Abstract
Chromatin remodelling is essential for cardiac development. Interestingly, the role of histone chaperones has not been investigated in this regard. HIRA is a member of the HUCA (HIRA/UBN1/CABIN1/ASF1a) complex that deposits the variant histone H3.3 on chromatin independently of replication. Lack of HIRA has general effects on chromatin and gene expression dynamics in embryonic stem cells and mouse oocytes. Here we describe the conditional ablation of Hira in the cardiogenic mesoderm of mice. We observed surface oedema, ventricular and atrial septal defects and embryonic lethality. We identified dysregulation of a subset of cardiac genes, notably upregulation of troponins Tnni2 and Tnnt3, involved in cardiac contractility and decreased expression of Epha3, a gene necessary for the fusion of the muscular ventricular septum and the atrioventricular cushions. We found that HIRA binds GAGA rich DNA loci in the embryonic heart, and in particular a previously described enhancer of Tnni2/Tnnt3 (TTe) bound by the transcription factor NKX2.5. HIRA-dependent H3.3 enrichment was observed at the TTe in embryonic stem cells (ESC) differentiated toward cardiomyocytes in vitro. Thus, we show here that HIRA has locus-specific effects on gene expression and that histone chaperone activity is vital for normal heart development, impinging on pathways regulated by an established cardiac transcription factor.
Introduction
The heart is the first organ to be formed that is vital for embryogenesis. In the post-gastrulation embryo at embryonic day (E) 6.5, the mesodermally derived cardiac crescent appears and undergoes a series of regulated morphological changes leading to a linear heart tube [1]. After heart looping at E9.5, the four chambers are progressively septated. By E15.5 the heart is fully functional. Cardiovascular development is tightly regulated by dynamic gene expression. Epigenetic modifications such as post-translational histone modification have been described to influence development and differentiation [2]. However, little is known of the role of histone chaperone functions during cardiac development.
HIRA is a component of the HUCA complex that deposits the variant histone H3.3 into chromatin independently of cellular replication [3], influencing transcription [3–5], genome integrity [6], fertilization [7], cellular senescence [8], and genome reprogramming [9]. In mouse embryonic stem cells (ESCs), HIRA deposits H3.3 predominantly in genic regions, but also at a subset of enhancer and intergenic regions [4]. Hira null embryos display a range of developmental defects during and subsequent to gastrulation [10]. A small proportion of these mutants survived to E10.5 and showed abnormal heart looping and substantial pericardial oedema amongst other defects including abnormal placentation suggesting that the heart defects may have been a secondary effect.
In order to assess the role of HIRA in cardiovascular development, we used a conditional allele of Hira in mice in conjunction with various relevant cardiac relevant CRE recombinases to bypass the early lethality of Hira null embryos. Mesp1 is the earliest known marker of cardiac progenitors which give rise to cardiomyocytes, endothelial cells (ECs), epicardial derived cells and smooth muscle cells. We employed Mesp1Cre to target Hira in these early cardiac progenitors, and then used Nkx2.5Cre, Mef2cCre and Tie2Cre drivers to refine requirements of HIRA in the second heart field (SHF) and endothelial lineages.
We show here that HIRA plays a major role in the cardiogenic mesoderm. Mesp1 conditionally mutant Hira embryos presented with generalised oedema and cardiac malformations such as ventricular septal defect (VSD), atrial septal defect (ASD), thin ventricular wall and constricted pulmonary trunk (PT). Using RNAseq we report that, of the most significantly changed genes, absence of HIRA impacts troponins known to be relevant for regulation of muscle contractility, and Epha3 required for the endothelial to mesenchymal transition (EMT) taking place in the atrioventricular cushions prior to septation and valve formation. Quantitative Chromatin Immunoprecipitation (qChIP) and ChIP followed by sequencing (ChIPseq) analyses show that HIRA is strongly enriched at the common enhancer of troponins Tnni2 and Tnnt3 (the TTe site) in E12.5 Wild Type (WT) hearts. ESCs differentiated towards cardiomyocytes confirmed this specific HIRA enrichment at the TTe site, associated with HIRA-dependent H3.3 deposition. The TTe site has been shown to be bound by NKX2.5, as determined by previous ChIP in embryonic hearts [11], and DamID experiments using HL-1 cells [12]. In summary we provide the first indication that histone chaperone complexes have a role in cardiovascular development and suggest that HIRA complexes directly regulate a subset of genes vital for cardiovascular morphogenesis.
Methods
Mouse lines
Animal maintenance, husbandry and procedures were carried out in accordance with British Home Office regulations. Hira knockout mice have been described previously [10]. The Hira pre-conditional allele was generated by the Wellcome Trust Sanger Institute: Hiratm1a(EUCOMM) Wtsi, MGI:4431679. The Cre line used were: Mesp1Cre (MGI:2176467; Mesp1tm2(cre)Ysa), Mef2CCre (MGI:3639735 Tg(Mef2c-cre)2Blk), Wnt1Cre (MGI:2386570; Tg(Wnt1-cre)11Rth), Nkx2.5Cre (MGI:2654594; Nkx2-5tm1(cre)Rjs), Tie2Cre (MGI: 2450311,Tg(Tek-cre)1Ywa). All lines were maintained on a CD1-ICR background.
Optical projection tomography
Embryos were fixed overnight in 4% PFA/PBS and mounted in low-melting agarose (Life Technologies). Up to E14.5, whole embryos were processed. At E15.5, the trunks were opened and cartilage from the rib cage was discarded to help with the subsequent scanning since cartilage does not clear completely. Samples were then trimmed to remove the excess of agarose and washed in 100% methanol followed by clearing in benzyl alcohol:benzyl benzoate (BABB). Scanning was undertaken using a Bioptonics OPT Scanner 3001M (MRC Technology, Edinburgh, UK). NRecon software (Skyscan NV) was used for image reconstruction from projections using a back-projection algorithm. FIJI (Image J) and Volocity were used for image analysis and 3D reconstruction.
RNA extraction and sequencing
RNA extraction was done in triplicate from Mesp1Cre;Hira-/fl and Mesp1Cre;Hira+/fl embryonic hearts at E11.5 and E12.5 using the QIAGEN RNeasy mini kit (74104). RNA QC was performed by a 2100 bioanalyzer. RNAseq was processed by Illumina NextSeq 500, and paired ends reads were produced. Reads were aligned and normalised using BOWTIE and DEseq R package. Strand NGS 2.5 software, which uses the DEseq algorithm, was used to incorporate additional downstream analysis such as Gene Ontology. The Mann Whitney unpaired test and Benjamini Hochberg False discovery rate (FDR) were applied. The genes were sorted using the following settings: adjusted p-value ≤ 0.05 and absolute fold change ≥ 1.5. We found 95% of similar results between the two analysis methods.
Reverse transcription and quantitative real time PCR
The High-Capacity RNA-to-cDNA™ Kit (Thermo fisher 4387406) was used to obtain cDNA from the RNA (see above) for the qRT-PCR experiments, according to the manufacturer’s instructions. Primers for qRT-PCR were designed using primer-blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) with the following option: primers must span an exon-exon junction and be separated by at least one intron, thus ensuring amplification of cDNA and not possible gDNA contamination. The PCR product size was set to be between 80 and 160 bp. The standard curve method was performed using SYBR green and results normalised to Gapdh. The CFX96 Touch™ Real-Time PCR Detection System was used. Following the reaction, melting curves were checked and samples were run on an agarose gel to verify the amplimer size.
In situ hybridisation on paraffin sections
The following plasmids were used: Tnni2, Image clone 1448494, Epha3 Pblu2KSP. They were kindly provided by Tim Mohun and Jeffrey Bush respectively. Briefly, plasmids were linearized using EcoRI and XhoI respectively and RNA synthesised using T3 and T7 RNA polymerase respectively. RNA was extracted from a 1% agarose gel using the QIAquick Gel Extraction Kit (Qiagen). 1 μg of linearised plasmid was used for in vitro transcription of probes using a DIG RNA labelling kit (Roche). Probes were purified by precipitation with the addition of 2 μl 0.5M EDTA (pH 8), 5 μl 4 M LiCl and 150 μl ethanol to the reaction and centrifugation of the precipitates.
Paraffin sections were prepared as follow. Briefly, slides were incubated in 20 μg/ml Proteinase K (Sigma-Aldrich) for 8 minutes, washed in 2 mg/ml glycine then PBS, then fixed in 4% PFA/PBS for 20 minutes. Following further PBS washes they were incubated for 1 hour at 70°C in a humidified chamber in hybridisation buffer (50% formamide, 5X SSC pH 4.5, 50 μg/ml yeast RNA, 1% SDS, 50 μg/ml heparin) followed by overnight incubation in hybridisation buffer containing between 1 and 2 μg/ml of antisense RNA probe. Slides were then rinsed twice in 2X SSC buffer pH4.5, followed by three washes at 65°C in Solution I (50% formamide, 5X SSC pH4.5, 1% SDS), two washes in Solution II (50% formamide, 2X SSC pH4.5) and finally two washes at RT in MABT (0.1 M maleic acid, 0.15 M NaCl, 0.01% Tween-20, 2 mM Levamisole (Sigma-Aldrich), pH7.5). Slides were then incubated in blocking solution (2% Boehringer Blocking Reagent (Roche), 10% sheep serum in MABT) for 1 hour followed by overnight incubation at 4°C with an alkaline-phosphatase (AP) conjugated anti-DIG antibody (Roche) diluted 1:2000 in blocking buffer. Following further washes in MABT and AP buffer (100 mM Tris, pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween-20, 2 mM Levamisol), AP activity was detected using BM Purple (Roche) for at least 24 hours.
HIRA qChIP
30 to 40 E12.5 WT hearts were pooled, washed in PBS and cross-linked for 45 min with 1.5 mM of EGS (Sigma, E3257), followed by 15 min of 1% formaldehyde (from a freshly made filtered stock at 18.5%) at 37°C. The reaction was quenched by the addition of 125 mM of Glycine, left for 15 min at RT. Hearts were lysed in 50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton, 1X anti-protease cocktail (Roche, 04693132001), 1 mM PMSF for 10 min at 4°C (LB1). The hearts were briefly spun down and resuspended in 10 mM Tris-HCL pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1X anti-protease cocktail, 1 mM PMSF for 10 min at 4°C (LB2). Finally the hearts were resuspended in10mM Tris-HCL pH 8.0, 100mM NaCl, 1mM EDTA, 0.5mM EGTA, 0.1% DOC, 0.5% N-Lauroylsarcosine, 1X anti-protease cocktail and 1mM PMSF (LB3) rolling O/N at 4°C. 5 min sonication at 5 μA with Soniprep 150 MS was completed 30 times, with 5 min on ice in between. Antibodies were coupled to magnetic beads (Dynabeads, InVitrogen, 112.03D) for at least 4 hours at 4C and washed 3 times in LB3. 10% of input was isolated and protein–DNA complexes were immunoprecipitated using WC15 antibody against HIRA, rolling overnight. Beads were then washed once with 20 mM tris pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton, 2 mM EDTA (WB1), once with 20 mM Tris pH 8.0, 500 mM NaCl, 0.1% SDS, 1% Triton, 2mM EDTA (WB2), once with 10 mM Tris pH 8.0, 150 mM LiCl, 1% NP-40, 1% DOC, 1 mM EDTA, then TE 10:1, 50 mM NaCl (WB3), and finally in TE 10:1. Samples were treated overnight with 50 mM Tris pH 8.0, 10 mM EDTA and 1% SDS at 65°C (EB), then with RNAse (Qiagen, 19101) for an hour at 25°C and with PK for 2h at 56°C. DNA was purified with QIAGEN’s PCR purification kit (28104). Purified DNA was quantified by quantitative PCR, using the purified input chromatin as a positive control. ChIP enrichment was calculated by normalisation to the Input signal (= 100%).
NKX2.5 qChIP
E12.5 embryos were dissected in PBS. Hearts were flash frozen and stored at -80°C during genotyping. 20 WT hearts and 20 Mesp1CreHirafl/ hearts were pooled respectively and fixed for 15 min in 1% formaldehyde at 37°C then quenched by 125 mM of Glycine for 15 min at RT. Hearts were lysed in RIPA buffer (50 mM Tris pH7, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 50 mM NaF, 0.5% DOC, 0.1% SD. 2 μM sodium orthovanadate, protease inhibitor cocktail 1X (Roche) and PMSF 1 mM were added prior to the experiment. The hearts in lysis buffer were placed on a rotating wheel at 4°C overnight. A syringe (25G) was used to finish the lysis. The lysates were then sonicated (10 rounds of 1 min of sonication at 5 μA with Soniprep 150 MS, with 1 min on ice in between). The protein G beads were incubated overnight at 4°C in PBS with 10 μg of NKX2.5 antibody (N-19 Santa Cruz) and washed 3x in the previous RIPA buffer. They were then incubated with the sonicated chromatin at 4°C O/N. The following day, the beads were washed 2X for 5min in WB1 (10 mM HEPES pH 7.6, 1 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100) and 2X for 5 min in WB2 (10 mM HEPES pH 7.6, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.01% Triton X-100). The samples were then processed the same way as for HIRA qChIP.
Library preparation, Sequencing and Analysis for HIRA ChIPseq
Libraries were prepared using the NEB DNA Ultra kit, with a selection of fragments size of ~200bp. They were sequenced on the Illumina NextSeq 500, v2 chemistry and produced paired ends. Alignment was done using bowtie2 with mm10. Peak detection and consensus sequence discovery was undertaken using Strand NGS software 2.5, which includes the algorithm of MACS1.4 (p≤10−4, other settings left as default) (Model-based Analysis for ChIPseq), after removal of poor quality and duplicate sequences normalisation was done using RPKM. Lists of genes within +/- 5Kb of the TSS and TES was generated using Strand NGS. Bedtools intersect tool was used to define the overlapping peaks between different BED files. In addition, PAPST (Peak Assignment and Profile Search Tool) software was used to overlap regions of interest and establish genome wide enrichment patterns of HIRA ChIPseq [13].
Embryonic stem cell culture and differentiation to cardiomyocytes
H3.3-HA tagged wild type (W9.5) and Hira-null (Clone 104) mESCs have been previously described [5]. They were maintained in an undifferentiated state on 0.1% gelatin coated flasks in Knockout™ D-MEM (GIBCO, 10829), supplemented with 15% ES-FCS (Millipore ES-009B), 1X Glutamax (GIBCO 35050–038), 1X Penicillin/Streptomycin (GIBCO 15140), 1X MEM NEAA (GIBCO 11140–035), 0.1 mM 2-β-mercaptoethanol (SIGMA M-7522) and 103 Units/ml LIF (Millipore, ESG-1106) at 37°C and 5% CO2. These cells were differentiated using the well-described hanging-drop method [14] at a concentration of 25 cells/μl in DMEM (GIBCO 61965–026), complemented with 15% ES-FCS (Millipore ES-009B), 1x Penicillin/Streptomycin (GIBCO 15140), 1x MEM NEAA (GIBCO 11140–035), 0.1mM 2-mercaptoethanol (SIGMA M-7522). Cells were detached and plated on regular gelatin coated TC plates at day 4 of differentiation.
Immunofluorescence
Paraffin sections were rinsed in PBS and permeabilised with 0.5% Triton X-100 for 10 mins at RT. Then rinsed twice in PBS. Blocking was accomplished with 1% BSA, 10% sheep serum and 0.1% Triton X-100 for 1 hour at RT. Slides were incubated with primary antibody (1:100 of Troponin C: ab30807) diluted in block buffer, then washed 3X in PBS + 0.1% Triton X-100 followed by 2 rinses in PBS. Incubation with secondary antibody was done diluted in block (1:200) for 1hr at RT. Finally the slides were rinsed 3 times (5–10 minutes) in PBS + 0.1% Triton X-100 (including DAPI in the final wash, and then rinsed twice in PBS) and mounted. Slides were captured on the confocal using Tilescan and Zstack on a 63x objective and merged with FIJI.
Co-immunoprecipitation
20 E14.5 hearts, and 30 E12.5 hearts, were dissected from WT embryos in cold PBS and flash frozen in liquid nitrogen before being digested in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 50mM NaF, 0.5% deoxycholic acid, sodium orthovanate 20 nM, anti-protease cocktail 1X, 1 mM PMSF). Sequential syringes with gradually smaller needles (19G, 23G, 25G) were used to help the lysis. Magnetic beads coated with sheep anti-mouse IgG (AB 11201D) were incubated overnight at 4°C with either the supernatant recovered from HIRA hybridomas (WC15) or the mouse IgG1k monoclonal isotype control antibody (AB 18447, lot GR 53099–6). The beads were then washed 3x with the previous RIPA buffer the next day. After the final wash, the protein lysate was incubated with the beads overnight rolling at 4°C. 100 μl of 1 ml was saved and stored at -80°C to load as the input. The next day the beads were washed in RIPA and resuspended in Laemli buffer (3X Laemli: 120 mM Tris pH6.8, 3% SDS, 5% Glycerol, 0.01% Bromophenol, 1.5% β-mercaptoethanol) and boiled for 15min to separate the beads from the antibodies and proteins. Pre-cast gradient gels (Mini-PROTEAN TGX 4%-15%, Biorad 456–1083) were used. 28 μl was loaded per well. The proteins were then transferred onto a PVDF membrane (Biorad 162–0177) using wet transfer. The membrane was blocked with 5% milk-TBST 1X (0.5% tween) for 1 hour at RT, washed 3 times with TBST then incubated with 1/100 of anti-WHSC1 (Atlas HPA015801), or 1/4 of WC119 hybridoma supernatant in 1% milk- TBST 1X overnight at 4°C. Next the membrane was washed 3 times with TBST, incubated with a secondary antibody (Amersham NA93310V) for 45 min at RT then washed 3 times with TBST and finally revealed with ECL (Amersham RPN2209, 28906837) with the appropriate secondary antibody (Amersham NA93340V or NA93310V). The hybridomas WC15 and WC119 were kindly given by Peter Adams.
Database Deposition
The RNAseq data is deposited at the Gene Expression Omnibus database with accession GSE79937, and ChIPseq data with accession GSE79826.
Results
HIRA is required in the developing heart
Hira is ubiquitously expressed from E8.5 during mouse embryogenesis [15]. Consistent with this, we detected expression throughout the embryonic heart at E13.5 by using the β-galactosidase cassette present in the preconditional Hira allele (S1A Fig). We then generated the Hira conditional allele using a FLPase transgenic cross (S1B Fig). This Hira conditional allele has been previously used in the literature [9], and when combined with an ubiquitous ActinCre driver, we observed the same gastrulation phenotypes as in our constitutive Hira null embryos (data not shown) [10]. Using our existing constitutive Hira null allele (Hira-), we bred Mesp1Cre to Hira-/+ mice to generate Mesp1Cre;Hira-/+ mice. Next we mated Mesp1Cre;Hira-/+ with Hirafl/fl mice to examine the role of HIRA in cardiac progenitors (the single null allele was included so only a conditional allele would be recombined, maximizing Cre-mediated production of Mesp1Cre lineage Hira null cells). We validated the recombination by PCR (S1C Fig), lack of exon 4 representation in the RNAseq (S1D Fig), and HIRA expression by western blotting (S1E Fig). Of 4 litters born, we did not identify any live Mesp1Cre;Hira-/fl pups (Table 1), indicating that cardiogenic mesodermal ablation of Hira from E6.5 is embryonically lethal.
Table 1. Embryonic phenotype resulting from the conditional ablation of Hira.
| Mesp1Cre;Hira-/fl | Nkx2.5Cre;Hira-/fl | Mef2cCre;Hira-/fl | Tie2Cre;Hira-/fl | Wnt1Cre;Hira-/fl | ||||
|---|---|---|---|---|---|---|---|---|
| STAGE | E10.5 | E12.5 | E13.5 | E15.5 | E15.5 | ADULT | E15.5 | E18.5 |
| VSD | N/A | N/A | N/A | 12/12 | 5/9 | N/A | 0/5 | 0/10 |
| ASD | N/A | N/A | N/A | 10/12 | 4/9 | N/A | 0/5 | 0/10 |
| Thin ventricular wall | 0/4 | 0/9 | 0/12 | 3/12 | 0/9 | N/A | 0/5 | 0/10 |
| Constricted PT | 0/4 | 0/9 | 0/12 | 1/12 | 3/9 | N/A | 0/5 | 0/10 |
| Oedema | 0/4 | 0/9 | 3/12 | 12/12 | 0/9 | N/A | 0/5 | 0/10 |
| Haemorrhage | 0/4 | 0/9 | 6/12 | 12/12 | 0/9 | N/A | 0/5 | 0/10 |
| Exencephaly | 0/4 | 1/9 | 0/12 | 0/12 | 0/9 | N/A | 0/5 | 0/10 |
| VIABLE by P10 | NO (0/12) | YES (1/9) | YES (4/4) | YES (3/6) | NO (0/40) | |||
Number of embryos observed for the indicated phenotype at the indicated stage related to the total number of embryos collected. N/A indicates none observed. The number of collected and thus viable embryos are also indicated at 10 days post-birth (P10) related to the number of expected embryos.
We detected a small proportion of exencephaly and haemorrhage at E12.5 (Fig 1A, Table 1). All Mesp1Cre;Hira-/fl embryos presented with a severe oedema at E15.5 (Fig 1A, Table 1). We observed a fully penetrant VSD in the heart of all mutants (n = 12); whilst the muscular ventricular septum completely separated the two ventricles in their littermate WT embryos at E15.5. Some embryos had atrioventricular septal defects with a common atrioventricular junction (Fig 1B, S1 Video). The large interventricular communication observed in Mesp1Cre;Hira-/fl embryos is not likely compatible with life during the late stage of embryogenesis [16]. In 83% of mutants, we detected a deficiency of the flap valve of the oval fossa, a derivative of the primary atrial septum, resulting in an ASD. None was observed in their WT littermates (Fig 1B) (n = 10). Haematoxylin and Eosin (H&E) staining revealed that some E12.5 Mesp1Cre;Hira-/fl embryos displayed abnormally shaped atrioventricular cushions, whilst mesenchymal tissue normally swells in the atrioventricular canal as a result of EMT in their WT littermates (Fig 1C). In normal development, the rightward tubercles of the atrioventricular cushions form the membranous septum [17, 18]. We have observed deficiency of the muscular part of the ventricular septum, since the point of contact between the septum and the atrioventricular cushions was mispositioned in the mutants compared to controls (Fig 1C). Furthermore, cushions were crescent shaped in the wild type but straighter in the mutants (Fig 1C). Thus, HIRA is required for normal cardiovascular development.
Fig 1. HIRA is required in the developing heart.
A. Lateral view of littermate embryos with the indicated genotype. Mesp1Cre;Hira-/fl embryos had a low penetrance external phenotype at E12.5: exencephaly (E) and light haemorrhage (H) are indicated in the mutants. At E15.5, all mutants showed severe oedema (O) as indicated. B. Transverse OPT reconstructions followed by virtual reslicing of E15.5 embryo trunks with the indicated genotype. VSD (V) and ASD (A) are indicated in the mutants. H&E of transverse sections of E14.5 embryos with the indicated genotype also showed a common atrioventricular junction (CJ) in the mutants as indicated. C. H&E staining of transverse sections from E12.5 embryos reveals a disruption of the endocardial cushion (EC) fusion (two controls and two littermate mutants shown). The muscular septum is deficient (*, top) and the relatively flat rather than crescentic cushion shape in the mutant are indicated (arrows). Scale bars represent 2mm (A), 0.5mm (B-C). D. Transverse sections of E12.5 embryonic hearts of the indicated genotype immunostained with DAPI and Troponin C captured on confocal showing disrupted sarcomeric structure in the mutant ventricular free wall. Scale bar: 10μm.
HIRA is required in the Nkx2.5, but not in the Mef2c or Wnt1 lineages
We then refined the requirement of HIRA in cardiac lineages with Nkx2.5Cre and Mef2cCre drivers. The expression of Nkx2.5Cre is restricted to cardiomyocytes, cardiac endothelium and pharyngeal endoderm [19, 20], and appears 24 hours later than Mesp1Cre expression. We observed that 33% (n = 3) of Nkx2.5Cre;Hira-/fl embryos had a hypoplastic pulmonary trunk (PT) (Fig 2A). 56% (n = 5) of mutants displayed a large VSD (n = 5) (Fig 2A) but did not show any sign of oedema. The survival rate of mutant pups was extremely low (1 mutant out of 9 expected) (Table 1).
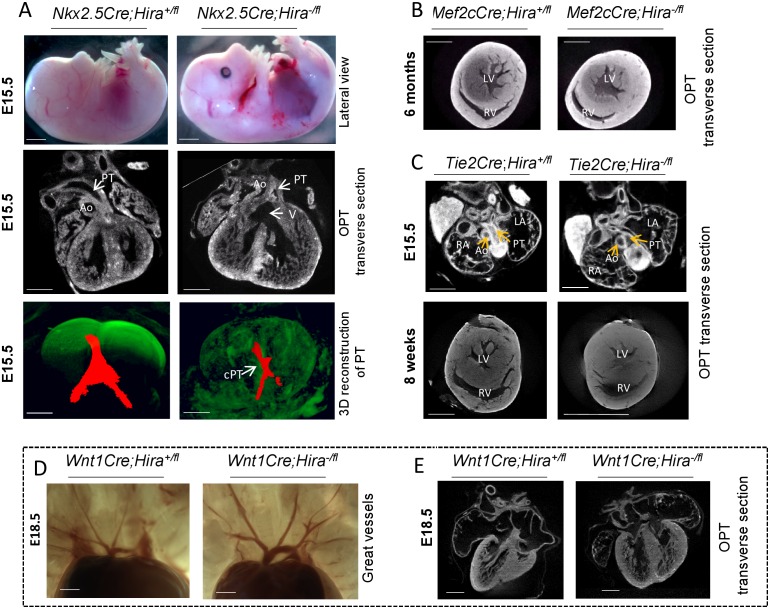
Fig 2. Hira requirements in distinct lineages contributing to the heart and vessels.
A. E15.5 Nkx2.5Cre;Hira-/fl embryos and their littermate control embryos were examined as indicated. Lateral view of Nkx2.5Cre;Hira-/fl mutants revealed a normal external appearance. OPT transverse section revealed a VSD (V) and an overriding of the aortic root of the muscular septum in as well as a constricted pulmonary trunk PT (cPT) in some mutants by 3D reconstruction. B&C. OPT transverse section of the heart of a 6 months old Mef2cCre;Hira-/fl (right/left ventricle: RV/LV) (B) or of Tie2Cre;Hira-/fl embryo and adult (C) with their respective littermate control. Mutant embryos did not show any defect and their adult heart only showed proportional size reduction to the body size. The Aorta (Ao), the PT, and the right and left atrium (RA/LA) are indicated. D&E. Great vessel structure (D) and OPT transverse section (E) on E18.5 Wnt1Cre;Hira-/fl and their control littermate hearts. No heart defects were observed in these mutants. Scale bars represent 2mm in whole embryo and 0.5mm in transverse sections.
We next mated Mef2cCre;Hira+/fl with Hirafl/fl mice to test whether HIRA was required in the SHF. Mef2c is expressed from E7.5 in the progenitors of the right ventricle, outflow tract, and ventricular septum. Mef2cCre driven ablation of Hira had no noticeable effect on development and postnatal life (Table 1 and Fig 2B).
A requirement for Hira has been described both in endothelial cells [21] and in neural crest cells [22] (in chick), we tested the role of Hira in these lineages using Tie2Cre and Wnt1Cre drivers, respectively. The Tie2Cre mutants were fully viable and had no detectable vessel defects at E15.5 (Fig 2C). Nevertheless, adult mutants were smaller at 8 weeks of age, with hearts smaller but in proportion relative to their reduced body size (Fig 2C), and without apparent structural abnormalities by OPT (n = 3). Wnt1Cre;Hira-/fl embryos demonstrated a perinatal lethality, not due to a cardiac malformation since they presented with a normal heart structure and no apparent vessel malformations at E18.5 (Fig 2D & 2E).
Absence of HIRA dysregulates cardiac gene expression
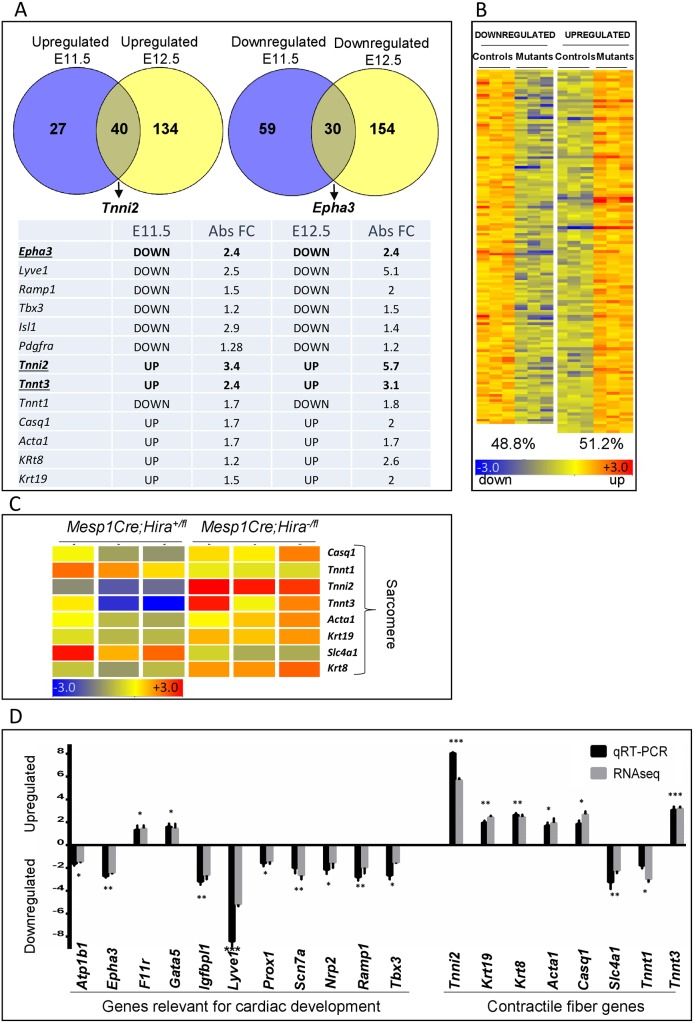
The full penetrance of Mesp1Cre Hira-conditional mutants allowed us to investigate the transcriptional changes underlying the cardiovascular defects observed in the absence of HIRA. We chose the E11.5 and E12.5 stages as they were prior to the appearance of the major phenotypes. At E11.5 and E12.5 stages there were 156 and 360 coding transcripts respectively with significantly altered expression in the mutant hearts (Mann Whitney unpaired test, Benjamini Hochberg FDR, p ≤ 0.05, FC ≥ 1.5) (Fig 3A, S1 and S2 Tables), with no trend towards up- or down-regulation of global transcription (48.8% down and 51.2% up; Fig 3B). We performed a gene ontology (GO) analysis with the differentially expressed genes and observed a link with myocyte contractility (S3 Table). Four GO terms included the same eight genes related to contractility and sarcomeric structure (Fig 3C), including the most up-regulated gene Tnni2. We next used quantitative real-time PCR (qRT-PCR) to validate expression differences for 11 changed genes that are relevant to cardiac development, and the eight genes related to cardiac contractility (Fig 3D).
Fig 3. HIRA is required for the regulation of a subset of genes expressed during cardiac morphogenesis.
A. Venn diagram indicating the number of genes whose expression is up or downregulated in Mesp1Cre;Hira-/fl compared to Mesp1Cre;Hira+/fl hearts at E11.5 and E12.5. The table indicates the level of fold change observed at E11.5 and at E12.5 on a subset of genes. B. Heatmap displaying the differential gene expression in mutant (Mesp1Cre;Hira-/fl) vs control (Mesp1Cre;Hira+/fl) in triplicates at E12.5. C. Heatmap of the gene ontology analysis revealed a trend in upregulation of sarcomeric contractile fibre genes (aside from Tnnt1 and Slc4a1 which were downregulated). D. qRT-PCR and RNASeq of the indicated genes within E12.5 hearts displayed as the fold induction in the mutants compared to their WT littermates (n = 3). Unpaired t-test: p<0.05 *, p<0.01 **, p<0.001 ***.
We examined selected genes known for their role in cardiac development and displaying the highest expression change in our mutants in both E11.5 and E12.5 mutants (Fig 3A). The expression of Epha3, a receptor tyrosine kinase required for EMT in the atrioventricular cushions [18], was downregulated of 2.7 fold in the heart of the mutants compared to their control littermates (Fig 3D). In agreement, in situ hybridization (ISH) revealed that Epha3 expression was greatly diminished in the cushions and the membranous part of the ventricular septum in the mutants compared to their control littermates (Fig 4A). We also found that Tnni2, a fast twitch skeletal muscle gene, was the most upregulated gene in the mutant hearts by both RNA-seq (5.9 fold) and qRT-PCR (7 fold) (Fig 3D). ISH confirmed Tnni2 overexpression in the mutants (Fig 5A), suggesting heart contractility could be affected by alteration of sarcomeric components. Indeed, staining with Troponin C on transverse sections to examine sarcomeric organisation demonstrated a lack of typical parallel organisation of the filament structures in the mutants (Fig 1D). There were no significant transcription changes in Hira+/- hearts at E12.5 (S4 Fig).
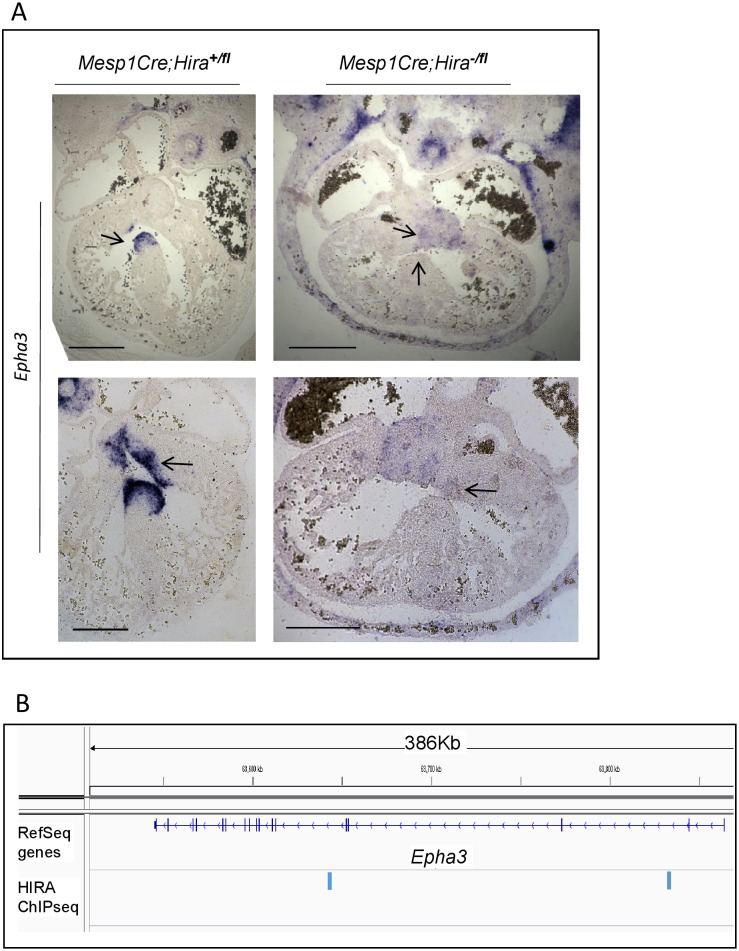
Fig 4. Epha3 is downregulated when HIRA is conditionally deleted, and HIRA binds to the gene body of Epha3.
A. In situ hybridization of Epha3 on transverse sections of E12.5 embryonic hearts with the indicated genotypes (n = 2). Hira-conditional mutant hearts displayed low expression of Epha3 in both the membranous portion of the septum (upper panels) and the endocardial cushions (lower panels) compared to littermate controls as indicated by a black arrow. Scale bar: 100μm B. IGV profile showing two HIRA enriched loci in intronic regions of Epha3.
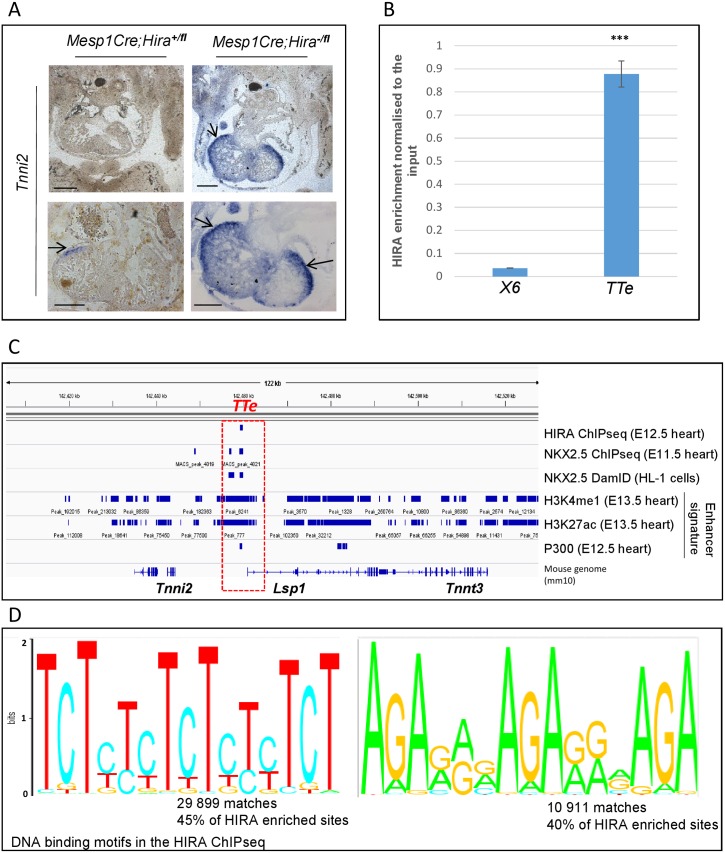
Fig 5. HIRA is enriched in the vicinity of Tnni2/Tnnt3, which is upregulated in conditionally deleted Hira mutants, and binds GAGA rich DNA.
A. In situ hybridization Tnni2 on transverse sections of E12.5 embryos with the indicated genotype, revealed an abnormally increased expression of Tnni2 particularly in the ventricular free wall of the mutant hearts (arrows). Scale bar: 100μm B. qChIP of the HIRA enrichment at the TTe locus. An intergenic region on chromosome 6 was used as a negative control. Data is normalized to the input (= 100) and is representative of 2 independent experiments. Errors bars represent the min/max from technical duplicates. Unpaired t-test: p<0.001 *** C. IGV profile showing the HIRA ChIP seq peak present at the Tnni2/Lsp1/Tnnt3 (TTe) site (situated 17Kb downstream of Tnni2 and 34Kb upstream of Tnnt3), co-localizing with NKX2.5 ChIPseq peaks (E11.5 heart [11]), NKX2.5 DamID peaks (HL-1 cell line [12]), and the active enhancer marks H3K4me1 and H3K27Ac ChIPseq peaks on WT E13.5 hearts (encode database ENCSR663VWL) and P300 (WT E12.5 heart [23]). D. The Matrix-based nucleotide profiles display here the motifs that HIRA binds most frequently. 45% of HIRA enriched sites contained a GAGA/TCTC motif.
HIRA is enriched in the vicinity of the Tnni2/Tnnt3 gene loci
To assess whether HIRA was directly regulating gene expression, we performed a HIRA ChIPseq of WT E12.5 hearts. We found 6625 peaks using Model-based Analysis for ChIPseq (MACS) analysis (p≤10−4) which mostly covered distal intergenic regions of the genome (74%) and introns (22.5%) (S2A Fig). Interestingly, 45% of the peaks contained the consensus motif GAGAGAGA (Fig 5D) that in Drosophila melanogaster is known to bind the GAGA factor.
Two significant HIRA-bound regions were identified in intronic regions of Epha3 (Fig 4B), of which one contained a GAGA motif. One GAGA motif was observed within a significant HIRA-bound region 17Kb downstream of Tnni2, 38Kb upstream of Tnnt3, and 1 kb of Lsp1 (Fig 5B). We subsequently refer to this peak as the Tnni2-Tnnt3-enh (TTe) site (see below). The expression of both Tnni2 and Tnnt3 was strongly upregulated in our mutant model however, we did not observe any dysregulation of Lsp1 expression. This is strikingly similar to what is observed in Nkx2.5 hypomorphs, which show Tnni2/Tnnt3, but not Lsp1, overexpression in E11.5 mutant versus wild type hearts [11] (S2B Fig). Moreover, it has been shown that the Tnni2 and Tnnt3 genes are directly repressed by NKX2.5 via direct binding to the TTe site (E11.5 hearts), and that the TTe sequence can direct reporter expression in the HL-1 (cardiomyocyte) cell line [11]. We found a 25-fold enrichment of HIRA at the TTe compared to a negative intergenic control region by qChIP (Fig 5B). We next examined active enhancer marks by overlapping HIRA E12.5 ChIPseq peaks with the histone modification H3K4me1and H3K27Ac ChIPseq peaks at E13.5, as well as P300 in embryonic hearts at E12.5 [23] at the TTe (Fig 5C) and genome wide (S2C Fig). Of the 78 sites containing overlaps of HIRA and NKX2.5, 63 (80.7%) overlapped with both H3K4me1 and H3K27Ac, including TTe. We interrogated these histone marks at the TTe in ESCs differentiated to cardiomyocytes and in their originating ESCs [24] (S3 Fig). The TTe locus became enriched for H3K4me1 and H3K27Ac in differentiated cardiomyocytes compared to ESCs, in agreement with the TTe-reporter data indicating that TTe is an active enhancer in the heart [11]. We next tested whether absence of HIRA affected NKX2.5 occupancy at the TTe by performing a NKX2.5 qChIP. At E12.5, NKX2.5 binding was moderately but significantly reduced to 75% (Fig 6) in Mesp1Cre;Hirafl/- hearts compared to WT hearts. In addition to Tnni2 and Tnnt3, we examined three other genes dysregulated in both Mesp1Cre;Hirafl/- and Nkx2.5 hypomorphic E11.5 hearts: Clcnkb, Abca4 and Slc9a3r1. Similarly to Tnni2 and Tnnt3, all three were upregulated by 2.9, 1.72 and 1.53 fold respectively in Hira mutant hearts. A reduced NKX2.5 binding at the Slc9a3r1 locus was also observed in the hearts of Hira conditional null mutants (Fig 6).
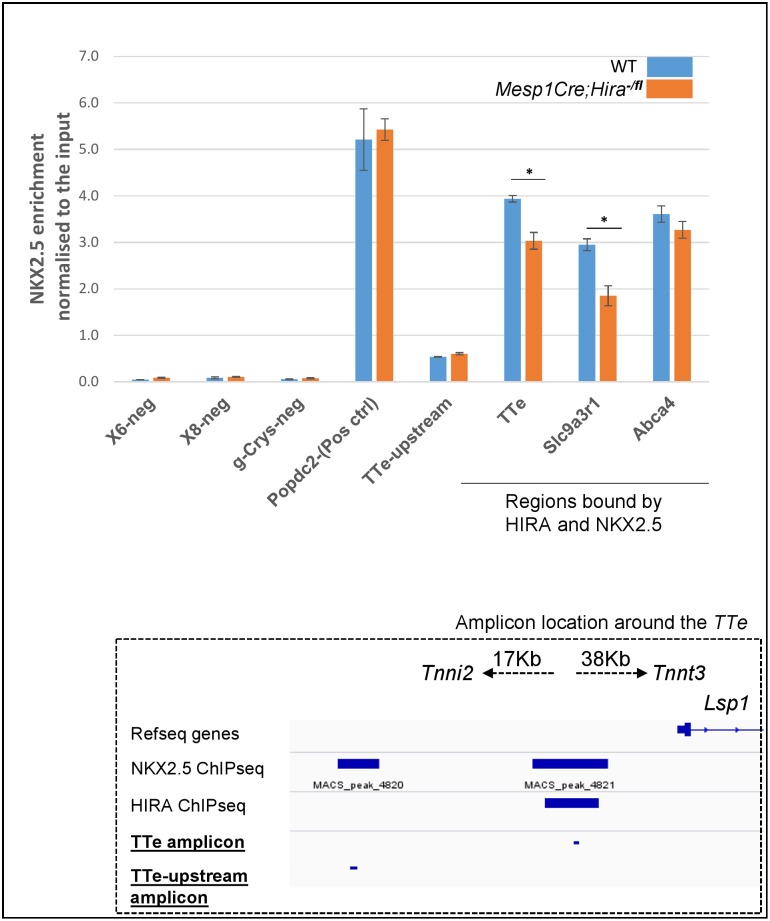
Fig 6. NKX2.5 shows diminished binding at the TTe locus in conditionally HIRA depleted hearts.
NKX2.5 qChIP on 20 WT and Mesp1Cre;Hira-/fl E12.5 hearts. Enrichment at the TTe was confirmed and found to be reduced in mutant hearts. Slc9a3r1 and Abca4 were enriched for both HIRA and NKX2.5 in WT hearts and displayed an increased expression in both Hira and Nkx2.5 mutant hearts. SLc9a3r1, but not Abca4, showed a significantly reduced binding of NKX2.5 in the absence of HIRA. Three different negative regions were tested. A region known to be enriched for NKX2.5, but not HIRA, binding (Popdc2) was used as a NKX2.5 qChIp positive control. Data is normalized to the input (= 100). Errors bars represent the min/max from technical duplicates. Unpaired t-test: p<0.05 *. Subpanel represents the different ChIPseq peaks along with the PCR amplicons used in the different qChIp at TTe, and at a locus bound by NKX2.5, but not by HIRA.
As NKX2.5 interacts with the WHSC1 histone methyltransferase to repress target genes during heart development [25], and in HeLa cells HIRA interacts with WHSC1 [26], we tested whether there was a HIRA-WHSC1 interaction in mouse embryonic hearts using co-immunoprecipitation. We detected an interaction between HIRA and WHSC1 at E12.5 (S5 Fig) and at E14.5 (Fig 7A).
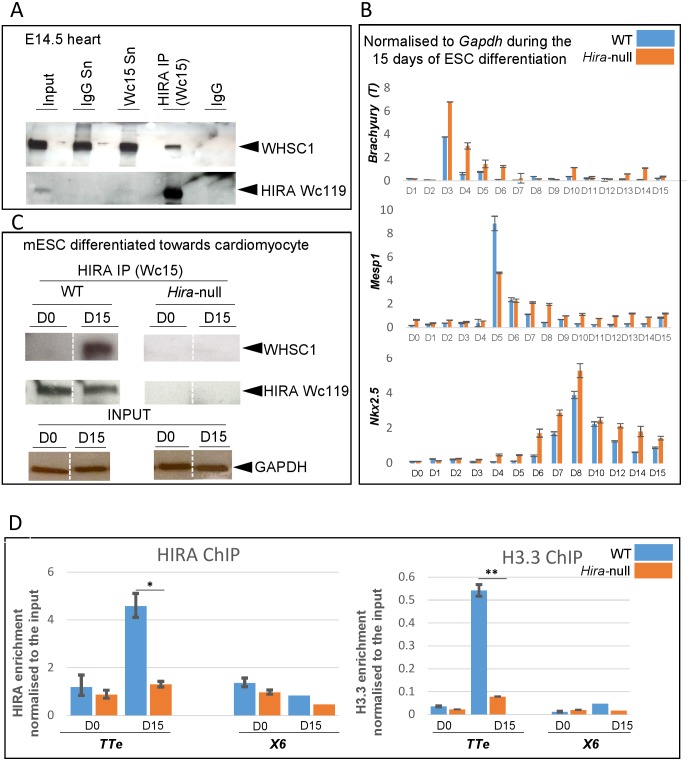
Fig 7. HIRA interacts with WHSC1 and binds to the TTe in the cardiac lineage and heart, and is required for differentiation-associated H3.3 enrichment at this locus.
A. 20 E14.5 hearts from WT embryos were isolated, pooled and immunoprecipitated with HIRA Wc15 antibody and immunoblotted with WHSC1. Presence of HIRA in the IP was verified using Wc119 antibody. B. Mouse ESCs (control and Hira-/- ES cells) were differentiated towards cardiomyocytes over 15 days in LIF free medium following the hanging drop method, and were subjected to RNA and protein isolation. qRT-PCR of Brachyury (T), Mesp1 and Nkx2.5 over the 15 days are presented and demonstrate sequential expression of the early mesoderm, cardiogenic mesoderm, and cardiac markers respectively. Data are shown as levels of mRNA normalized to Gapdh and are representative of five independent experiments. Errors bars represent the standard deviation of technical triplicates. C. Protein isolated from undifferentiated ESCs and 15 days differentiated control and Hira-/- ESCs were immunoprecipitated with anti-HIRA Wc15 antibody and immunoblotted with anti-WHSC1 antibody. The results shown are all from the same western membrane but separated lanes are brought into proximity and separated by dashed bars. GAPDH is used as a control for input. D. HIRA and H3.3 qChIP on differentiating ESCs shows an enrichment of HIRA at the TTe at day 15. H3.3 enrichment at the TTe increases significantly in WT but not in Hira null ESCs following this differentiation. X6 is an intergenic negative control region. Unpaired t-test was used: p<0.05 *, p<0.01 **.
We then used previously reported WT and Hira-null ESCs carrying an HA-tag knock in of H3.3 [4] to investigate potential interactions during the early stages of cardiac differentiation. The cells were differentiated for fifteen days using the aggregation method in order to obtain cardiogenic mesoderm and primitive cardiomyocytes. qRT-PCR was performed to assess the sequential gene expression of specific mesodermal and cardiac markers. Brachyury, Mesp1 and Nkx2.5 were expressed sequentially at days 3, 5 and 8 of differentiation respectively (Fig 7B), and spontaneously beating cardiomyocytes were observed (S1 Video). At day 15 of differentiation towards cardiomyocytes, the HIRA-WHSC1 interaction was observed (Fig 7C).
HIRA regulates H3.3 deposition at the TTe in differentiated embryonic stem cells
Using the same ESC culture conditions as above, we then tested HIRA and H3.3-HA enrichments at the TTe by qChIP at D0 and D15 of differentiation. We found enrichment of HIRA and H3.3 at the TTe in WT, but not Hira null, differentiated cells (Fig 7D).
Discussion
Modification of chromatin is well established as an important mechanism of genetic regulation and is usually associated with covalent modification of histones or deposition of variant histones, or both [27]. In this work we examine the role of the replication independent histone chaperone component HIRA, a known modulator of gene expression [28]. Previous work suggested HIRA acts during vascular EC development [21] and in cardiac neural crest cells [29]. Hira knockout embryos had abnormal cardiac structure and function [10], but as these embryos were delayed with yolk sac vascular anomalies it was not known whether these defects were secondary events.
We successfully bypassed early lethality of Hira-/- embryos using a Hira conditional allele, demonstrating HIRA plays a crucial role in the cardiogenic mesoderm. At E15.5 all Mesp1Cre;Hira-/fl embryos presented with whole body oedema and large VSDs, the majority of which were accompanied by ASDs, defects likely incompatible with life. No live born mutants of this genotype were recovered. Nkx2.5Cre is mainly expressed in ventricular cardiomyocytes of the FHF and SHF, although the extent and degree of expression in the SHF appears to be reporter gene and genetic background dependent [30, 31]. In Nkx2.5Cre;Hira-/fl embryos we detected an overriding aorta, a VSD, and a constriction of the PT in some mutants. The lack of phenotype in Mef2c conditional knockouts suggests that Hira is required in the FHF and ablation in the SHF has no effect or acts to exacerbate the phenotype of FHF mutants. We note that the Nkx2.5Cre used here is a knock-in allele and thus there is the potential for interaction between hemizygous levels of NKX2.5 and complete absence of HIRA. However, we did not observe any phenotype in Nkx2.5Cre;Hira+/fl embryos.
As the Mesp1Cre lineage includes vascular ECs we investigated whether Hira was required for vessel development within this population using Tie2Cre, which is active from E7.5 [32]. Moreover, HIRA has been previously shown to mediate the response to angiogenic signals by upregulating Vegfr1 in yolk sac ECs [21]. The authors of this work concluded that HIRA was required for growth of new blood vessels. However, we found that Tie2Cre;Hira-/fl mutants suffered only from partially penetrant proportionate growth reduction, with no embryonic lethality despite efficient recombination of the conditional allele [21]. The absence of vessel defects and embryo oedema suggests that the oedema observed in Mesp1Cre mutants is not secondary to an EC defect and is therefore more likely due to the septal defects and any disturbance of contractility secondary to alterations in troponin gene expression (see below).
Experiments in chick, using antisense morpholinos directed against Hira, suggested that Hira may play a role in the cardiac neural crest [29] and is required for outflow tract septation. The absence of cardiac and great vessel abnormalities in the Wnt1Cre;Hira-/fl embryos up to E18.5 argues against an autonomous role of HIRA in cNCCs during cardiovascular development. The perinatal death that was observed in Wnt1Cre;Hira-/fl mutants could rather be the result of a neurological problem or a cleft palate defect interfering with swallowing [33], since Wnt1Cre is expressed in craniofacial NCCs. In considering these results it is worth noting the recent discussion of the fact that various murine mutations of genes expressed in NCCs produce different phenotypes to those of other vertebrates [34].
We next asked how absence of HIRA affected the gene expression profile in the developing heart. We chose to analyse Mesp1 conditional knockouts since this was a setting with a 100% penetrant phenotype (by E15.5). In total, less than 2% of the genes expressed in the heart at E12.5 were significantly upregulated or downregulated. Several genes critical for heart development were unaffected by the ablation of Hira, e.g. Myh6/7, Gata4, Nkx2.5 and Tbx5, implying that HIRA has a specific effect on transcription at a small number of target genes. In order to understand the relevant molecular mechanism we performed a HIRA ChIPseq on E12.5 hearts. We detected HIRA enrichment at 6625 loci mapped to 2515 genes, with mapping defined as the presence of a peak in the gene body and/or within 5Kb upstream of the TSS and downstream of TES. There were 47 such genes that had a dysregulated expression in the absence of HIRA by RNA-seq, although this was not a statistically significant increase over chance. Further work, such as chromatin conformation capture, would be required to corroborate peak-gene relationships. It should also be borne in mind that, as for the TTe site discussed below, relevant enhancer binding sites may be >5kb from a target gene. Nevertheless, we reason HIRA acts directly at a relatively small subset of genes, and indirectly modulates expression via more general effects on chromatin structure acting over longer ranges. This is consistent with our finding that just over 7.5% of HIRA peaks at E12.5 were found to overlap with both H3K4me1 and H3K27Ac marks (at E13.5, the closest stage available).
Examination of HIRA binding sites for over representation of sequences that might be responsible for HIRA recruitment revealed that the most frequent sequence had a core consensus site GAGAGAGA, which is equivalent to that identified as the binding motif for the Drosophila melanogaster transcriptional regulator GAGA factor [35]. GAGA factor interacts with FACT, and intriguingly GAGA/FACT associates with Drosophila HIRA. Together, these proteins direct H3.3 replacement to establish chromatin boundaries e.g. at HOX loci [36].
Interestingly, HIRA enrichment at the TTe locus in the wild type situation correlates with an upregulation of cardiac Tnni2 (7 fold) and Tnnt3 (3.2 fold) following conditional ablation in the cardiogenic lineage. The encoded proteins are classified as fast skeletal muscle troponins [37]. The troponin complex binds calcium ions, and through its association with actin and tropomyosin is involved in the regulation of striated muscle contraction. While various troponins are expressed in developing heart at E12.5 [38], skeletal troponins have low expression in cardiac muscle, and upregulation of Tnnt3 has been associated with reduced cardiac contractility [39]. In a Notch1 gain of function mutant, structural defects of the myocardium were attributed to elevated expression of Tnni2 [40]. GO analysis of the RNAseq revealed that several sarcomeric genes (also under the GO term myofibril) were dysregulated. Tnni2, Tnnt3, Casq1, Acta1, Krt8, Krt19 [41] were upregulated and the anion exchanger Slc4a1 and Tnnt1 were downregulated. The disorganised sarcomeric structure we observed within mutant embryonic cardiomyocytes likely contributes to diminished efficiency of contraction [42]. The mouse heart starts beating at E8.5; reduced cardiac output can lead to whole body oedema at E14.5 [43]. Thus, the haemorrhage and oedema we observed at E15.5 could be a consequence of both morphological abnormalities (septal defects) and compromised cardiomyocyte function.
The dysregulation of Tnni2/Tnnt3 was investigated further in the light of recently published work demonstrating that both in the HL-1 cardiomyocyte cell line (DamID screening) and E11.5 whole hearts (ChIPseq) NKX2.5 bound a sequence 17kb downstream of Tnni2 and 38kb upstream of Tnnt3 coincident with our HIRA binding site (Fig 5C) [11, 12]. In hearts of E11.5 and E14.5 embryos with a hypomorphic allele of Nkx2.5 there was upregulation of Tnni2 and Tnnt3 [11], recapitulating what we observed in Hira conditionals, raising the possibility that HIRA and NKX2.5 might co-regulate this locus. Absence of HIRA resulted in a diminution of NKX2.5 binding at the TTe.
The literature suggested a further potential mechanism whereby NKX2.5- and HIRA-regulation might converge. In fibroblasts HIRA was previously described to interact with the histone-lysine N-methyltransferase WHSC1, regulating the H3.3 deposition and H3K36me3 marks at interferon-regulated genes [26]. WHSC1 is a gene haploinsufficient in the Wolf-Hirschhorn syndrome, a human birth defect where congenital heart defects, usually atrial and/or ventricular septal defects, occur in 30–45% of cases [44]. Whsc1-null mice also have atrial and ventricular septal defects [25]. Whereas no Nkx2.5+/- or Whsc1+/- mice had septal defects, such abnormalities were found in one third of double heterozygotes [25]. In support of such a connection we found HIRA and WHSC1 co-immunoprecipitated from embryonic hearts and cardiomyocytes. It will therefore be interesting to investigate whether WHSC1 and HIRA co-regulate Tnni2 and other targets during heart development analysing compound mutants and histone modifications in such mice. Both NKX2.5 and WHSC1 are implicated in human genetic haploinsufficiency resulting in congenital heart defect. However, to our knowledge, no mutations of HIRA have been described in exome sequencing analyses of patients with congenital heart defect, although one study of a Chinese cohort discovered a genetic association between a variant in the 3’ UTR region of HIRA and the diagnosis of tetralogy of Fallot [45].
As HIRA is known to deposit the H3.3 variant within chromatin we examined H3.3 levels at the TTe site in the presence and absence of HIRA. We utilized a cell line model akin to that used by Wamstad and colleagues [24] to examine H3.3 deposition in ESC-derived cardiomyocytes, making use of an HA-knockin to H3.3. We confirmed HIRA enrichment at the TTe in differentiated ESCs (equivalent to Wamstad stages 3 and 4 based upon marker expression and the presence of beating cardiomyocytes) versus undifferentiated ESCs, and observed a 7 fold increase of HIRA-dependent H3.3 deposition at the TTe.
We also examined one of the most significantly down-regulated genes at both E11.5 and E12.5 which had a known role in heart development: Epha3. Epha3 is expressed in the atrioventricular cushion and mesenchymal cap of the developing septum primum from E10.5 [18]. Accordingly, we examined expression in E12.5 Mesp1Cre;Hira-/fl mutants hearts and observed a consistent downregulation of Epha3 in precisely those regions of mutants versus the controls. Interestingly, Epha3-null embryos are perinatal lethals with ASD and cardiac failure, and have earlier atrioventricular cushion defects [18]. This mirrors several defects seen at E12.5 in Mesp1Cre;Hira-/fl mutant hearts. Thus, we reason diminished expression of Epha3 is likely to contribute to the septal defects in Hira mutants. Epha3 expression was not significantly altered in Nkx2.5 hypomorphs [11], nor was there evidence of NKX2.5 binding close to Epha3 in either the DamID [12] or NKX2.5 ChIPseq datasets [11]. There were two HIRA binding regions at Epha3, one of them containing the GAGA consensus site. Thus, we anticipate other HIRA partners might be involved in transcription activation at this locus. The NKX2.5-WHSC1 complex is thought to repress targets [25], so the absence of NXK2.5 binding sites at the down-regulated Epha3 is not unexpected.
In summary, our data provide the first in vivo demonstration in vertebrates of a post-gastrulation requirement for the histone chaperone complex protein HIRA: conditional mutagenesis in cardiogenic mesoderm resulting in severe structural defects and evidence of heart failure leading to embryonic lethality. HIRA was shown to bind to the same enhancer of Tnni2 as the cardiogenic transcription factor NKX2.5, suggesting regulation of specific loci as well as more general effects on chromatin represent an important subset of the pleiotropic functions of HIRA. This work also emphasises HIRA can act in repressive as well as activating situations, as has recently been shown in ESCs and plants [5, 46]. It has previously been shown that the WD repeats and LXXLL motifs are necessary for HIRA-mediated repression in vitro [22]. In ESCs, only a third of enhancer sites bound by HIRA had significantly reduced H3.3 levels in the absence of HIRA [5]. Thus, it will be interesting to determine the role of other H3.3 chaperone associated proteins, such as DAXX, during heart development. Moreover, as a member of a replication independent histone chaperone complex, HIRA might have a particular role in post-mitotic tissues later in life, for instance in the aging of cardiomyocytes and neurones, and in their response to injury.
Supporting Information
A. β-Galactosidase assay on E10.5 and E13.5 HiraSanger/+ and Hira+/+ embryos, either on whole mount or on heart sections as indicated, showing an ubiquitous expression of Hira. B. Schematic representation of the different Hira alleles used in the manuscript. C. Recombination of the floxed allele by Mesp1 driven CRE recombinase was assessed by PCR using DNA from the anterior limb which contains Mesp1 positive cells, detection of the various alleles is shown. D. Recombination was also demonstrated at the mRNA level by the reduced number of reads in RNAseq (22 in control and 3 in mutant). E. Reduced protein level of HIRA in Mesp1Cre;Hira-/fl hearts. This experiment was done by comparing pools of 3 Hira+/fl and 3 Mesp1Cre;Hira-/fl E13.5 hearts. Remaining protein is likely to originate from non-Mesp1-expressing cardiac linages (e.g. circulating cells and ingressing neural crest).
(TIFF)
A. Table summarising the genic distribution of HIRA binding sites and the GAGA motifs across the genome. 45% of HIRA peaks were found to have a GAGA motif. B. Venn diagram displaying the number of genes upregulated in hearts of E12.5 Mesp1Cre;Hira-/fl embryos and in E11.5 Nkx2.5 hypomorph hearts [11]. C. Venn diagram displaying the number of overlaps between HIRA ChIPseq peaks at E12.5 (this study), enhancer signatures H3K4me1 and H3K27Ac at E13.5 (encode database ENCSR663VWL) and NKX2.5 ChIPseq peaks in E11.5 hearts [11] as indicated. D. a. Venn diagram displaying the overlap between genes whose expression is dysregulated in E12.5 Mesp1Cre;Hira-/fl hearts and genes which are enriched for HIRA in WT E12.5 hearts. The enrichment includes any peak in the gene body and/or within 5Kb upstream of the TSS or downstream of TES. b. The equation for the calculation of the probability of having 47 genes (x) in common between two independent groups: 2515 (b, HIRA ChIPseq genes) and 360 (a, RNAseq data) in the mouse genome which has approximately 22 000 genes (n). The result of this hypergeometric probability calculation is not significant: p(x > = 47) = 0.184.
(TIFF)
The enrichment of H3K4me1 and H3K27ac and the lack of repressive H3K27me3 modifications represent a distinct chromatin pattern observed in active enhancers (black box around the TTe). The development of this pattern mirrors the upregulation of Tnni2 seen during the differentiation process and supports the association of the TTe enhancer with its expression. Histone ChIpseq in ESC derived cardiomyocytes and ESCs were obtained from Wamstad and colleagues [24] (Gnomex accession numbers 44R and 7R2).
(TIFF)
A subset of genes affected in Mesp1Cre;Hira-/fl hearts was quantified in Hira+/- mutant hearts using real time PCR. No significant changes were detected in Hira heterozygotes.
(TIFF)
30 embryonic hearts from WT embryos were isolated at E12.5, pooled and immunoprecipitated (IP) with anti-HIRA Wc15 antibody then immunoblotted with anti-WHSC1 antibody. Presence of HIRA in the IP was verified using anti-HIRA Wc119 antibody.
(TIFF)
(TIFF)
Presented here are genes with the lowest p-value. The fold change is presented here as an absolute value. Test applied: Mann-Whitney unpaired, Benjamini Hochberg FDR, p ≤ 0.05, FC ≥ 1.5. The complete list can be accessed with at the GEO database under accession number GSE79937.
(TIFF)
GO analysis of the genes significantly dysregulated in Mesp1Cre;Hira-/fl embryonic hearts that have an over-representation of one or more GO terms that pass the cut-off p-value of 10−4. Terms relating to contractility and myofibril structure are highlighted in orange.
(TIFF)
(TIFF)
(TIFF)
1: Transverse reconstruction of an OPT scan of Mesp1Cre;Hira+/fl and Mesp1Cre;Hira-/fl embryonic trunks at E15.5 with the VSD indicated. 2: 3D reconstruction of the PT in Nkx2.5Cre;Hira+/fl and Nkx2.5Cre;Hira-/fl embryonic hearts at E15.5. 3: Beating ESCs observed in differentiation experiment.
(7Z)
Acknowledgments
We thank Laura Banaszynski and David Allis for the knockin H3.3 ESC and the 12AC5 hybridoma lines, Amelie Calmont for technical assistance, Chela James for bioinformatics advice, Bertrand Vernay and Angela d’Esposito for precious help with OPT. Special thanks to Tony Brooks from UCL Genomics for carrying out RNAseq and ChIPseq. We also thank the Wellcome Trust Sanger Institute and the EUCOMM Consortium for providing the ES lines and mice.
Abbreviations
- ASD
Atrial Septal Defect
- AVSD
Atrioventricular septal defect
- ChIP
Chromatin Immunoprecipitation
- ChIPseq
ChIP followed by sequencing
- ESC
Embryonic stem cell
- EC
Endothelial cells
- EMT
Endothelial to mesenchymal transition
- FHF
First heart field
- GO
Gene ontology
- H&E
Haematoxylin and Eosin
- HUCA
HIRA/UBN1/CABIN1/ASF1a complex
- ISH
In situ hybridization
- cNCC
Cardiac neural crest cell
- OFT
Outflow tract
- OPT
Optical projection tomography
- PT
Pulmonary trunk
- qChIP
Quantitative Chromatin Immunoprecipitation
- qRT-PCR
Quantitative real-time PCR
- RT
Room temperature
- SHF
Second heart field
- TES
Transcriptional end site
- TSS
Transcriptional start site
- TTe
Tnni2-Tnnt3 enhancer
- VSD
Ventricular septum defect
Data Availability
The RNAseq data is deposited at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with accession GSE79937, and ChIPseq data with accession GSE79826.
Funding Statement
This research was funded by British Heart Foundation (grants RG/15/14/31880; FS/11/83/29333 (PJS); and FS/10/031/28395 (ALAC)), Fondation Leducq (FR) grant TNE 15CVD01 (PJS), and supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Moorman A, Webb S, Brown NA, Lamers W, Anderson RH. DEVELOPMENT OF THE HEART: (1) FORMATION OF THE CARDIAC CHAMBERS AND ARTERIAL TRUNKS. Heart. 2003;89(7):806–14. PMC1767747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annual review of physiology. 2012;74:41–68. Epub 2011/11/01. 10.1146/annurev-physiol-020911-153242 . [DOI] [PubMed] [Google Scholar]
- 3.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Molecular cell. 2011;44(6):928–41. Epub 2011/12/27. 10.1016/j.molcel.2011.12.006 . [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–91. Epub 2010/03/10. 10.1016/j.cell.2010.01.003 ; PubMed Central PMCID: PMCPmc2885838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155(1):107–20. Epub 2013/10/01. 10.1016/j.cell.2013.08.061 ; PubMed Central PMCID: PMCPmc3838450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson HE, Wardle J, Korkut ŞV, Murton HE, López-Maury L, Bähler J, et al. The Fission Yeast HIRA Histone Chaperone Is Required for Promoter Silencing and the Suppression of Cryptic Antisense Transcripts. Molecular and cellular biology. 2009;29(18):5158–67. 10.1128/MCB.00698-09. PMC2738286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS genetics. 2007;3(10):1991–2006. Epub 2007/10/31. 10.1371/journal.pgen.0030182 ; PubMed Central PMCID: PMCPmc2041997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rai TS, Cole JJ, Nelson DM, Dikovskaya D, Faller WJ, Vizioli MG, et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes & development. 2014;28(24):2712–25. 10.1101/gad.247528.114. PMC4265675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Developmental cell. 2014;30(3):268–79. Epub 2014/08/05. 10.1016/j.devcel.2014.06.022 ; PubMed Central PMCID: PMCPmc4134436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, et al. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Molecular and cellular biology. 2002;22(7):2318–28. Epub 2002/03/09. ; PubMed Central PMCID: PMCPmc133693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupays L, Shang C, Wilson R, Kotecha S, Wood S, Towers N, et al. Sequential Binding of MEIS1 and NKX2-5 on the Popdc2 Gene: A Mechanism for Spatiotemporal Regulation of Enhancers during Cardiogenesis. Cell Rep. 2015;13(1):183–95. Epub 2015/09/29. 10.1016/j.celrep.2015.08.065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouveret R, Waardenberg AJ, Schonrock N, Ramialison M, Doan T, de Jong D, et al. NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets. 2015;4 10.7554/eLife.06942 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bible PW, Kanno Y, Wei L, Brooks SR, O’Shea JJ, Morasso MI, et al. PAPST, a User Friendly and Powerful Java Platform for ChIP-Seq Peak Co-Localization Analysis and Beyond. PLoS ONE. 2015;10(5):e0127285 10.1371/journal.pone.0127285. PMC4430287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Yang P. In vitro differentiation of mouse embryonic stem (mES) cells using the hanging drop method. Journal of visualized experiments: JoVE. 2008;(17). Epub 2008/12/11. 10.3791/825 ; PubMed Central PMCID: PMCPmc3253613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilming LG, Snoeren CA, van Rijswijk A, Grosveld F, Meijers C. The murine homologue of HIRA, a DiGeorge syndrome candidate gene, is expressed in embryonic structures affected in human CATCH22 patients. Human molecular genetics. 1997;6(2):247–58. Epub 1997/02/01. . [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Shun MC, Dickson AK, Engelman AN. Embryonic Lethality Due to Arrested Cardiac Development in Psip1/Hdgfrp2 Double-Deficient Mice. PLoS One. 2015;10(9):e0137797 Epub 2015/09/15. 10.1371/journal.pone.0137797 ; PubMed Central PMCID: PMCPmc4569352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson RH, Brown NA, Mohun TJ. Insights regarding the normal and abnormal formation of the atrial and ventricular septal structures. Clinical Anatomy. 2015:n/a–n/a. 10.1002/ca.22627 [DOI] [PubMed] [Google Scholar]
- 18.Stephen LJ, Fawkes AL, Verhoeve A, Lemke G, Brown A. A critical role for the EphA3 receptor tyrosine kinase in heart development. Dev Biol. 2007;302(1):66–79. Epub 2006/10/19. 10.1016/j.ydbio.2006.08.058 . [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Nomura-Kitabayashi A, Sultana N, Cai W, Cai X, Moon AM, et al. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Developmental Biology. 2014;390(1):68–79. 10.1016/j.ydbio.2014.02.023. 10.1016/j.ydbio.2014.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Developmental Biology. 2008;323(1):98–104. 10.1016/j.ydbio.2008.08.013. 10.1016/j.ydbio.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, et al. Regulation of Angiogenesis by Histone Chaperone HIRA-mediated Incorporation of Lysine 56-acetylated Histone H3.3 at Chromatin Domains of Endothelial Genes. Journal of Biological Chemistry. 2010;285(53):41567–77. 10.1074/jbc.M110.190025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad A, Takami Y, Nakayama T. WD dipeptide motifs and LXXLL motif of chicken HIRA are necessary for transcription repression and the latter motif is essential for interaction with histone deacetylase-2 in vivo. Biochemical and biophysical research communications. 2003;312(4):1266–72. Epub 2003/12/04. . [DOI] [PubMed] [Google Scholar]
- 23.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq Identification of Weakly Conserved Heart Enhancers. Nature genetics. 2010;42(9):806–10. 10.1038/ng.650. PMC3138496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, et al. Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell. 2012;151(1):206–20. 10.1016/j.cell.2012.07.035. PMC3462286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460(7252):287–91. Epub 2009/06/02. 10.1038/nature08086 . [DOI] [PubMed] [Google Scholar]
- 26.Sarai N, Nimura K, Tamura T, Kanno T, Patel MC, Heightman TD, et al. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. The EMBO journal. 2013;32(17):2392–406. Epub 2013/08/08. 10.1038/emboj.2013.176 ; PubMed Central PMCID: PMCPmc3770338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–89. 10.1038/nrm3941 [DOI] [PubMed] [Google Scholar]
- 28.Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell reports. 2013;3(4):1012–9. 10.1016/j.celrep.2013.03.026. PMC3974909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrell MJ, Stadt H, Wallis KT, Scambler P, Hixon RL, Wolfe R, et al. HIRA, a DiGeorge syndrome candidate gene, is required for cardiac outflow tract septation. Circulation research. 1999;84(2):127–35. Epub 1999/02/05. . [DOI] [PubMed] [Google Scholar]
- 30.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis (New York, NY: 2000). 2001;31(4):176–80. Epub 2002/01/10. . [DOI] [PubMed] [Google Scholar]
- 31.Ramsbottom SA, Sharma V, Rhee HJ, Eley L, Phillips HM, Rigby HF, et al. Vangl2-regulated polarisation of second heart field-derived cells is required for outflow tract lengthening during cardiac development. PLoS genetics. 2014;10(12):e1004871 Epub 2014/12/19. 10.1371/journal.pgen.1004871 ; PubMed Central PMCID: PMCPmc4270488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–42. Epub 2001/02/13. 10.1006/dbio.2000.0106 . [DOI] [PubMed] [Google Scholar]
- 33.Trainor PA. Craniofacial Birth Defects: The Role of Neural Crest Cells in the Etiology and Pathogenesis of Treacher Collins Syndrome and the Potential for Prevention. American journal of medical genetics Part A. 2010;0(12):2984–94. 10.1002/ajmg.a.33454. PMC3686507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barriga EH, Trainor PA, Bronner M, Mayor R. Animal models for studying neural crest development: is the mouse different? Development (Cambridge, England). 2015;142(9):1555–60. Epub 2015/04/30. 10.1242/dev.121590 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adkins NL, Hagerman TA, Georgel P. GAGA protein: a multi-faceted transcription factor. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2006;84(4):559–67. Epub 2006/08/29. 10.1139/o06-062 . [DOI] [PubMed] [Google Scholar]
- 36.Nakayama T, Shimojima T, Hirose S. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development (Cambridge, England). 2012;139(24):4582–90. Epub 2012/11/09. 10.1242/dev.083246 . [DOI] [PubMed] [Google Scholar]
- 37.Ochala J. Thin filament proteins mutations associated with skeletal myopathies: defective regulation of muscle contraction. Journal of molecular medicine (Berlin, Germany). 2008;86(11):1197–204. Epub 2008/06/25. 10.1007/s00109-008-0380-9 . [DOI] [PubMed] [Google Scholar]
- 38.Franco D, Lamers WH, Moorman AF. Patterns of expression in the developing myocardium: towards a morphologically integrated transcriptional model. Cardiovascular research. 1998;38(1):25–53. Epub 1998/07/31. . [DOI] [PubMed] [Google Scholar]
- 39.Huang QQ, Feng HZ, Liu J, Du J, Stull LB, Moravec CS, et al. Co-expression of skeletal and cardiac troponin T decreases mouse cardiac function. American journal of physiology Cell physiology. 2008;294(1):C213–22. Epub 2007/10/26. 10.1152/ajpcell.00146.2007 . [DOI] [PubMed] [Google Scholar]
- 40.Watanabe Y, Kokubo H, Miyagawa-Tomita S, Endo M, Igarashi K, Aisaki K, et al. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development (Cambridge, England). 2006;133(9):1625–34. Epub 2006/03/24. 10.1242/dev.02344 . [DOI] [PubMed] [Google Scholar]
- 41.Stone MR, O'Neill A, Lovering RM, Strong J, Resneck WG, Reed PW, et al. Absence of keratin 19 in mice causes skeletal myopathy with mitochondrial and sarcolemmal reorganization. Journal of cell science. 2007;120(Pt 22):3999–4008. Epub 2007/11/01. 10.1242/jcs.009241 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Risebro CA, Searles RG, Melville AA, Ehler E, Jina N, Shah S, et al. Prox1 maintains muscle structure and growth in the developing heart. Development (Cambridge, England). 2009;136(3):495–505. Epub 2008/12/19. 10.1242/dev.030007 ; PubMed Central PMCID: PMCPmc2655234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng X, Wu X, Druso JE, Wei H, Park AY, Kraus MS, et al. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(18):6638–43. Epub 2008/05/02. 10.1073/pnas.0802319105 ; PubMed Central PMCID: PMCPmc2373317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends in genetics: TIG. 2005;21(3):188–95. Epub 2005/03/01. 10.1016/j.tig.2005.01.008 . [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Zhang J, Cao Y, Diao L, Wang H, Ma X, et al. [Sequence analyses of HIRA gene 3'UTR region and related microRNA]. Zhonghua yi xue za zhi. 2014;94(16):1223–6. Epub 2014/06/14. . [PubMed] [Google Scholar]
- 46.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MCP. Maize rough sheath2 and Its Arabidopsis Orthologue ASYMMETRIC LEAVES1 Interact with HIRA, a Predicted Histone Chaperone, to Maintain knox Gene Silencing and Determinacy during Organogenesis. The Plant cell. 2005;17(11):2886–98. 10.1105/tpc.105.035477. PMC1276017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. β-Galactosidase assay on E10.5 and E13.5 HiraSanger/+ and Hira+/+ embryos, either on whole mount or on heart sections as indicated, showing an ubiquitous expression of Hira. B. Schematic representation of the different Hira alleles used in the manuscript. C. Recombination of the floxed allele by Mesp1 driven CRE recombinase was assessed by PCR using DNA from the anterior limb which contains Mesp1 positive cells, detection of the various alleles is shown. D. Recombination was also demonstrated at the mRNA level by the reduced number of reads in RNAseq (22 in control and 3 in mutant). E. Reduced protein level of HIRA in Mesp1Cre;Hira-/fl hearts. This experiment was done by comparing pools of 3 Hira+/fl and 3 Mesp1Cre;Hira-/fl E13.5 hearts. Remaining protein is likely to originate from non-Mesp1-expressing cardiac linages (e.g. circulating cells and ingressing neural crest).
(TIFF)
A. Table summarising the genic distribution of HIRA binding sites and the GAGA motifs across the genome. 45% of HIRA peaks were found to have a GAGA motif. B. Venn diagram displaying the number of genes upregulated in hearts of E12.5 Mesp1Cre;Hira-/fl embryos and in E11.5 Nkx2.5 hypomorph hearts [11]. C. Venn diagram displaying the number of overlaps between HIRA ChIPseq peaks at E12.5 (this study), enhancer signatures H3K4me1 and H3K27Ac at E13.5 (encode database ENCSR663VWL) and NKX2.5 ChIPseq peaks in E11.5 hearts [11] as indicated. D. a. Venn diagram displaying the overlap between genes whose expression is dysregulated in E12.5 Mesp1Cre;Hira-/fl hearts and genes which are enriched for HIRA in WT E12.5 hearts. The enrichment includes any peak in the gene body and/or within 5Kb upstream of the TSS or downstream of TES. b. The equation for the calculation of the probability of having 47 genes (x) in common between two independent groups: 2515 (b, HIRA ChIPseq genes) and 360 (a, RNAseq data) in the mouse genome which has approximately 22 000 genes (n). The result of this hypergeometric probability calculation is not significant: p(x > = 47) = 0.184.
(TIFF)
The enrichment of H3K4me1 and H3K27ac and the lack of repressive H3K27me3 modifications represent a distinct chromatin pattern observed in active enhancers (black box around the TTe). The development of this pattern mirrors the upregulation of Tnni2 seen during the differentiation process and supports the association of the TTe enhancer with its expression. Histone ChIpseq in ESC derived cardiomyocytes and ESCs were obtained from Wamstad and colleagues [24] (Gnomex accession numbers 44R and 7R2).
(TIFF)
A subset of genes affected in Mesp1Cre;Hira-/fl hearts was quantified in Hira+/- mutant hearts using real time PCR. No significant changes were detected in Hira heterozygotes.
(TIFF)
30 embryonic hearts from WT embryos were isolated at E12.5, pooled and immunoprecipitated (IP) with anti-HIRA Wc15 antibody then immunoblotted with anti-WHSC1 antibody. Presence of HIRA in the IP was verified using anti-HIRA Wc119 antibody.
(TIFF)
(TIFF)
Presented here are genes with the lowest p-value. The fold change is presented here as an absolute value. Test applied: Mann-Whitney unpaired, Benjamini Hochberg FDR, p ≤ 0.05, FC ≥ 1.5. The complete list can be accessed with at the GEO database under accession number GSE79937.
(TIFF)
GO analysis of the genes significantly dysregulated in Mesp1Cre;Hira-/fl embryonic hearts that have an over-representation of one or more GO terms that pass the cut-off p-value of 10−4. Terms relating to contractility and myofibril structure are highlighted in orange.
(TIFF)
(TIFF)
(TIFF)
1: Transverse reconstruction of an OPT scan of Mesp1Cre;Hira+/fl and Mesp1Cre;Hira-/fl embryonic trunks at E15.5 with the VSD indicated. 2: 3D reconstruction of the PT in Nkx2.5Cre;Hira+/fl and Nkx2.5Cre;Hira-/fl embryonic hearts at E15.5. 3: Beating ESCs observed in differentiation experiment.
(7Z)
Data Availability Statement
The RNAseq data is deposited at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with accession GSE79937, and ChIPseq data with accession GSE79826.