are green). Non-supplemented media (Main) and white adipose tissue stimulation media (WAT = Main + insulin, IBMX, dexamethasone and indomethacin) showed similar trends in secretion values (A), but responded to TNFα differently (B). See Figure S2 for a comparison to explant conditions.
All of the cells seeded in scaffolds secreted important adipokines regardless of media supplementation (Figure 5A) including: leptin R, adiponectin (ACRP30), adipsin (synthesized by adipocytes and circulates in the bloodstream[31]), inflammatory interleukins; including interleukin-6 (IL-6), IL-6 receptor subunit alpha, and interleukin-8 (IL-8), as well as tissue inhibitor of metalloproteinases (TIMP) 1, interferon gamma-induced Protein (IP10, secreted by mature and differentiating adipocytes[32]), angiopoietin (ANGPT) - 2, plasminogen activator inhibitor 1 (PAI-1) and oncostatin-M (OSM). However, some proteins such as resistin and macrophage colony-stimulating factor (M-CSF), varied depending on the patient. While adiponectin and leptin are primarily secreted by adipocytes, the majority of secreted cytokines, including the inflammatory interleukins, are secreted by the other cell types found in adipose tissue.[33, 34] Therefore, patient specific variations in resident percentages of different cell types[34, 35] could account for differences seen in levels of secreted proteins. In fact, those patient specific differences are likely to play an important role in discovering why some obese patients develop type 2 diabetes and others do not.[36]
3.5 Stimulation with an acute inflammatory mediator
An increasing amount of evidence suggests that low-grade chronic inflammation links excess adipose tissue to metabolic disorders.[37] To test out the applicability of this model to respond to an acute inflammatory stimulus the media was supplemented with tumor necrosis factor alpha (TNFα) and the secretion patterns were compared to values without supplementation (Figure 5B). When scaffold cultures in the Main group were treated with TNFα there was an increase in the secretion of proteins that are higher in insulin-resistant obese subjects[38] including: proinflammatory IL-1α, angiogenic osteoprotegerin (OPG) and TIMP2, anti-inflammatory macrophage stimulatory protein alpha (MSPα) and chemoattractant macrophage inflammatory protein 1-beta (MIP-1β). Additionally, increased regulated-on-activation-normal-T-Cell-expressed-and-secreted (RANTES), IL-6 and IL-8 levels were elevated with the acute inflammatory stimulus, which is consistent with elevated levels in type 2 diabetic patients.[39] This model also confirmed that PAI-1 production is correlated with TNFα stimulation, reinforcing the idea that there is a possible local contribution of TNFα in the regulation of PAI-1 production by human adipose tissue.[40] Likewise, TGFβ1 levels were increased after TNFα administration, consistent with findings that TGF-β1 induces PAI-1 expression.[41] Finally, there were elevated levels of OSM in the Main group treated with TNFα, which is a protein secreted by cells in the stromal vascular fraction and stimulates adipocytes to secrete PAI-1 and IL-6.[42] However, the large variability in responses of the three different samples reinforces the need for patient specific models of disease mechanisms.
3.6 Differences in secreted proteins between media conditions
The cytokine array demonstrated that scaffolds in the Main group consistently secreted a number of proteins that were not secreted in all of the samples cultured in the WAT group (Figure 5A). For example, all of the samples in the Main group secreted leptin, a hormone that is higher in plasma and serum levels of obese patients and signals the hypothalamus to decrease food intake and increase energy expenditure.[43] Interestingly, all of the samples in the Main group also secreted Agouti-related protein (AGRP), which is modulated by leptin,[43] and increases food intake and decreases energy expenditure.[44] Pro-inflammatory cytokines were also secreted consistently in this group and not consistently in the WAT group including: interleukin-1 beta (IL-1β), a cytokine that is suspected to mediate the damaging effects of macrophages on insulin signal transduction in adipocytes;[45] monocyte chemoattractant protein 1 (MCP1), a cytokine that regulates macrophage recruitment to sites of inflammation;[46] and the receptor for interleukin-1 receptor-like 1 (IL-1 R-L1, ST2), which is a cytokine mainly expressed by stromal endothelial cells and is associated with the polarization of macrophages in adipose tissues.[47] Angiogenic proteins that are upregulated in obese subjects: OPG, TIMP2 and epithelial-derived neutrophil-activating peptide 78 (ENA78);[38] and insulin-like growth factor-binding protein 2 (IGFBP 2), which is increased in obese children (and associated with increased adiposity and decreasing insulin sensitivity [48]), were also consistently upregulated in this group and not consistently in the WAT group. On the other hand, stimulated cells (WAT) consistently secreted many proteins that weren't secreted in all of the samples cultured in non-supplemented media (Main) including (Figure 5A): insulin-like growth factor-binding protein 3 (IGFBP3), pulmonary and activation-regulated chemokine (PARC), platelet-derived growth factor (PDGF) AB, PDGF AA, PDGF BB, and Vascular endothelial growth factor (VEGF). As opposed to IGFBP-2 (which was always expressed in the Main group), IGFBP-3 is involved in glucose homeostasis and is decreased in obesity.[49] PARC protein levels, however, were also secreted and are upregulated in obese patients.[38] The PDGF polypeptides and VEGF belong to a family of structurally and functionally similar growth factors that function in hematopoietic development, neurogenesis, and neuroprotection.[50] The differences in protein secretion levels between media groups is likely related to different populations of cells proliferating and differentiating.
3.7 Evaluation of long term culture capability
An important criterion for an adipose tissue model is the ability to maintain the model for extended time frames to enable studies associated with chronic disease signaling. Of significance, explants could not be sustained in long term culture (3 months) due to their fragility. Explants broke apart at variable points during culture, with some breaking apart before the 14 day timepoint and others after it. Thus, the last endpoint comparing explant and scaffold cultures was chosen at 14 days (and extra explants were cut to ensure the endpoint had sufficient comparisons to scaffold cultures). Scaffold aided adipose tissues remained intact and were evaluated for DNA content and lipid metabolism at 3 months. Consistent with earlier time points, DNA content varied depending on whether the media was supplemented (Figure 6). Triglyceride content was maintained from day 0 to 3 months in the Main group and increased in the WAT group. Glycerol secretion was also maintained with no significant differences from initial values to values obtained at 3 months. Since DNA content, triglyceride content, and glycerol secretion were all maintained, these results indicate that lipid laden cells can be sustained for long term culture (>3 months) in the silk scaffolds and maintain lipolytic function. In addition, scanning electron microscopy (SEM) analysis demonstrated that the pores were filled with cells and tissue at 3 months (Figure S3). Long term lipid metabolism in vitro makes this technique not only amendable to studying metabolic transitions associated with type II diabetes, but also for studying certain infectious diseases that affect adipose tissue[51] and different cancers affected by obesity;[52] including breast cancer which affects the mammary adipose tissue.[53] All of these diseases are chronic conditions that will likely require long term culturing to manifest accurately. It also has potential as an approach for autologous soft tissue regeneration, especially where adipose tissue grafts and natural biomaterials resorb too quickly to maintain tissue regeneration.[54]
Figure 6. At 3 months glycerol secretion was maintained, while DNA content and triglyceride content depended on media supplementation.
DNA content varied depending on whether the media was supplemented at 3 months (A). There was a significant decrease in DNA content in the supplemented group (WAT) compared to day 0 (p<0.001). However, there was no significant change in DNA content between day 0 and 3 months for scaffolds cultured in non-supplemented media (Main) media (p=0.093). Triglyceride content (B) was maintained from day 0 to 3 months in the Main group (p=0.992), and increased in the WAT group (p<0.001). Additionally, glycerol secretion (C) was maintained with no significant differences from initial values to values obtained at 3 months (p=0.267), with no differences between media conditions (p=0.370). * indicates significant difference from day 0 (unpaired sample t test). Error bars represent standard error of the mean. Assays were performed with two patients with 5 samples from each patient (run in duplicate) for each of the experiments.
3.8 Validating the scaffold assisted method for use with lipoaspirate samples
To validate that this technique could be used with small volumes of lipoaspirate samples, two lipoaspirate patient samples were obtained (Figure S4) and compared to the prior results with blended adipose tissue (results repeated from Figure 4). The lipoaspirate seeded scaffolds showed no differences in the amount of DNA due to the seeding method. Additionally, both seeding methods demonstrated the same trends with media supplementation where supplementing the media (WAT) enhanced triglyceride accumulation per cell and glycerol secretion without affecting the DNA content, while media without supplements (Main) increased DNA content without affecting the lipid metabolism. Similarity in trends between the two seeding methods indicates that this technique is amendable to a patient specific approach where a small volume of lipoaspirate can be obtained from an individual patient.
3.9 Material choice
Silk scaffolds were used in this study since they can be extended for months in culture[18] avoiding premature collapse of structures. Silk scaffolds provide biocompatibility, porous features for transport, robust mechanical properties, and retain size with slow proteolytic biodegradation, and require no chemical or photo crosslinking for stability and function.[55] Matrix molecules such as collagen provide structural support and important matrix-mediated cell signaling, however, these systems degrade too rapidly to serve the functional goals of tissue models[24] for long term models. Crosslinking can be considered, however, cell signaling is impacted and crosslinking agents are often cytotoxic.[56] While silk was an ideal material for providing the structural integrity required to sustain the fragile and buoyant adipose tissue, other materials could be considered that have been used for long term cultures including: poly (e-caprolactone),[57] poly (ethylene terephthalate),[58] polyglycolic acid scaffolds,[59] and poly (lactide-co-glycolide).[60]
4. Conclusion
This study evaluated the potential for maintaining human mature adipocytes in vitro in a non-signaling, cell compatible and mechanically tunable 3D silk scaffold system that could support long term culture. Since liquefied adipose tissue was seeded directly into the scaffolds, it contained the relevant cell types (adipocytes, preadipocytes, endothelial cells, and smooth muscle cells) required to create a physiologically relevant system. Media supplementation was used to tune the lipid metabolism and proliferation of cells in the culture systems. Additionally, the scaffolds remained intact with similar numbers of cells and triglycerides after 3 months of culture, which could not be acheived in explant cultures. Most importantly, soaking silk scaffolds directly from a small volume of liquified adipose tissue makes this technique amendable to the study of patient-specific disease mechanisms and drug responses, where a small volume of lipoaspirate can be obtained from a pateint.
5. Experimental Section
5.1 Materials
Bombyx mori silkworm cocoons were acquired from Tajima Shoji Co (Yokohama, Japan). All cell culture supplies were purchased from Invitrogen (Carlsbad, CA) unless otherwise noted. In addition, calcein, ethidium, phalloidin, 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) and Picogreen kits were obtained from Invitrogen. Human recombinant insulin, 3-isobutyl-1-methylxanthine (IBMX), dexamethasone, indomethacin, bovine serum albumin (BSA), goat serum, and TNFα were obtained from Sigma–Aldrich (St. Louis, MO). AdipoRed was purchased from Lonza (Walkersville, MD). Primary and secondary antibodies were purchased from Sigma-Aldrich and BD Biosciences (San Diego, CA) as noted. Triglyceride and Glycerol quantification kits were obtained from BioAssay Systems (Hayward, CA). The human obesity arrays were obtained from RayBiotech (Norcross, GA). Polyethylene molds (Catalog number: 03-338-1E) were acquired from Fisher Scientific (Waltham, MA).
5.2 Silk scaffold fabrication
To generate silk scaffolds (see Figure S3A for an SEM of a scaffold without cells), silk solution was extracted from B. mori silkworm cocoons as described previously.[61] The solution was lyophilized, re-solubilized in a 17% w/v hexafluoro-2-propanol (HFIP) solution, and poured over 6.8 grams of salt (particles were sieved to only include particles in the range of 500–600 μm therefore creating pores in that range[62]) in a polyethylene vial. The vials were sealed and left in a fume hood for 2 days to ensure even distribution of the HFIP, silk and salt. The vials were opened for 1 day to allow the HFIP to evaporate and were then immersed in methanol overnight. To allow the methanol to evaporate the vials were removed from the methanol and left open in the hood for 1 day. Finally, the vials were immersed in water to leach out the salt particles. After multiple wash steps, the scaffolds were removed from the containers, cut to size (cylinders, 2 mm height × 4 mm diameter), and autoclaved. The scaffolds were then soaked overnight in media (DMEM/F12, 10% fetal bovine serum, 1X Antibiotic-Antimycotic) prior to cell seeding.
5.3 Culturing of explants and adipose soaked scaffolds
On the same day of surgery subcutaneous adipose tissue was acquired from elective abdominoplasty procedures. Five different patient samples were obtained for comparisons between explants and scaffolds (Table S2). Blunt dissection was used to separate the adipose tissue from the skin and the fascia of Scarpa. Explants were cut to the same size as the scaffolds (2 mm height × 4 mm diameter) and cultured in the same media. The remaining adipose tissue was liquefied in a blender by successive short pulses until the tissue had the viscosity of lipoaspirate. Media was then aspirated from the soaking scaffolds and the scaffolds were added directly to the liquefied adipose tissue in 50 mL falcon tubes. The tubes were placed in an incubator for one hour (37°C, 5% CO2). The scaffolds were then separated from the excess tissue and placed into 24 well plates for 2 hours (37°C, 5% CO2) without media to allow the cells to attach to the scaffold. Maintenance media or white adipose tissue (WAT) stimulation media (DMEM/F12, 10% fetal bovine serum, 1X antibiotic-antimycotic, 1 μM insulin, 0.5 mM IBMX, 1 μM dexamethasone, 0.05 mM indomethacin) was then added and changed twice a week. Scaffolds and explants were cultured in different media conditions (Figure 1) referred throughout as Main (maintenance media) or WAT (white adipose tissue stimulation media). Endpoint analyses were performed at day 0, day 4, day 14, and in some cases at 3 months. In an additional set of experiments, scaffolds were soaked in lipoaspirate (Table S2 for patient info) to compare lipoaspirate samples to blended abdominoplasty samples.
5.4 Immunostaining
To visualize the distribution of live and dead cells in the explants and scaffolds, brightfield and fluorescent images were taken on a macroscope (Olympus MVX10) after 4 days of culture. The constructs were stained for 30 minutes at room temperature with 2 μM calcein AM and 4 μM ethidium homodimer-1 prior to imaging.
Cellular morphology in the explants and scaffolds was evaluated in samples that were fixed with 10% neutral buffered formalin for 30 minutes. Samples were washed twice with phosphate buffered saline (PBS) containing 0.2% Triton-X100 and stained for 1 hour with AdipoRed (1:35), DAPI (1:1000), and Alexa flour 488 phalloidin (1:40). Following the staining process, the samples were rinsed with PBS twice. Samples were imaged with either an inverted Leica DMIRE2 confocal microscope or a Leica SP5X Laser Scanning Confocal Microscope at the DFCI Microscopy Core using Leica LAS acquisition software. Scaffolds were imaged with 10x dry or 20x water-immersion objectives using 488nm Argon, 405nm UV diode, or white light lasers (470nm-670nm). PMTs (photomultiplier tubes) collected fluorescence signal from DAPI (405nm/420-440nm), Phalloidin (488nm/500-520nm), AdipoRed (488/564-616nm), and scaffolds (which are naturally-autofluorescent and visible in all of the PMTs). Pseudocolors were assigned to each: DAPI/Blue, Phalloidin/Green, AdipoRed/Red, and Scaffold which appeared as violet/turquoise. Z-stack images were acquired and processed using LeicaLite or LeicaLAS software to create single maximum projection 3D-like images. The final images were analyzed with ImageJ (NIH) to determine unilocular lipid size (3 different patient samples were used and at least 50 cells measured for each condition, 1 patient is shown in Figure 3 for clarity as each patient had different lipid sizes, likely related to patient specific differences in BMI, age and other factors[63]).
Prior to staining for cell type markers, samples were fixed with 10% neutral buffered formalin for 30 minutes at room temperature and blocked in a buffer for an hour (1% goat serum, 0.2% BSA and 0.2% Triton-X100 in PBS). The primary antibodies, human cluster of differentiation 31 (CD31, BD 555444, 1:50) produced in mouse, human α-smooth muscle actin (αSMA, Sigma A2547, 1:400) produced in mouse, and human preadipocyte factor 1 (PREF1, Sigma HPA053879, 4 μg/ml) antibody produced in rabbit, were then applied to the samples in the buffer solution for 1 hour, followed by three 10 minute washes with the buffer. The secondary antibodies; goat anti-mouse (BD555988, 1:200), and goat anti-rabbit (Sigma F0382, 1:80), were diluted in the buffer, and counterstained with DAPI (1:1000) for 1 hour, followed by three 10 minute washes with the buffer. All staining steps were performed at room temperature. Three different patient samples were imaged for each group, with separate samples used for each marker. The samples were imaged with the macroscope (Olympus MVX10) to determine overall distribution of cell types in the explants and scaffolds.
5.5 DNA and Triglyceride Content
Scaffolds and explants were lysed in a Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5), shredded using micro-dissection scissors and stored at −20°C until the assays were performed. After thawing, the samples were immediately assayed according to the manufacturer's protocol for DNA content and triglyceride content with the Picogreen assay and the EnzyChromTM Triglyceride Assay Kit, respectively. Assays were performed with two or more patients with 5 samples from each patient (run in duplicate) for each of the experiments. Triglyceride content was normalized by DNA content to account for potential variations in cell numbers.
5.6 Protein secretion
To quantify secretion of different proteins, media samples with or without stimulation by 10 ng/mL TNFα for 24 hours were stored at −20 °C until the assays were performed. After thawing, the samples were immediately assayed according to the manufacturer's protocol with either the EnzyChromTM Adipolysis Assay Kit or the Human Obesity Array C1. The glycerol assay was performed on two or more patients with 5 samples from each patient (run in duplicate) for each of the experiments and normalized by DNA content.
Human sandwich-based obesity arrays containing 60 different target proteins (for a full list of proteins and abbreviations see Table S1) were run according to the manufacturer's protocol. Briefly, the antibody coated arrays were blocked for 30 minutes, and incubated with 1 mL of the supernatant samples overnight at 4°C on a shaker. Then, the arrays were washed and incubated with 1mL of biotin-conjugated antibodies overnight at 4°C on a shaker. Next, the arrays were washed and incubated with streptavidin-conjugated horseradish peroxidase for 2 hours. Finally, the arrays were washed and the chemiluminescence signal was detected with a Syngene G Box imager. Data was extracted using ImageJ software where images were inverted and analyzed with the built in analysis for gels. Lanes were plotted and averages of peaks were determined for each dot. A media control was subtracted from each array value to account for proteins in the media. Additionally, background was subtracted based on negative control intensity values for each array. On each array the proteins were run in duplicate and were averaged. For comparison between patients, media samples from three patient samples were run for each group at the 14 day time point. Technical replicates within the same patient were all done at the same time from the same tissue sample. Glycerol secretion was normalized by DNA content.
5.7 Scanning Electron Microscopy
Silk scaffolds with and without cells were cross-linked with a 2.5% glutaraldehyde solution and progressively dehydrated in a graded series of ethanols (30%, 50%, 75%, 95% and twice in 100%, 30 minutes at each concentration). The samples were critical point dried with a liquid CO2 dryer (AutoSamdri-815, Tousimis Research Corporation, Rockville, MD). Prior to imaging with a scanning electron microscope (Zeiss UltraPlus SEM or Zeiss Supra 55 VP SEM, Carl Zeiss SMT Incorporation, Peabody, MA) the samples were coated with a thin layer (10 nm thick) of Pt/Pd at a voltage of 2 ~ 3 kV using a sputter coater (208HR, Cressington Scientific Instruments Incorporation, Cranberry Township, PA).
5.8 Statistics
Statistics were performed with GraphPad Prism software (GraphPad, CA, USA). A two way Analysis of Variance (ANOVA) was performed for unilocular lipid size, DNA content, triglyceride content and glycerol secretion, where the factors were culture condition (explant vs scaffold) and media supplementation at different time points (Day 0, Day 4 Main, Day 4 WAT, Day 14 Main, Day 14 WAT). To test significant differences between day 0 and normalized DNA content, a one sample t test for each group was performed with a theoretical mean of 1. Significant differences from seeding values in DNA content, and triglyceride accumulation at 3 months were determined with an unpaired sample t test. Differences in glycerol secretion from initial values and 3 month values were determined with a two way ANOVA. To determine if there were significant differences between lipoaspirate soaked scaffolds and the scaffolds soaked with liquefied adipose tissue a two way ANOVA was used. When there was a significant effect of a factor in the two way ANOVA tests a Tukey post-hoc test was performed between the different groups. When there was a significant interaction between the two factors in the two-way ANOVA all of the groups were compared individually with a Tukey post-hoc test. Significant differences were always defined as p<0.05.
5.9 Study Approval
Human adipose tissue samples were obtained with institutional review board approval (Protocol #0906007). Informed signed consent was obtained from either the patient or from the next of kin.
Supplementary Material
Acknowledgements
The authors wish to thank Dr. Sonal Pandya for providing the surgical specimens and Dr. Kelly Burke for helpful discussions. The authors would also like to thank Dr. Irene Georgakoudi for the use of her confocal microscope. This work was funded by the NIH Tissue Engineering Resource Center (P41 EB002520), AFIRM (W81XWH-08-2-0032), and the Department of Defense Grant W81XWH-05-1-0390.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Fantuzzi G. The Journal of allergy and clinical immunology. 2005;115:911. doi: 10.1016/j.jaci.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 2.Sugihara H, Yonemitsu N, Miyabara S, Toda S. Journal of lipid research. 1987;28:1038. [PubMed] [Google Scholar]
- 3.Jensen MD. The Journal of clinical endocrinology and metabolism. 2008;93:S57. doi: 10.1210/jc.2008-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shulman GI. The Journal of clinical investigation. 2000;106:171. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]; Danforth E., Jr. Nature genetics. 2000;26:13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]; Unger RH. Endocrinology. 2003;144:5159. doi: 10.1210/en.2003-0870. [DOI] [PubMed] [Google Scholar]
- 5.Eyre H, Kahn R, Robertson RM, Clark NG, Doyle C, Hong Y, Gansler T, Glynn T, Smith RA, Taubert K, Thun MJ. Stroke; a journal of cerebral circulation. 2004;35:1999. doi: 10.1161/01.CIR.0000133321.00456.00. [DOI] [PubMed] [Google Scholar]
- 6.Serlachius M, Andersson LC. Experimental cell research. 2004;296:256. doi: 10.1016/j.yexcr.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 7.MacDougald OA, Hwang CS, Fan H, Lane MD. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9034. doi: 10.1073/pnas.92.20.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Z, Dong Z, Chang Q, Zhan W, Zeng Z, Zhang S, Lu F. Tissue engineering. Part C, Methods. 2014;20:875. doi: 10.1089/ten.tec.2013.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang KH, Liao HT, Chen JP. Acta biomaterialia. 2013;9:9012. doi: 10.1016/j.actbio.2013.06.046. [DOI] [PubMed] [Google Scholar]; Liu W, Yang X, Yan X, Cui J, Liu W, Sun M, Rao Y, Chen F. Stem cells international. 2014;2014:423635. doi: 10.1155/2014/423635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YH, Wu JY, Chou PJ, Chen CH, Wang CZ, Ho ML, Chang JK, Yeh ML, Chen CH. Cytotherapy. 2014;16:485. doi: 10.1016/j.jcyt.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Zhang HH, Kumar S, Barnett AH, Eggo MC. The Journal of endocrinology. 2000;164:119. doi: 10.1677/joe.0.1640119. [DOI] [PubMed] [Google Scholar]
- 11.Shen JF, Sugawara A, Yamashita J, Ogura H, Sato S. International journal of oral science. 2011;3:117. doi: 10.4248/IJOS11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugihara H, Yonemitsu N, Miyabara S, Yun K. Differentiation; research in biological diversity. 1986;31:42. doi: 10.1111/j.1432-0436.1986.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 13.Jaikumar D, Sajesh KM, Soumya S, Nimal TR, Chennazhi KP, Nair SV, Jayakumar R. International journal of biological macromolecules. 2014 doi: 10.1016/j.ijbiomac.2014.12.037. [DOI] [PubMed] [Google Scholar]; Wu I, Nahas Z, Kimmerling KA, Rosson GD, Elisseeff JH. Plastic and reconstructive surgery. 2012;129:1247. doi: 10.1097/PRS.0b013e31824ec3dc. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cheung HK, Han TT, Marecak DM, Watkins JF, Amsden BG, Flynn LE. Biomaterials. 2014;35:1914. doi: 10.1016/j.biomaterials.2013.11.067. [DOI] [PubMed] [Google Scholar]; Wittmann K, Storck K, Muhr C, Mayer H, Regn S, Staudenmaier R, Wiese H, Maier G, Bauer-Kreisel P, Blunk T. Journal of tissue engineering and regenerative medicine. 2013 doi: 10.1002/term.1830. [DOI] [PubMed] [Google Scholar]; Korurer E, Kenar H, Doger E, Karaoz E. Journal of biomedical materials research. Part A. 2014;102:2220. doi: 10.1002/jbm.a.34901. [DOI] [PubMed] [Google Scholar]; Wang L, Johnson JA, Zhang Q, Beahm EK. Acta biomaterialia. 2013;9:8921. doi: 10.1016/j.actbio.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; Strassburg S, Nienhueser H, Bjorn Stark G, Finkenzeller G, Torio-Padron N. Journal of tissue engineering and regenerative medicine. 2013 doi: 10.1002/term.1769. [DOI] [PubMed] [Google Scholar]; Li SL, Liu Y, Hui L. Journal of tissue engineering and regenerative medicine. 2013 doi: 10.1002/term.1695. [DOI] [PubMed] [Google Scholar]
- 14.Luo B, Choong C. Journal of biomaterials applications. 2015;29:903. doi: 10.1177/0885328214548881. [DOI] [PubMed] [Google Scholar]; Fan X, Tian C, Fu Y, Li X, Deng L, Lu Q. Zhongguo xiu fu chong jian wai ke za zhi = Zhongguo xiufu chongjian waike zazhi = Chinese journal of reparative and reconstructive surgery. 2014;28:377. [PubMed] [Google Scholar]
- 15.Haug V, Torio-Padron N, Stark GB, Finkenzeller G, Strassburg S. Microvascular research. 2015;97:159. doi: 10.1016/j.mvr.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Bouillon R, Carmeliet G, Lieben L, Watanabe M, Perino A, Auwerx J, Schoonjans K, Verstuyf A. Nature reviews. Endocrinology. 2014;10:79. doi: 10.1038/nrendo.2013.226. [DOI] [PubMed] [Google Scholar]
- 17.Kang JH, Gimble JM, Kaplan DL. Tissue engineering. Part A. 2009;15:2227. doi: 10.1089/ten.tea.2008.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choi JH, Gimble JM, Vunjak-Novakovic G, Kaplan DL. Tissue engineering. Part C, Methods. 2010;16:1157. doi: 10.1089/ten.tec.2009.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellas E, Marra K, Kaplan DLP. Tissue engineering. Part C, Methods. 2013;19:745. doi: 10.1089/ten.tec.2012.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach JC, Lin YC, Brayfield CA, Minteer DM, Li H, Rubin JP, Marra KG. Tissue engineering. Part C, Methods. 2012;18:54. doi: 10.1089/ten.tec.2011.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toda S, Uchihashi K, Aoki S, Sonoda E, Yamasaki F, Piao M, Ootani A, Yonemitsu N, Sugihara H. Organogenesis. 2009;5:50. doi: 10.4161/org.5.2.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sugihara H, Yonemitsu N, Toda S, Miyabara S, Funatsumaru S, Matsumoto T. Journal of lipid research. 1988;29:691. [PubMed] [Google Scholar]
- 21.Wang Y, Kim HJ, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2006;27:6064. doi: 10.1016/j.biomaterials.2006.07.008. [DOI] [PubMed] [Google Scholar]; Altman GH, Horan RL, Lu HH, Moreau J, Martin I, Richmond JC, Kaplan DL. Biomaterials. 2002;23:4131. doi: 10.1016/s0142-9612(02)00156-4. [DOI] [PubMed] [Google Scholar]; Meinel L, Hofmann S, Karageorgiou V, Kirker-Head C, McCool J, Gronowicz G, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2005;26:147. doi: 10.1016/j.biomaterials.2004.02.047. [DOI] [PubMed] [Google Scholar]
- 22.Choi JH, Bellas E, Gimble JM, Vunjak-Novakovic G, Kaplan DL. Tissue engineering. Part A. 2011;17:1437. doi: 10.1089/ten.tea.2010.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ward A, Quinn KP, Bellas E, Georgakoudi I, Kaplan DL. PloS one. 2013;8:e55696. doi: 10.1371/journal.pone.0055696. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bellas E, Panilaitis BJ, Glettig DL, Kirker-Head CA, Yoo JJ, Marra KG, Rubin JP, Kaplan DL. Biomaterials. 2013;34:2960. doi: 10.1016/j.biomaterials.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abbott RD, Raja WK, Wang RY, Stinson JA, Glettig DL, Burke KA, Kaplan DL. Methods. 2015;84:84. doi: 10.1016/j.ymeth.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Frazier TP, Bowles A, Lee S, Abbott R, Tucker HA, Kaplan D, Wang M, Strong A, Brown Q, He J, Bunnell BA, Gimble JM. Stem cells. 2016 doi: 10.1002/stem.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]; Girandon L, Kregar-Velikonja N, Bozikov K, Barlic A. Folia biologica. 2011;57:47. [PubMed] [Google Scholar]; Altman AM, Yan Y, Matthias N, Bai X, Rios C, Mathur AB, Song YH, Alt EU. Stem cells. 2009;27:250. doi: 10.1634/stemcells.2008-0178. [DOI] [PubMed] [Google Scholar]
- 23.Bellas E, Marra K, Kaplan DLP. Tissue engineering. Part C, Methods. 2013 doi: 10.1089/ten.tec.2012.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauney JR, Nguyen T, Gillen K, Kirker-Head C, Gimble JM, Kaplan DL. Biomaterials. 2007;28:5280. doi: 10.1016/j.biomaterials.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang SJ, Fu RH, Shyu WC, Liu SP, Jong GP, Chiu YW, Wu HS, Tsou YA, Cheng CW, Lin SZ. Cell transplantation. 2013;22:701. doi: 10.3727/096368912X655127. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Kim KA, Kim JH, Sul HS. The Journal of nutrition. 2006;136:2953. doi: 10.1093/jn/136.12.2953. [DOI] [PubMed] [Google Scholar]
- 27.Skalli O, Pelte MF, Peclet MC, Gabbiani G, Gugliotta P, Bussolati G, Ravazzola M, Orci L. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1989;37:315. doi: 10.1177/37.3.2918221. [DOI] [PubMed] [Google Scholar]
- 28.Newton JP, Hunter AP, Simmons DL, Buckley CD, Harvey DJ. Biochemical and biophysical research communications. 1999;261:283. doi: 10.1006/bbrc.1999.1018. [DOI] [PubMed] [Google Scholar]
- 29.Im GI. Tissue engineering. Part B, Reviews. 2014;20:545. doi: 10.1089/ten.TEB.2013.0731. [DOI] [PubMed] [Google Scholar]
- 30.Festuccia WT, Laplante M, Berthiaume M, Gelinas Y, Deshaies Y. Diabetologia. 2006;49:2427. doi: 10.1007/s00125-006-0336-y. [DOI] [PubMed] [Google Scholar]; Arner P. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1995;19:435. [PubMed] [Google Scholar]; Vermette M, Trottier V, Menard V, Saint-Pierre L, Roy A, Fradette J. Biomaterials. 2007;28:2850. doi: 10.1016/j.biomaterials.2007.02.030. [DOI] [PubMed] [Google Scholar]; Choi JH, Bellas E, Vunjak-Novakovic G, Kaplan DL. Methods in molecular biology. 2011;702:319. doi: 10.1007/978-1-61737-960-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]; Turner PA, Tang Y, Weiss SJ, Janorkar AV. Tissue engineering. Part A. 2015;21:1837. doi: 10.1089/ten.TEA.2014.0531. [DOI] [PubMed] [Google Scholar]
- 31.Napolitano A, Lowell BB, Damm D, Leibel RL, Ravussin E, Jimerson DC, Lesem MD, Van Dyke DC, Daly PA, Chatis P, et al. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1994;18:213. [PubMed] [Google Scholar]
- 32.Herder C, Hauner H, Kempf K, Kolb H, Skurk T. International journal of obesity. 2007;31:403. doi: 10.1038/sj.ijo.0803432. [DOI] [PubMed] [Google Scholar]
- 33.Fain JN. Vitamins and hormones. 2006;74:443. doi: 10.1016/S0083-6729(06)74018-3. [DOI] [PubMed] [Google Scholar]; Fain JN, Madan AK, Hiler ML, Cheema P, Bahouth SW. Endocrinology. 2004;145:2273. doi: 10.1210/en.2003-1336. [DOI] [PubMed] [Google Scholar]
- 34.Curat CA, Wegner V, Sengenes C, Miranville A, Tonus C, Busse R, Bouloumie A. Diabetologia. 2006;49:744. doi: 10.1007/s00125-006-0173-z. [DOI] [PubMed] [Google Scholar]
- 35.van Harmelen V, Skurk T, Rohrig K, Lee YM, Halbleib M, Aprath-Husmann I, Hauner H. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2003;27:889. doi: 10.1038/sj.ijo.0802314. [DOI] [PubMed] [Google Scholar]
- 36.Vistisen D, Witte DR, Tabak AG, Herder C, Brunner EJ, Kivimaki M, Faerch K. PLoS medicine. 2014;11:e1001602. doi: 10.1371/journal.pmed.1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumeng CN, Saltiel AR. The Journal of clinical investigation. 2011;121:2111. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]; Odegaard JI, Chawla A. Science. 2013;339:172. doi: 10.1126/science.1230721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skopkova M, Penesova A, Sell H, Radikova Z, Vlcek M, Imrich R, Koska J, Ukropec J, Eckel J, Klimes I, Gasperikova D. Obesity. 2007;15:2396. doi: 10.1038/oby.2007.285. [DOI] [PubMed] [Google Scholar]
- 39.Herder C, Haastert B, Muller-Scholze S, Koenig W, Thorand B, Holle R, Wichmann HE, Scherbaum WA, Martin S, Kolb H. Diabetes. 2005;54(Suppl 2):S11. doi: 10.2337/diabetes.54.suppl_2.s11. [DOI] [PubMed] [Google Scholar]; Pickup JC, Mattock MB, Chusney GD, Burt D. Diabetologia. 1997;40:1286. doi: 10.1007/s001250050822. [DOI] [PubMed] [Google Scholar]
- 40.Wajchenberg BL. Endocrine reviews. 2000;21:697. doi: 10.1210/edrv.21.6.0415. [DOI] [PubMed] [Google Scholar]; Samad F, Uysal KT, Wiesbrock SM, Pandey M, Hotamisligil GS, Loskutoff DJ. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6902. doi: 10.1073/pnas.96.12.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samad F, Yamamoto K, Pandey M, Loskutoff DJ. Molecular medicine. 1997;3:37. [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez-Infantes D, White UA, Elks CM, Morrison RF, Gimble JM, Considine RV, Ferrante AW, Ravussin E, Stephens JM. The Journal of clinical endocrinology and metabolism. 2014;99:E217. doi: 10.1210/jc.2013-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klok MD, Jakobsdottir S, Drent ML. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2007;8:21. doi: 10.1111/j.1467-789X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 44.Small CJ, Liu YL, Stanley SA, Connoley IP, Kennedy A, Stock MJ, Bloom SR. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2003;27:530. doi: 10.1038/sj.ijo.0802253. [DOI] [PubMed] [Google Scholar]
- 45.Gao D, Madi M, Ding C, Fok M, Steele T, Ford C, Hunter L, Bing C. American journal of physiology. Endocrinology and metabolism. 2014;307:E289. doi: 10.1152/ajpendo.00430.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rollins BJ, Walz A, Baggiolini M. Blood. 1991;78:1112. [PubMed] [Google Scholar]
- 47.Miller AM. Journal of inflammation. 2011;8:22. doi: 10.1186/1476-9255-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picone O, Laigre P, Fortun-Lamothe L, Archilla C, Peynot N, Ponter AA, Berthelot V, Cordier AG, Duranthon V, Chavatte-Palmer P. Theriogenology. 2011;75:287. doi: 10.1016/j.theriogenology.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 49.Murphy LJ. Experimental diabesity research. 2003;4:213. doi: 10.1155/EDR.2003.213. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim HS. Annals of pediatric endocrinology & metabolism. 2013;18:9. doi: 10.6065/apem.2013.18.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fredriksson L, Li H, Eriksson U. Cytokine & growth factor reviews. 2004;15:197. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]; Andrae J, Gallini R, Betsholtz C. Genes & development. 2008;22:1276. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desruisseaux MS, Nagajyothi ME, Trujillo HB, Tanowitz PE. Scherer, Infection and immunity. 2007;75:1066. doi: 10.1128/IAI.01455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renehan AG, Frystyk J, Flyvbjerg A. Trends in endocrinology and metabolism: TEM. 2006;17:328. doi: 10.1016/j.tem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Khan S, Shukla S, Sinha S, Meeran SM. Cytokine & growth factor reviews. 2013;24:503. doi: 10.1016/j.cytogfr.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Patrick CW., Jr. The Anatomical record. 2001;263:361. doi: 10.1002/ar.1113. [DOI] [PubMed] [Google Scholar]
- 55.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]; Wang Y, Rudym DD, Walsh A, Abrahamsen L, Kim HJ, Kim HS, Kirker-Head C, Kaplan DL. Biomaterials. 2008;29:3415. doi: 10.1016/j.biomaterials.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim HJ, Kim UJ, Kim HS, Li C, Wada M, Leisk GG, Kaplan DL. Bone. 2008;42:1226. doi: 10.1016/j.bone.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lucero HA, Kagan HM. Cellular and molecular life sciences : CMLS. 2006;63:2304. doi: 10.1007/s00018-006-6149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wissink MJ, van Luyn MJ, Beernink R, Dijk F, Poot AA, Engbers GH, Beugeling T, van Aken WG, Feijen J. Thrombosis and haemostasis. 2000;84:325. [PubMed] [Google Scholar]
- 57.Abrahamsson CK, Yang F, Park H, Brunger JM, Valonen PK, Langer R, Welter JF, Caplan AI, Guilak F, Freed LE. Tissue Eng Pt A. 2010;16:3709. doi: 10.1089/ten.tea.2010.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao F, Ma T. Biotechnol Bioeng. 2005;91:482. doi: 10.1002/bit.20532. [DOI] [PubMed] [Google Scholar]
- 59.Liu K, Zhou GD, Liu W, Zhang WH, Cui L, Liu X, Liu TY, Cao YL. Biomaterials. 2008;29:2183. doi: 10.1016/j.biomaterials.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 60.Saha S, Kirkham J, Wood D, Curran S, Yang XB. Cell and tissue research. 2013;352:495. doi: 10.1007/s00441-013-1586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, Kaplan DL. Nature protocols. 2011;6:1612. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim UJ, Park J, Kim HJ, Wada M, Kaplan DL. Biomaterials. 2005;26:2775. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 63.Ray H, Pinteur C, Frering V, Beylot M, Large V. Lipids in health and disease. 2009;8:58. doi: 10.1186/1476-511X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hirsch J, Gallian E. Journal of lipid research. 1968;9:110. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.