Time-resolved x-ray solution scattering reveals the conformational signaling mechanism of a bacterial phytochrome.
Abstract
Phytochromes are light sensor proteins found in plants, bacteria, and fungi. They function by converting a photon absorption event into a conformational signal that propagates from the chromophore through the entire protein. However, the structure of the photoactivated state and the conformational changes that lead to it are not known. We report time-resolved x-ray scattering of the full-length phytochrome from Deinococcus radiodurans on micro- and millisecond time scales. We identify a twist of the histidine kinase output domains with respect to the chromophore-binding domains as the dominant change between the photoactivated and resting states. The time-resolved data further show that the structural changes up to the microsecond time scales are small and localized in the chromophore-binding domains. The global structural change occurs within a few milliseconds, coinciding with the formation of the spectroscopic meta-Rc state. Our findings establish key elements of the signaling mechanism of full-length bacterial phytochromes.
INTRODUCTION
Phytochromes are red-light sensors, which allow bacteria, plants, and fungi to respond to the environment. Phytochromes exist in two (meta-) stable states termed Pr and Pfr, which absorb red and far-red light, respectively. They are photochromic and can be switched reversibly by red (Pr→Pfr) or far-red (Pfr→Pr) illumination. Most phytochromes have Pr as their resting state, but some bacterial variants relax to Pfr in the dark (1).
Most phytochromes are dimeric proteins, and all exhibit a modular domain architecture. A photosensory core is conserved between phytochromes in plants, bacteria, and fungi. It usually consists of Per/Arndt/Sim (PAS), cyclic guanosine monophosphate phosphodiesterase/adenyl cyclase/FhlA (GAF), and phytochrome-specific (PHY) domains (Fig. 1A). The bilin chromophore is attached to the PAS or GAF domain, and different output domains are found on the C-terminal side of the photosensory core (2). In cyanobacteria and bacteria, the output module commonly consists of histidine kinase domains, and the protein is the first part of a two-component signaling mechanism (3, 4). The histidine kinase module comprises a dimerization and histidine-containing phosphotransfer (DHp) domain and a catalytic adenosine 5′-triphosphate–binding (CA) domain. Plant phytochromes have C-terminal domains that resemble those of histidine kinases but lack the crucial histidine for phosphorylation. Instead, they function as serine-threonine kinases in vitro (5), which is just one of several biochemical output mechanisms in vivo (6–8).
Fig. 1. Phytochrome structure and photoconversion.
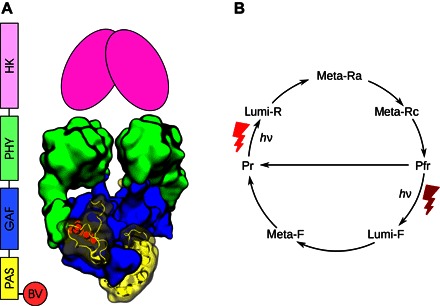
(A) Sequential and structural representation of the D. radiodurans phytochrome, based on PDB entry 4Q0J (33). The putative histidine kinase (HK) has not been described by crystallography and is shown schematically. BV, biliverdin chromophore. (B) Simplified photocycle for bacterial phytochromes. The number of intermediates varies from phytochrome to phytochrome, as discussed in the text.
The discovery of bacterial homologs led to the determination of a series of three-dimensional structures of phytochrome fragments (9–16). Figure 1 provides a representative illustration of the domain sequence and structure of bacterial phytochromes. The PAS and GAF domains assume a tightly packed, globular structure, which is stabilized by a knot formed by the backbone (9). The PHY domain is connected to the GAF domain through a long helical spine and through the so-called PHY “arm” or “tongue,” which caps the chromophore on the outside of the dimeric protein (10, 11). The PHY tongue is folded as a β sheet in Pr and as an α helix in Pfr (10, 11, 14). There are no published three-dimensional structures of phytochromes with their histidine kinase output domains included.
Phytochrome photoconversion has been studied by optical spectroscopy since the discovery of the protein in the 1950s. Initially, the plant phytochrome now called PhyA was subjected to flash photolysis measurements (17–21). Figure 2 summarizes representative time constants found by kinetic analyses. Several spectroscopic intermediate species were resolved, together describing a complex reaction mechanism leading from Pr to Pfr. Similar conversion behavior was observed in the cyanobacterial homolog Cph1 from Synechocystis sp. PCC 6803 (22, 23), although fewer time constants were observed. The bacterial phytochrome Agp1 from Agrobacterium tumefaciens has also been studied by flash spectroscopy (24, 25), and three intermediates have been identified. For Cph1 and Agp1, transient proton release to the solvent has been observed spectroscopically (see Fig. 2) (23, 24).
Fig. 2. Spectroscopic photoconversion in plant, cyanobacterial, and bacterial phytochromes.
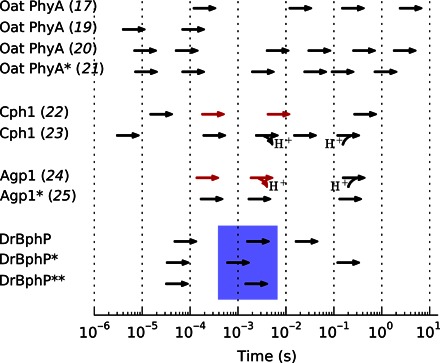
Proton release and uptake are marked. Red arrows signify a kinetic H/D isotope effect larger than 3/2. Asterisks mark N-terminal fragments (**PAS-GAF, *PAS-GAF-PHY); all other samples are full-length. DrBphP refers to the results of the present study (see figs. S1 and S2), and the blue box indicates the steps found to involve the main structural transformation.
The summary of spectroscopic time constants in Fig. 2 indicates that the complexity of the Pr→Pfr photoconversion process increases from bacteria to cyanobacteria to plants. Focusing on the simplest case of bacterial phytochromes, the photocycle can be described by three intermediates usually termed lumi-R, meta-Ra, and meta-Rc (Fig. 1B) (25). Lumi-R, the first ground state intermediate, forms on time scales shorter than those shown in Fig. 2 and converts into meta-Ra within microseconds. Then, a proton is released to the solvent to form the meta-Rc state within a few milliseconds, and finally, the photoproduct, Pfr, is formed in tens or hundreds of milliseconds. In the last step, partial proton uptake occurs (23, 24). We note that this photocycle is devised from spectroscopy, which is primarily sensitive to the state of the chromophore and only indirectly sensitive to structural changes in the protein. To detect the protein structural changes that accompany phytochrome photoconversion, alternative experimental approaches such as time-resolved x-ray scattering are needed (26).
Structurally, the tetrapyrrole chromophore is thought to isomerize within picoseconds of excitation with red light (27). There is now a broad consensus in the literature that this step involves the Z→E isomerization of the double bond between the 15,16-carbon atoms, which leads to a rotation of the chromophore D-ring (21, 28, 29). Much less is known about how this structural change couples to the remainder of the protein. On the basis of time-resolved x-ray scattering and crystal structures, we recently proposed a large-scale opening of the dimer in the PAS-GAF-PHY fragment from Deinococcus radiodurans in Pfr and found that this change was caused by the refolding of the PHY tongue (14). The latter result has now been corroborated with spectroscopy (30–32).
Little is known about how this structural signal affects the full-length protein. On the one hand, electron micrographs of the phytochrome from D. radiodurans indicated that some of the dimers open up at the histidine kinase, but the electron density of the output domains was only partially observed in the ensemble-averaged reconstructions (33, 34). We have recently found that for mutated variants of the D. radiodurans phytochrome the dimerization interface between sister output domains can, in principle, be broken by light absorption (35). Also, the only crystal structure of a near–full-length phytochrome reveals an open conformation (12), but it should be noted that this phytochrome is not a histidine kinase and that its output domain was truncated. Thus, the breaking of the dimer interface at the output domains should be considered as a possible mode of action for bacterial phytochromes. On the other hand, all known crystal structures of bacterial histidine kinases and phosphatases indicate that the dimer interface stays intact upon activation of the output domains (36). Thus, the evidence at this stage is not conclusive. It remains unclear how light absorption modulates the activity of the output domains of phytochromes.
Here, we describe how the structure of the bacterial phytochrome from D. radiodurans evolves during the course of photoconversion. We present time-resolved x-ray scattering and optical absorption signals from the full-length protein as well as from two shorter fragments. In addition, we also present ab initio structural models derived from the x-ray scattering measurements of the full-length protein. The results reveal how the histidine kinase domains are rearranged during photoconversion. By comparing with time-resolved spectroscopy, we show that this structural transition coincides with the transition into the meta-Rc state.
RESULTS
We performed nano- to millisecond transient absorption spectroscopy with visible light for PAS-GAF, PAS-GAF-PHY, and full-length samples. The data (figs. S1 and S2) establish that the D. radiodurans phytochrome has a photocycle similar to that of the A. tumefaciens homolog (Fig. 1B) (24, 25). The derived time constants (53 μs and 3.5 ms for PAS-GAF; 55 μs, 1.0 ms, and 200 ms for PAS-GAF-PHY; and 80 μs, 2.6 ms, and 27 ms for full-length samples) are indicated in Fig. 2.
Figure 3 shows the time-resolved x-ray scattering signal from all three samples. Unlike the transient absorption spectra (fig. S1), the x-ray data show very little response on microsecond time scales. This is remarkable considering that time-resolved x-ray scattering can resolve conformational changes of less than 1 Å (root mean square deviation), provided that secondary structural elements move (37–41). We therefore conclude that the structural rearrangements in the early intermediates are small.
Fig. 3. Time-resolved x-ray scattering data.
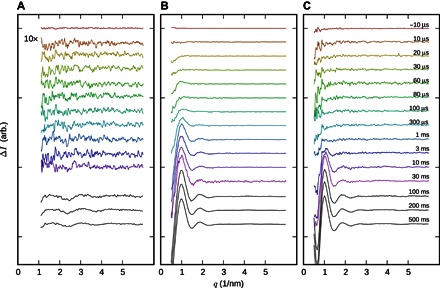
(A) PAS-GAF fragment. (B) PAS-GAF-PHY fragment. arb., arbitrary. (C) Full-length protein. Delay times after red-light (671 nm) excitation are color-coded and marked in (C). Black curves are from a separate experiment with 10-ms time resolution. The scattering vector modulus is defined as q = 4πsin(θ)/λ.
On the millisecond time scale, a significant change in the x-ray scattering is observed for all three samples. Figure 4 shows the time dependence of this signal, which was extracted from the data by matrix decomposition (see Materials and Methods). The spectroscopic evolution is superimposed. By comparing the structural and spectroscopic kinetics, it becomes clear that the major structural change is associated with the second-to-last spectroscopic conversion. In terms of the photocycle shown in Fig. 1B, this corresponds to the formation of the meta-Rc state. For the full-length protein, the scattering signal evolves for tens of milliseconds into its final form and thus extends into the last transition of the photocycle.
Fig. 4. Kinetics of photoconversion.
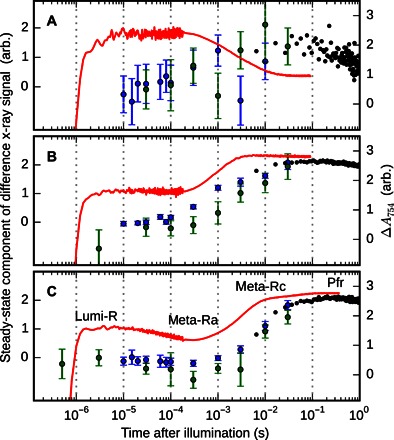
(A to C) Traces for PAS-GAF (A), PAS-GAFPHY (B), and full-length samples (C). For the transient absorption data, the absorbance at 754 nm is shown (red lines). For the x-ray data, the similarity of the time-resolved data to the “steady-state” component (the response between 500 ms and 1 s) is shown (circles). This is extracted as described in the Materials and Methods. Error bars represent 95% confidence intervals based on individual detector images. Blue, green, and black circles represent independent measurements performed at ID09b, BioCARS, and cSAXS, respectively. The half-times for the structural transitions are approximately 1, 2, and 6 ms for the PAS-GAF, PAS-GAF-PHY, and full-length constructs, respectively.
A comparison of the shape of the difference x-ray scattering profiles for millisecond delay times reveals that the light-induced x-ray scattering of the PAS-GAF-PHY fragment (Fig. 3B) and that of the full-length protein (Fig. 3C) have a similar positive peak at q = 1/nm but have different shapes for q < 1/nm. For the PAS-GAF-PHY fragment, difference scattering curves with a positive peak at q = 1/nm have been interpreted as an opening of the PHY domains (14). We note that the reappearance of this peak in the full-length phytochrome does not necessarily imply a similar motion. This is because the signals reflect changes in the pair distance distribution function of the proteins (42), and this function is expected to be significantly different for the PAS-GAF-PHY and full-length molecules.
To describe the structural change in the full-length phytochrome, we evaluated the radius of gyration (Rg) and the structural parameter Vc = I0[∫qI(q)dq]− 1. Vc is a small-angle scattering invariant that depends on the particle volume per self-correlation length (43), and I0 denotes the forward scattering. The parameters were computed from small-angle x-ray scattering (SAXS) data of the phytochrome in the Pr and Pfr states (fig. S3A). The SAXS data in Pr were taken from the study of Takala et al. (35), whereas the Pfr data were reconstructed from the Pr data combined with the difference x-ray scattering signal at millisecond delay times (Fig. 3) (38). This procedure overcomes the difficulty of incomplete photoconversion and helps in avoiding potential experimental artifacts such as radiation damage and monomerization of the proteins, which may arise in separate SAXS experiments in the Pr and Pfr states. In the reconstruction, the difference data are weighted by the photocoversion yield [α = [Pfr]∕([Pr] + [Pfr])]. We experimentally estimated α to be 0.38 ± 0.06 by titrating the laser power in the difference x-ray scattering experiment (see Fig. 5 and Materials and Methods).
Fig. 5. Photoconversion efficiency as function of laser energy density.
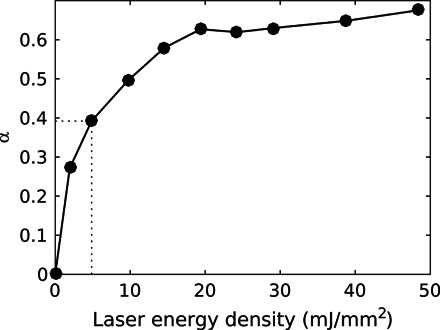
The photoconversion yield was estimated for each laser energy density by considering that the plateau yield, as given by the relative absorptions of the pure Pr and Pfr states, is 64%. The dotted line indicates the laser energy density used to acquire the data.
Considering the expected approximate size and shape of the dimeric protein, the SAXS analysis yielded reasonable values for Rg and Vc in the Pr state (Table 1 and table S1). By transition to the Pfr state, Rg did not change significantly, and Vc changed by <1% compared to the Pr state (see α = 0.38 in table S1). We verified that Rg and Vc are invariable in the range 0.26 < α < 0.45, covering the margin of error for α (see table S1). We also measured the difference x-ray scattering for a concentration series and found that ΔRg and ΔI0 converged to stable values for low concentrations (fig. S3B, inset). The small change of the SAXS invariants from Pr to Pfr shows that the conformational change, albeit global, does not grossly alter the protein shape.
Table 1. SAXS parameters.
Parameters pertaining to the Pr state were derived from absolute scattering data. The Pfr parameters were derived from reconstructed scattering curves, obtained by adding the difference curves (Fig. 3) at various assumed levels of conversion. The parameters Rg and I0 were estimated using conventional Guinier analysis. See table S1 for a more extensive version of this table.
| Rg (Å) | I (q = 0) | Vc (Å2) | |
| Pr | 53.25 (±0.05) | 37.6 | 1139 |
| Pfr (α = 0.45) | 53.32 (±0.05) | 36.7 | 1131 |
| Pfr (α = 0.38) | 53.27 (±0.05) | 36.5 | 1129 |
| Pfr (α = 0.32) | 53.19 (±0.1) | 36.3 | 1127 |
| Pfr (α = 0.26) | 53.23 (±0.1) | 36 | 1125 |
| Pfr (α = 0.19) | 53.01 (±0.1) | 35.4 | 1118 |
| Pfr (α = 0.13) | 52.69 (±0.1) | 34.2 | 1107 |
To gain more detailed structural insight, we performed ab initio structural modeling against absolute SAXS data corresponding to the Pr and Pfr states. The models were computed using DAMMIN (44) and aligned with DAMAVER (45). Table S2 summarizes the ab initio modeling. In Fig. 6, the models are seen to resemble the expected shape of a full-length phytochrome. A thicker part at the bottom of the electron density contains the PAS and GAF domains. A slimmer part represents the PHY domains and connects to a thicker part at the top of the figure where the DHp and CA domains of the histidine kinase are expected. Overlaying the models with our previously proposed solution structure for the PAS-GAF-PHY fragment from D. radiodurans (14) and a homology model based on the cytoplasmic part of a histidine kinase from Thermotoga maritima [Protein Data Bank (PDB) code 2C2A] shows good agreement.
Fig. 6. Pr and Pfr shape reconstructions.
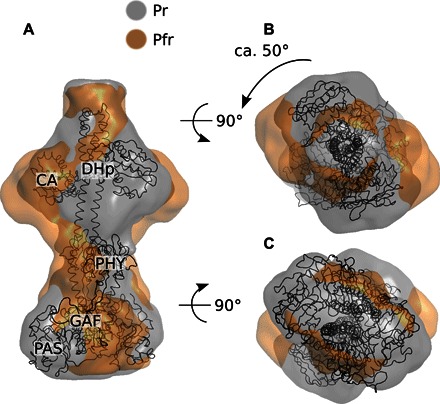
(A to C) Side (A), top (B), and bottom (C) views of the homology model of the full-length phytochrome from D. radiodurans (shown as cartoons) and the ab initio models generated by DAMMIN (shown as surfaces). Illumination induces a rotation in the kinase domain relative to the photosensory domain.
The models indicate that the quaternary structure of the protein does not change when going from Pr (gray) to Pfr (orange). We found that none of the generated ab inito models of the Pfr state shows an opening of the dimer at the DHp interface. This agrees with the observed small changes of the invariants Rg and Vc. Instead, the dimerization interface of the kinase domain is kept intact, and a rotation of the output domain with respect to the PAS/GAF domains is observed. The rotational motion is robust across the confindence range of α and was observed, to different extents, for all values with the exception of α = 0.13.
DISCUSSION
Phytochromes modulate their biochemical output activity when absorbing light. It is currently unclear how this is achieved structurally (46). It has been speculated that bacterial phytochromes break the dimer interface between the output domains when photoactivated (35, 47). The idea was supported by the crystal structures of PAS-GAF-PHY fragments (14, 47) and by light-induced monomerization, detected for a phytochrome mutant (35). These studies used truncated and mutated phytochromes. Here, the model-independent descriptors ΔRg and ΔVc and the ab initio reconstructions show that this opening of the dimer does not occur in the full-length protein. This is reasonable considering that additional dimer interface area is present between the DHp domains of the full-length phytochrome.
Instead, our x-ray solution scattering data suggest a rotational motion. This rearrangement of the output domains is supported by limited proteolysis experiments (48) and by measurements of the susceptibility for cleavage by tobacco etch virus protease (49), indicating light-induced changes close to the phosphoaccepting histidine in the phytochrome from D. radiodurans. Transduction of the signal by a rotational motion is also supported by the quaternary arrangement of the protein. The two monomers are coiled around each other, and it is perceivable that light modifies the degree of coiling. This is reminiscent of linker modules in histidine kinases, which are often coiled coils or domains with long helical spines (36). Functional data on the engineered histidine kinase YF1 (50) show that the angular orientation of the coiled coil linker is crucial for kinase functionality (51). Moreover, histidine kinases are typically dimeric and the CA domains change binding site when activated (36). Light-induced rearrangements of the DHp domains may control the balance of this equilibrium. Molecular predictions based on genome analysis of many histidine kinases have revealed movements of the CA domains (52), which appear to be very similar to the structural rearrangements that we observe (fig. S5).
This argument demonstrates that bacterial phytochromes behave similarly to other histidine kinases. Phytochromes that do not have histidine kinase output domains, such as plant phytochromes (53), may have other gating mechanisms. However, some shared features are expected considering the high sequence homology and the highly conserved PAS-GAF-PHY or GAF-PHY domain architecture of the photosensory core (3).
The time-resolved x-ray scattering data show that the structural changes before formation of the meta-Rc state are below the detection limit. Even very small displacements of secondary structural elements, unfolding or refolding of parts of the protein, and domain movements would likely result in a detectable signal (37–39, 41, 54). Thus, the early rearrangement must be small. We believe that motions of a few side chains, protons, water molecules, or the chromophore are possible candidates.
Although the magnitude and shape of the time-resolved x-ray signal vary markedly between PAS-GAF and the larger fragments, the main signal from each sample evolves on approximately the same time scale (Fig. 4). We therefore suggest that the rate-limiting step of the structural conversion is internal to the PAS-GAF domains. If so, the millisecond process in the PAS-GAF moiety controls the larger-scale transformation of the rest of the molecule. The process is only slightly retarded by the attached PHY or PHY–histidine kinase modules, from 1 to 2 and 6 ms, respectively. Because the main structural change coincides with the formation of the meta-Rc state and the same transition has been associated with the transfer of a proton to the surrounding solution (23, 24), we suggest that the deprotonation step ultimately controls the biochemical activity of phytochromes.
For the full-length protein, a final process is detected spectroscopically, which overlaps with the tail of the evolution of the time-resolved x-ray data. The present data do not provide direct information on the nature of this process. However, given the general direction of signal propagation, it is plausible that this process represents final rearrangements of the output domains, which weakly feed back onto the chromophore. Because the functional role of the D. radiodurans phytochrome is uncertain (3), there is a possibility that this last process corresponds to more significant structural changes in other phytochromes.
In the main structural transition, the PHY, the DHp, and the CA domains react in unison (Figs. 3 and 4). The data do not indicate any intermediate states, where, for example, the PHY domains but not the DHp or CA domains have rearranged. The role of the PHY domain is that of a mechanical linker, similar to a transmission shaft, which tightly couples the output and sensory domains. This is consistent with the fact that the output domains alter the energetics of the back reaction (31, 35).
In conclusion, the results presented in this study establish that the structural rearrangements in the excited state of the chromophore, in lumi-R, and in meta-Ra, are small in scale (Fig. 3). Changes within the chromophore-binding domains of the protein likely control the entire structural photoconversion process (Fig. 4). An effective rotation in the output domains is the end product of structural photoconversion (Fig. 6), either via an actual rotation or through an altered binding geometry for the CA domains.
Together with previous findings, this establishes key elements of the structural photocycle of bacterial phytochromes. Small-scale motions close to the chromophore (this study) lead to refolding of the PHY tongue [Fig. 7, arrow A; identified by Takala et al. (14)]. The refolding of the PHY tongue has been assigned to the transition from meta-R to Pfr in a phytochrome from Rhodopseudomonas palustris (30) and in Cph2 from Synechocystis sp. (32). We find here that the structural activation of the entire phytochrome occurs at the same stage of the photocycle. In PAS-GAF-PHY, refolding of the PHY tongue leads to the opening of the dimer (14). In the full-length phytochrome, the changes in the PHY tongue instead lead to a screw motion and rotation of the output domains (Fig. 7, arrow B). All this is happening in one concerted rearrangement, demonstrating the tight coupling across the entire phytochrome.
Fig. 7. Schematic of structural change in the D. radiodurans phytochrome.
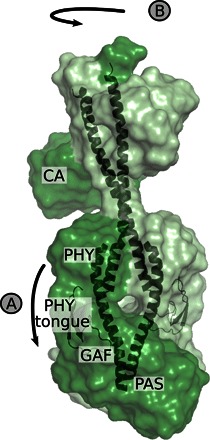
(A) Following the study by Takala et al. (14), light-induced changes in the photosensory domains cause the refolding of the PHY tongue. (B) Here, we found that this rearrangement is translated into rotation of the kinase domains and a change in position of the CA domains. The signal is relayed through the PHY domains and the central helices.
MATERIALS AND METHODS
Protein production, purification, and preparation
Phytochrome samples were produced and purified as detailed in Lehtivuori et al. (55). Large sample volumes were used in the time resolved x-ray experiments, typically with 500 μl loaded at a time. Protein concentrations varied around 25 ± 5 mg/ml, and a 30 mM tris buffer (pH, 8), with no additional components, was used throughout. Details of the sample delivery system are provided in Björling et al. (26). Concentration artifacts for these exact conditions were previously ruled out (26). For time-resolved spectroscopic experiments, the protein was diluted in the same buffer to 1.5 to 2.7 mg/ml. For conventional SAXS, samples were diluted as described below.
Time-resolved absorption spectroscopy
Flash spectroscopy at nanosecond time resolution was performed for the PAS-GAF, PAS-GAF-PHY, and full-length samples, as illustrated in figs. S1 and S2.
Measurements
The kinetics were measured at room temperature using a modified Luzchem laser flash photolysis system (mLFP111 prototype from Luzchem Co.) (56). The excitation pulses were at 700 nm, with power density of 1.2 mJ/cm2, duration of 10 ns, and repetition rate of 0.25 Hz. The pulses were generated by a tunable Ti:sapphire laser (Solar TII CF125 with built-in second harmonic generation), pumped by the second harmonic of a Nd:yttrium aluminum garnet laser (Solar TII LF-117). The continuous wave probe light was from a tungsten halogen lamp (AvaLight-HAL, Avantes). The time-resolved absorbance changes were monitored separately at 10 selected wavelengths ranging from 610 to 810 nm, in a random order to avoid systematic error. To improve the signal-to-noise ratio, interference filters (bandpass, ±15 nm) were inserted before and after the sample position and 100 measurements were averaged at each wavelength. The optical response was recorded with a photomultiplier tube (Hamamatsu, R7400U-20) and digitized in an oscilloscope (300 MHz; Tektronix, TDS3032B). The sample sat in a vertically mounted glass capillary with an inner diameter close to 1.1 mm (VITREX, microhematocrit) with an absorbance at 700 nm of about 0.2 to 0.4/mm. To avoid excessive sample degradation, the sample solution (volume, 400 μl) was cycled using a peristaltic pump (Ismatec, Reglo Digital) at a flow rate of 0.5 ml/min through a glass reservoir, the capillary, and connecting Teflon tubing (1-mm inner diameter). A far-red diode (750 ± 5 nm, 3 mW; Leading-Tech Laser Co.) was used to revert the sample to the Pr state by constant illumination through the Teflon tubing.
Kinetic analysis
Each time-resolved data set was analyzed using the program PygSpec (56). The time-dependent difference between the sample absorbance at time t and in the ground state was analyzed by approximating the data by a sum of exponentials convoluted with the Gaussian pulse. The expression
was used, where A0(λ) is the final difference spectrum, and Ai(λ) (with i = 1, …, N) is the transient component spectrum, each associated with a decay time τi. The parameter β is an empirical quantity that represents the nonexponential behavior often found in biological samples. The fitting expression is merely a convenient analytical expression, and there is no meaningful relation between β and the actual distribution. However, it serves as a reasonable mathematical tool for estimating the decay time constants, τi, from nonexponential decays. The minimum number of exponential components was determined by evaluation of fit residuals, and N was increased until the SD of weighted residuals did not improve further.
X-ray scattering data collection
SAXS measurements
Conventional SAXS data of the protein in Pr state were collected at BM29 at the European Synchrotron Radiation Facility (ESRF) at room temperature as described elsewhere (14, 35). The collected patterns were subjected to standard Guinier analysis to retrieve Rg and I0. Data recorded at low and high concentrations (1.3 and 22 mg/ml, respectively) were merged at q ≈ 1.5/nm.
Pump-probe time-resolved x-ray scattering
Time-resolved laser pump and x-ray–probe experiments were carried out at the BioCARS beamline 14-ID-B of the Advanced Photon Source and at beamline ID09B of the ESRF. Pink beams with peak energies of 12.0 and 14.0 keV were used at BioCARS and ID09B, respectively. The sample was excited using red lasers at 10 Hz, either a 5-ns pulse with a wavelength of 660 nm at an energy density of 2 mJ/mm2 (ID09B) or a 2-ps pulse at 680 nm at the same energy density (BioCARS). At both facilities, the sample was recovered after each red laser pulse using a shutter-controlled flash of a continuous laser (750 nm), at several times the energy density of the excitation pulse. Integrating CCD (charge-coupled device) detectors were used for all experiments, and multiple pump-probe shots were averaged onto the same image, to maximize the signal-to-noise ratio and the experimental duty cycle without saturating the detector. Hundreds of these images were then collected for each time point. Sample volumes of several hundred microliters were used to dilute any radiation-induced damage to the sample. The protein solution was pumped continuously to replace the sample volume for each laser exposure. These measurements were done at room temperatures.
The time delay was defined as the time between the arrivals of the laser and x-ray pulses. Positive delays were always interleaved with negative time points, which served as blank images. To avoid the influence of occasional air bubbles, images where the integrated scattering over 17/nm < q < 20/nm deviated with more than 2% from the median for each sample were rejected. All images were then normalized over the range 14/nm < q < 16/nm, where water heating displays an isosbestic point. Difference scattering curves for each time delay were produced by subtracting for each positive-delay image the appropriately interpolated average of the preceding and subsequent negative-delay images. Finally, difference scattering curves from heating induced by a 1470-nm laser were used to subtract out effects of laser-induced heating.
Time-resolved experiments using a rapid readout detector
The measurements were performed at the cSAXS beamline X12SA at the Swiss Light Source. The millisecond resolution data in Figs. 3 and 4 were collected as described by Takala et al. (14). The difference x-ray scattering data used for the structural analysis (fig. S3B) were collected as follows: an x-ray energy of 11.2 keV was used, and the detector was read out at 25 Hz over the course of a 5.5-s x-ray exposure. Excitation of the sample was achieved using a 671-nm laser pulse 1 s after the first x-rays hit the sample. The dependence of the difference scattering signal on excitation pulse energy was measured by varying the length of the laser pulses (Fig. 5). Most of the data were collected at an energy density of ca. 5 mJ/mm2, corresponding to a 10-ms laser pulse. To ensure that the protein was in Pr at the beginning of each experiment (0.5 s after the first x-rays), the sample was exposed to a 50-ms laser pulse at 789 nm, corresponding to an energy density of ca. 7 mJ/mm2. To obtain difference scattering patterns, subsequent x-ray exposures with the 671-nm laser were subtracted from exposures without the laser. For each “laser on” cycle, the neighboring “laser off ” cycles were then averaged and subtracted from the laser on cycle. The sample solution was stationary in the x-ray beam during each x-ray exposure, and a new sample was supplied by pumping the solution in between cycles. A reservoir solution of several hundred microliters of sample was kept at 20°C, and the capillary was kept at room temperature. As for the data recorded in pump-probe geometry, the scattering curves were normalized at 14/nm < q < 16/nm. Data deviating from the median integrated scattering in the region 17/nm < q < 20/nm by more than 5% were considered outliers and were rejected.
Analysis of x-ray scattering data
Kinetic analysis of time-resolved x-ray data
In Fig. 4, the structural process is represented by the in-growth of the steady-state signal. This information can be extracted by decomposing the overall signal into a set of N basis curves, each with an associated time evolution. Thus, ΔI (q, τ) is expressed as a matrix D, which is factored into a matrix S of basis curves and a matrix T of time evolutions
| (1) |
If either S or T is known, the other can be extracted from the data by solving Eq. 1 in the least-squares sense. For this particular case, two components were used (N = 2) to define S. The first was a constant offset, found to be almost insignificant, and the other was the steady-state signal, as approximated by the difference scattering over 500 ms < τ < 1 s. For the pump-probe data, each repeat (each individual detector image) was considered as its own observation, so that the spread in T for each delay time τ could be used to estimate the uncertainty. This was done by letting each repeat contribute an element to T, and by binning identical delay times only after solving Eq. 1. Thus, Fig. 4 also displays the 95% confidence intervals as error bars.
SAXS analysis software
The pair distance distribution functions of scattering curves was calculated using GNOM (57). Difference scattering data were processed using our own code, and conventional SAXS parameters for all values of α were computed using programs from the ATSAS package (58), with the exception of the SAXS invariant Vc and molecular mass that were calculated as described by Rambo and Tainer (43).
Determining ΔRg and ΔI0 from difference x-ray scattering data
Changes in Rg and I0 were evaluated with an adapted Guinier analysis where the difference scattering at low scattering angles was approximated as
similar to that of Levantino et al. (59). I0 (Pr) and Rg (Pr) were extracted from the Pr SAXS data, and the corresponding values for the Pfr state were recovered through minimizing the Euclidean distance between the data and the fit.
Reconstruction of Pfr SAXS curves
Low q difference x-ray scattering (q < 1/nm) measured at low concentration (7.2 mg/ml) was merged with high-concentration data (30 mg/ml) for higher q values. The absolute SAXS curve recorded at BM29 in the Pr state (35) was scaled and added to the difference data. This effectively yielded a reconstructed absolute SAXS curve representing the pure Pfr state. For this procedure, the photoconversion yield (α) had to be determined. We measured the magnitude of the difference x-ray scattering at q = 1/nm as a function of laser power (Fig. 5). It is known that the highest achievable yield of Pfr is approximately 64%, which defines the upper end of the y scale. From the figure, we estimated α = 0.38 ± 0.06. Absolute SAXS curves in Pfr were calculated for α = {0.13, 0.19, 0.26, 0.32, 0.38, 0.45}.
Ab initio modeling of difference x-ray scattering
To assess the overall structure of the Pr and Pfr states, ab initio models were generated using DAMMIN (44). Models were calculated in rounds. Twenty DAMMIN models were calculated for each scattering curve. Because phytochromes are expected to be elongated homodimeric proteins, the symmetry was fixed as P2, and the shape was assumed to be prolate along the protein. The assumption on twofold symmetry rests on the solved crystal structures of the D. radiodurans phytochrome photosensory domain [for example, PDB code 4Q0J (33)] as well as the near-symmetric arrangement seen in histidine kinases (36). The starting shape was a sphere. For each curve, the 20 generated models were aligned using DAMAVER (45). This gave a model that was used as input for a second DAMMIN round to generate 10 additional structures. The resulting models were compared using DAMSEL, and the model with least average normalized spatial discrepancy was chosen as the representative model.
Construction of homology models
A homology model for the full-length D. radiodurans phytochrome was constructed by combining known structures and homology information. The output domain was modeled using the SWISS-MODEL web interface (60), with the cytoplasmic portion of a histidine kinase from T. maritima (PDB code 2C2A) as a monomeric template. The model dimer was then derived using symmetry operations of the template structure, and the histidine kinase model extended in the N-terminal direction with an ideal helix containing the D. radiodurans phytochrome residues 492 to 501.
The model of the output domain was combined with our previously proposed solution structure of the Pr state of PAS-GAF-PHY (14) in a number of steps. First, to reconcile the registry mismatch between the two parts of the connecting helices from the PAS-GAF-PHY fragment and the model output domain, a series of molecular dynamics simulations were run, where the backbone of residues 492 to 501 on the output domain was restrained by progressively higher force constants to overlap with the corresponding residues on the PAS-GAF-PHY fragments. Next, after merging the models to form the full-length molecule, these restraints were progressively removed, and the new helical arrangement was found to be stable. Last, nonpolar contacts and salt bridges between the connecting helices of opposing monomers were manually formed in a short simulation where these residues (495-496, 496-495, 502-502, 509-509, and 516-516) were artificially bound together, after which they were stable in all subsequent runs.
The quality of the final homology model, although subjected to the choices made during its construction, was evaluated by running a 500-ns equilibrium simulation, with all restraints removed. The region connecting the PAS-GAF-PHY fragment with the output domain was seen to be entirely stable, whereas the positions of the loosely linked CA domains were incompletely sampled on this time scale.
Supplementary Material
Acknowledgments
We thank A. Liukkonen for preparing parts of the protein samples used in this study. Funding: The authors acknowledge funding from The Swedish Foundation for International Cooperation in Research and Higher Education. S.W. acknowledges funding from the European Research Council and the Foundation of Strategic Research, Sweden. J.A.I., H.T., and H.L. acknowledge funding from the Academy of Finland and Finnish Cultural Foundation. M.H. was funded by Marie Curie grant IEF-GA-2013-CHE-624864 ANISOPROTEINXRAY. Beamline access at ID09b and BM29 of the ESRF is acknowledged. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. The use of BioCARS was also supported by NIH National Institute of General Medical Sciences grant 1R24GM111072. The time-resolved setup at BioCARS was funded in part through a collaboration with P. Anfinrud (NIH/National Institute of Diabetes and Digestive and Kidney Diseases). Author contributions: A.B., O.B., H.T., J.A.I., and S.W. designed the study. A.B., O.B., H.L., H.T., A.J.H., M.P., M.H., S.N., L.H., R.H., I.K., A.M., G.N., D.K., M.W., J.A.I., and S.W. contributed significantly to the time-resolved x-ray scattering experiments. A.B. and O.B. analyzed the data and constructed the structural models. H.L., V.C., and N.V.T. performed the spectroscopic measurements and analyzed the data. S.W. and J.A.I. supervised all aspects of the project. A.B., O.B., and S.W. wrote the paper with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/8/e1600920/DC1
table S1. SAXS parameters.
table S2. Ab initio modeling.
fig. S1. Time-resolved absorption spectra.
fig. S2. Decay-associated spectra extracted from time-resolved absorption spectroscopy.
fig. S3. Concentration dependence of x-ray scattering data and Guinier fits.
fig. S4. SAXS data in Pr and Pfr.
fig. S5. Comparison of reconstructed envelopes and the structural model from the study of Dago et al. (52).
REFERENCES AND NOTES
- 1.Karniol B., Vierstra R. D., The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. U.S.A. 100, 2807–2812 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Möglich A., Yang X., Ayers R. A., Moffat K., Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 61, 21–47 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Auldridge M. E., Forest K. T., Bacterial phytochromes: More than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Bhoo S.-H., Davis S. J., Walker J., Karniol B., Vierstra R. D., Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414, 776–779 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Yeh K.-C., Lagarias J. C., Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. U.S.A. 95, 13976–13981 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin K. A., Quail P. H., Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes J., Phytochrome cytoplasmic signaling. Annu. Rev. Plant Biol. 64, 377–402 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Bae G., Choi G., Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281–311 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D., A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Essen L.-O., Mailliet J., Hughes J., The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X., Kuk J., Moffat K., Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellini D., Papiz M. Z., Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20, 1436–1446 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Anders K., Daminelli-Widany G., Mroginski M. A., von Stetten D., Essen L.-O., Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288, 35714–35725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., Westenhoff S., Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., Vierstra R. D., Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., Matysik J., Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linschitz H., Kasche V., Butler W. L., Siegelman H. W., The kinetics of phytochrome conversion. J. Biol. Chem. 241, 3395–3403 (1966). [PubMed] [Google Scholar]
- 18.Kendrick R. E., Spruit C. J. P., Phototransformations of phytochrome. Photochem. Photobiol. 26, 201–214 (1977). [DOI] [PubMed] [Google Scholar]
- 19.Aramendia P. F., Ruzsicska B. P., Braslavsky S. E., Schaffner K., Laser flash photolysis of 124-kilodalton oat phytochrome in water and deuterium oxide solutions: Formation and decay of the I700 intermediates. Biochemistry 26, 1418–1422 (1987). [Google Scholar]
- 20.Schmidt P., Gertsch T., Remberg A., Gärtner W., Braslavsky S. E., Schaffner K., The complexity of the Pr to Pfr phototransformation kinetics is an intrinsic property of native phytochrome. Photochem. Photobiol. 68, 754–761 (1998). [Google Scholar]
- 21.Robben U., Lindner I., Gärtner W., New open-chain tetrapyrroles as chromophores in the plant photoreceptor phytochrome. J. Am. Chem. Soc. 130, 11303–11311 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Remberg A., Lindner I., Lamparter T., Hughes J., Kneip C., Hildebrandt P., Braslavsky S. E., Gärtner W., Schaffner K., Raman spectroscopic and light-induced kinetic characterization of a recombinant phytochrome of the cyanobacterium Synechocystis. Biochemistry 36, 13389–13395 (1997). [DOI] [PubMed] [Google Scholar]
- 23.van Thor J. J., Borucki B., Crielaard W., Otto H., Lamparter T., Hughes J., Hellingwerf K. J., Heyn M. P., Light-induced proton release and proton uptake reactions in the cyanobacterial phytochrome Cph1. Biochemistry 40, 11460–11471 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Borucki B., von Stetten D., Seibeck S., Lamparter T., Michael N., Mroginski M. A., Otto H., Murgida D. H., Heyn M. P., Hildebrandt P., Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. J. Biol. Chem. 280, 34358–34364 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Borucki B., Seibeck S., Heyn M. P., Lamparter T., Characterization of the covalent and noncovalent adducts of Agp1 phytochrome assembled with biliverdin and phycocyanobilin by circular dichroism and flash photolysis. Biochemistry 48, 6305–6317 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Björling A., Berntsson O., Takala H., Gallagher K. D., Patel H., Gustavsson E., St. Peter R., Duong P., Nugent A., Zhang F., Berntsen P., Appio R., Rajkovic I., Lehtivuori H., Panman M. R., Hoernke M., Niebling S., Harimoorthy R., Lamparter T., Stojković E. A., Ihalainen J. A., Westenhoff S., Ubiquitous structural signaling in bacterial phytochromes. J. Phys. Chem. Lett. 6, 3379–3383 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Dasgupta J., Frontiera R. R., Taylor K. C., Lagarias J. C., Mathies R. A., Ultrafast excited-state isomerization in phytochrome revealed by femtosecond stimulated Raman spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 1784–1789 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X., Ren Z., Kuk J., Moffat K., Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mroginski M. A., Murgida D. H., Hildebrandt P., The chromophore structural changes during the photocycle of phytochrome: A combined resonance Raman and quantum chemical approach. Acc. Chem. Res. 40, 258–266 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Stojković E. A., Toh K. C., Alexandre M. T. A., Baclayon M., Moffat K., Kennis J. T. M., FTIR spectroscopy revealing light-dependent refolding of the conserved tongue region of bacteriophytochrome. J. Phys. Chem. Lett. 5, 2512–2515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velazquez Escobar F., Piwowarski P., Salewski J., Michael N., Fernandez Lopez M., Rupp A., Qureshi B. M., Scheerer P., Bartl F., Frankenberg-Dinkel N., Siebert F., Andrea Mroginski M., Hildebrandt P., A protonation-coupled feedback mechanism controls the signalling process in bathy phytochromes. Nat. Chem. 7, 423–430 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Anders K., Gutt A., Gärtner W., Essen L.-O., Phototransformation of the red light sensor cyanobacterial phytochrome 2 from Synechocystis species depends on its tongue motifs. J. Biol. Chem. 289, 25590–25600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgie E. S., Wang T., Bussell A. N., Walker J. M., Li H., Vierstra R. D., Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289, 24573–24587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Zhang J., Vierstra R. D., Li H., Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 10872–10877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takala H., Björling A., Linna M., Westenhoff S., Ihalainen J. A., Light-induced changes in the dimerization interface of bacteriophytochromes. J. Biol. Chem. 290, 16383–16392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhate M. P., Molnar K. S., Goulian M., DeGrado W. F., Signal transduction in histidine kinases: Insights from new structures. Structure 23, 981–994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson M., Malmerberg E., Westenhoff S., Katona G., Cammarata M., Wöhri A. B., Johansson L. C., Ewald F., Eklund M., Wulff M., Davidsson J., Neutze R., Structural dynamics of light-driven proton pumps. Structure 17, 1265–1275 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Kim T. W., Lee J. H., Choi J., Kim K. H., van Wilderen L. J., Guerin L., Kim Y., Jung Y. O., Yang C., Kim J., Wulff M., van Thor J. J., Ihee H., Protein structural dynamics of photoactive yellow protein in solution revealed by pump-probe x-ray solution scattering. J. Am. Chem. Soc. 134, 3145–3153 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Cammarata M., Levantino M., Schotte F., Anfinrud P. A., Ewald F., Choi J., Cupane A., Wulff M., Ihee H., Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering. Nat. Methods 5, 881–886 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho H. S., Dashdorj N., Schotte F., Graber T., Henning R., Anfinrud P., Protein structural dynamics in solution unveiled via 100-ps time-resolved x-ray scattering. Proc. Natl. Acad. Sci. U.S.A. 107, 7281–7286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnlund D., Johansson L. C., Wickstrand C., Barty A., Williams G. J., Malmerberg E., Davidsson J., Milathianaki D., DePonte D. P., Shoeman R. L., Wang D., James D., Katona G., Westenhoff S., White T. A., Aquila A., Bari S., Berntsen P., Bogan M., van Driel T. B., Doak R. B., Kjær K. S., Frank M., Fromme R., Grotjohann I., Henning R., Hunter M. S., Kirian R. A., Kosheleva I., Kupitz C., Liang M., Martin A. V., Nielsen M. M., Messerschmidt M., Seibert M. M., Sjöhamn J., Stellato F., Weierstall U., Zatsepin N. A., Spence J. C. H., Fromme P., Schlichting I., Boutet S., Groenhof G., Chapman H. N., Neutze R., Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat. Methods 11, 923–926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plech A., Wulff M., Bratos S., Mirloup F., Vuilleumier R., Schotte F., Anfinrud P. A., Visualizing chemical reactions in solution by picosecond x-ray diffraction. Phys. Rev. Lett. 92, 125505 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Rambo R. P., Tainer J. A., Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun D. I., Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volkov V. V., Svergun D. I., Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagano S., From photon to signal in phytochromes: Similarities and differences between prokaryotic and plant phytochromes. J. Plant Res. 129, 123–135 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Baker A. W., Forest K. T., Structural biology: Action at a distance in a light receptor. Nature 509, 174–175 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Takala H., Lehtivuori H., Hammarén H., Hytönen V. P., Ihalainen J. A., Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry 53, 7076–7085 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Burgie E. S., Zhang J., Vierstra R. D., Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Möglich A., Ayers R. A., Moffat K., Design and signaling mechanism of light-regulated histidine kinases, J. Mol. Biol. 385, 1433–1444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohlendorf R., Schumacher C. H., Richter F., Möglich A., Library-aided probing of linker determinants in hybrid photoreceptors. ACS Synth. Biol. 10.1021/acssynbio.6b00028 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Dago A. E., Schug A., Procaccini A., Hoch J. A., Weigt M., Szurmant H., Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 109, E1733–E1742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vierstra R. D., Zhang J., Phytochrome signaling: Solving the Gordian knot with microbial relatives. Trends Plant Sci. 16, 417–426 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Vaidya A. T., Top D., Manahan C. C., Tokuda J. M., Zhang S., Pollack L., Young M. W., Crane B. R., Flavin reduction activates Drosophila cryptochrome, Proc. Natl. Acad. Sci. U.S.A. 110, 20455–20460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehtivuori H., Rissanen I., Takala H., Bamford J., Tkachenko N. V., Ihalainen J. A., Fluorescence properties of the chromophore-binding domain of bacteriophytochrome from Deinococcus radiodurans. J. Phys. Chem. B 117, 11049–11057 (2013). [DOI] [PubMed] [Google Scholar]
- 56.N. V. Tkachenko, Optical Spectroscopy: Methods and Instrumentations (Elsevier, Amsterdam, 2006). [Google Scholar]
- 57.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992). [Google Scholar]
- 58.Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., Svergun D. I., New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45 (Pt. 2), 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levantino M., Schirò G., Lemke H. T., Cottone G., Glownia J. M., Zhu D., Chollet M., Ihee H., Cupane A., Cammarata M., Ultrafast myoglobin structural dynamics observed with an X-ray free-electron laser. Nat. Commun. 6, 6772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T., SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/8/e1600920/DC1
table S1. SAXS parameters.
table S2. Ab initio modeling.
fig. S1. Time-resolved absorption spectra.
fig. S2. Decay-associated spectra extracted from time-resolved absorption spectroscopy.
fig. S3. Concentration dependence of x-ray scattering data and Guinier fits.
fig. S4. SAXS data in Pr and Pfr.
fig. S5. Comparison of reconstructed envelopes and the structural model from the study of Dago et al. (52).


