Abstract
The insulin-like growth factor (IGF) system is a well-studied growth regulatory pathway implicated in breast cancer biology. Clinical trials testing monoclonal antibodies directed against the type I IGF receptor (IGF1R) in combination with estrogen receptor-α (ER) targeting have been completed, but failed to show benefits in patients with endocrine resistant tumors compared to ER targeting alone. We have previously shown that the closely related insulin receptor (InsR) is expressed in tamoxifen resistant breast cancer cells. Here we examined if inhibition of InsR affected tamoxifen-resistant (TamR) breast cancer cells. InsR function was inhibited by three different mechanisms: InsR shRNA, a small InsR blocking peptide, S961 and an InsR monoclonal antibody (mAb). Suppression of InsR function by these methods in TamR cells successfully blocked insulin-mediated signaling, monolayer proliferation, cell cycle progression and anchorage-independent growth. This strategy was not effective in parental cells likely due to the presence of IGFR/InsR hybrid receptors. Down-regulation of IGF1R in conjunction with InsR inhibition was more effective in blocking IGF- and insulin-mediated signaling and growth in parental cells compared to single receptor targeting alone. Our findings show TamR cells were stimulated by InsR and were not sensitive to IGF1R inhibition, whereas in tamoxifen-sensitive parental cancer cells, the presence of both receptors, especially hybrid receptors, allowed cross-reactivity of ligand-mediated activation and growth. To suppress the IGF system, targeting of both IGF1R and InsR is optimal in endocrine sensitive and resistant breast cancer.
Introduction
Approximately 75% of the breast cancer cases express estrogen receptor-α (ER), representing the most prevalent breast cancer subtype (1). Patients with ER-positive breast cancer can be treated by inhibiting ER function. This strategy has been successful in early stage and advanced breast cancer (2, 3), but a significant proportion of patients never responded to ER inhibition (de novo or primary resistance) or have progression after a prolonged period of therapy (acquired or secondary resistance) (4, 5). Endocrine resistance still poses a key clinical problem. Recently, targeting of mTORC1 and CDK4/6 have been used to treat ER-positive tumors (6, 7), but there is still a need for additional strategies, aiming to delay or ideally overcome resistance to endocrine therapy.
Insulin-like growth factor (IGF) signaling occurs through multiple receptors including the type I IGF receptor (IGF1R), insulin receptor (InsR), and hybrid IGF1R/InsR. This receptor system has been implicated in cancer development as well as crosstalk with ER, suggesting that it may contribute to the regulation of ER-positive breast cancer (8, 9). IGF1R is an estrogen regulated gene and enhances ER transcriptional activity, suggesting co-targeting of receptors might be clinical useful (10, 11). A number of anti-IGF inhibitors including anti-IGF1R monoclonal antibodies (mAbs), tyrosine kinases inhibitors (TKIs) and ligand neutralizing antibodies were developed primarily to target IGF1R and IGF ligands while leaving InsR unperturbed (12). Despite the hope that anti-IGF1R targeted therapies would provide clinical benefit in endocrine resistant breast cancer, we showed that tamoxifen resistant (TamR) cells lacked IGF1R expression (13). This finding was validated in women with breast cancers – recurrent endocrine treated tumors showed lower level of IGF1R compared to the pre-treated tumors (14, 15). Thus, it would be unlikely for anti-IGF1R mAbs to have clinical activity in endocrine resistant cells. These observations likely explain why the results of phase III clinical trials of anti-IGF1R mAbs tested in endocrine resistant population have been negative (16).
Unlike IGF1R, InsR is not an estrogen regulated gene and its level remained intact in TamR cells. InsR is closely related to IGF1R, sharing 84% similarity within catalytic domain, 45–65% in ligand-binding domain and more than 50% in the overall amino acid sequence (17). The highly homologous InsR activates almost identical downstream signaling cascades in a ligand-dependent fashion. On the loss of IGF1R function, osteoblasts shifted from IGF- to insulin-mediated growth and differentiation (18). Down-regulation of IGF1R in breast cancer increased sensitivity to insulin (19). In addition, a patient tumor developed an increased InsR gene copy number while being treated with, and eventually becoming resistant to endocrine therapy (20). Although InsR expression in cancer has been documented for several decades (21–24), InsR inhibition has been intentionally avoided because of concern over disrupting glucose homeostasis.
InsR inhibitors have been developed as dual IGF1R/InsR tyrosine kinase inhibitors: BMS-754807 and OSI-906. These two drugs have completed several clinical trials, including a phase II study against ER+ breast cancer resistant to aromatase inhibitors. The trial has completed but the results have not been disclosed (NCT01225172). Early clinical evidence suggests that TKIs are safer than originally anticipated. Although hyperglycemia was evident in patients treated with OSI-906, encouraging disease control was observed in patients (25, 26).
In this study, we determined that InsR signaling serves as a bypass pathway and compensates for the loss of IGF1R in TamR breast cancer cells. We suppressed InsR functions using three different mechanisms in TamR versus parental breast cancer cells. Our data showed that InsR inhibition alone blocked signaling and cell proliferation in TamR cells but not in the parental cells. When anti-IGF1R mAb was given in conjunction with InsR inhibitor, a complete suppression of insulin-stimulated growth in parental cells was observed, suggesting the involvement of hybrid receptors in the mediation of IGF/insulin in breast cancer cells. Thus, dual inhibition of IGF1R and InsR is necessary for optimal suppression of this signaling system.
Results
TamR cells were more sensitive to insulin treatment compared to their parental cells
MCF-7L and T47D are ER-positive human breast cancer cell lines and are estrogen sensitive and are inhibited by selective estrogen receptor modulators such as tamoxifen. We previously generated TamR cells from MCF-7L and T47D and showed reduced IGF1R expression levels and a lack of efficacy of anti-IGF1R monoclonal antibodies in TamR cells. However, AEW541, a dual TKI that targets both IGF1R and InsR was able to inhibit insulin- and IGF-stimulated signaling and growth (13).
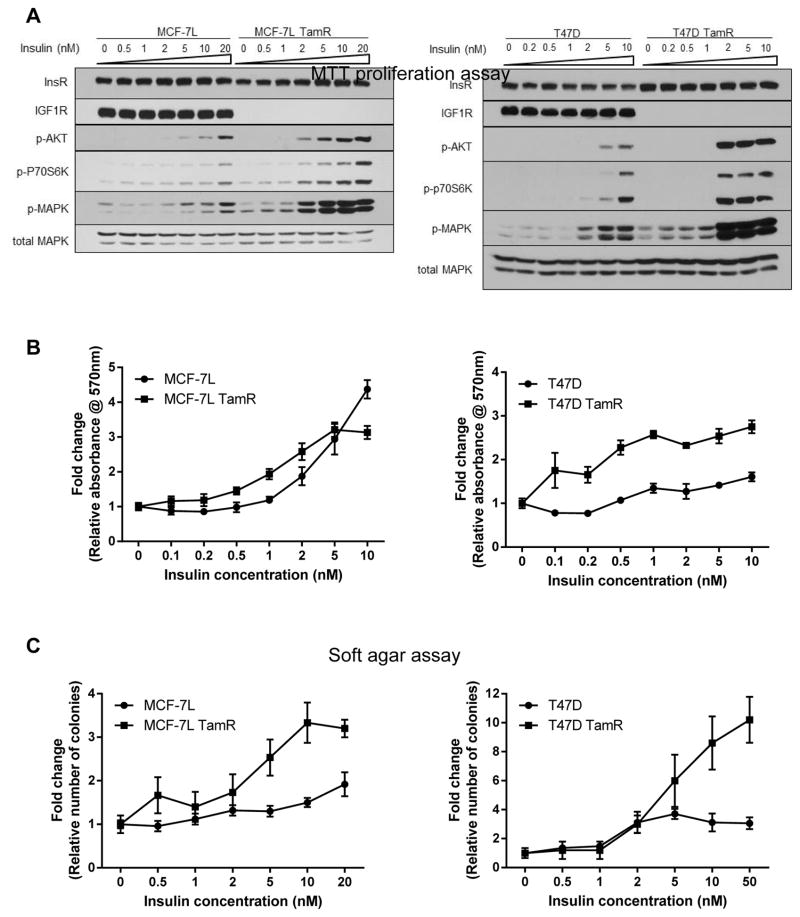
To better understand the role of insulin/InsR in TamR cells, we treated MCF-7L, T47D and their TamR cells with increasing concentrations of insulin for 15 minutes. As measured by AKT, P70S6K and MAPK phosphorylation, insulin signaling stimulated phosphorylation at lower levels of insulin in TamR cells compared to MCF-7L or T47D parental cells (Fig. 1A). Monolayer growth and soft agar assays showed greater proliferation and colony formation at lower concentrations of insulin in TamR cells compared to their parental cells (Fig. 1B and C).
Figure 1. TamR breast cancer cells were more sensitive to insulin treatment compared to their parental cells.
(A) MCF-7L and T47D cells were plated, serum starved for 24 hours and treated with increasing concentrations of insulin for 15 minutes. Whole cell lysates were collected, separated by SDS-PAGE and subjected to the indicated immunoblotting analyses. (B) Cell monolayer growth of MCF-7L and T47D were measured using MTT proliferation assay. Cells were serum starved for 24 hours and then treated with increasing concentrations of insulin. Readings were taken 5 days later. The results were normalized to untreated group. (C) Anchorage-independent growth assay was carried out on MCF-7L and T47D cells. Colonies formed were counted 14 days and 20 days later, respectively. Values were normalized to untreated group and were presented as fold change (mean ± SD, n=3).
Genetic knockdown of InsR reduced insulin-regulated signaling and growth in TamR cells, but not in the parental cells
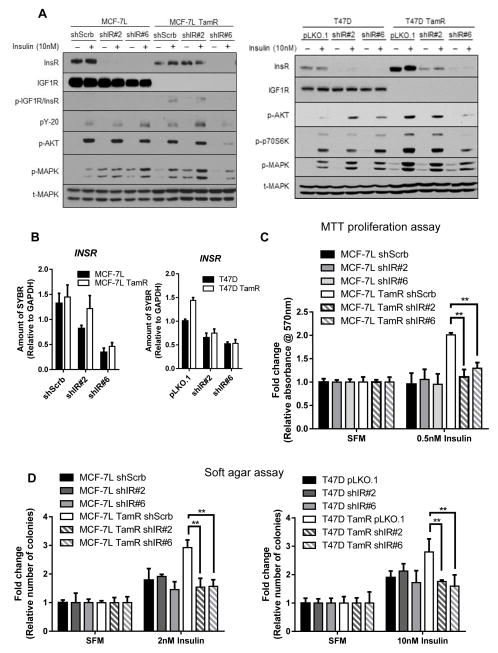
Stable InsR knockdown cell lines were generated in MCF-7L, T47D and their TamR cells using lentiviral shRNA. IGF1R level was unaffected in both MCF-7L and T47D parental cells. Compared to control shRNA, shIR#6 was a more efficient knockdown construct than shIR#2 as down-regulation of InsR protein and mRNA levels were greater in cells transduced with shIR#6, validated by immunoblotting and qRT-PCR (Fig. 2A and B). A reduction of insulin-mediated signaling was measured by IGF1R/InsR, AKT, MAPK, P70S6K and IRS (pY-20) phosphorylation after InsR knockdown in TamR cells especially in shIR#6 TamR cells. Surprisingly, this was not the case in parental cells even though the InsR level was significantly down-regulated (Fig. 2A).
Figure 2. Insulin receptor knockdown reduced insulin-regulated signaling and growth in TamR cells, but not in parental cells.
MCF-7L and T47D and their counter TamR cells were InsR knock-downed with lentiviral shRNA (shIR#2, shIR#6) or plasmid control (shSrcb or pLKO.1). InsR protein and mRNA expression levels were respectively determined using Western blot analyses as shown in lentiviral transduced (A) MCF-7L and T47D cells and qRT-PCR as shown in (B) MCF-7L and T47D cells. Cells were plated, serum started for 24 hours and treated with or without 10nM insulin for 15 minutes. Whole cell lysates were collected, separated by SDS-PAGE and subjected for indicated immunoblotting analyses. For qRT-PCR analysis, total RNA was collected from cells in full media. Data was normalized to housekeeping gene, GAPDH. Results represent mean ± SD of triplicates from three independent experiments. (C) Cell monolayer growth was determined using MTT assay. Transduced MCF-7L and TamR cells were serum starved for 24 hours and treated with or without insulin for 5 days. (D) Anchorage-independent growth of lentiviral transduced MCF-7L and T47D was measured after 19 days and 25 days, respectively. Values were normalized to untreated group and were represented in fold change (mean ± SD, n=3). Two-way ANOVA with Bonferroni comparison was performed to identify significance among untreated versus treated groups and shIR versus control groups. *, p<0.05; **, p<0.01.
It was notable that even with knockdown, some InsR biochemical activity could be seen. To further determine the biological significance of InsR knockdown, cells were studied by cell cycle analyses, monolayer growth, and anchorage-independent growth assays. Down-regulation of InsR abolished insulin-mediated proliferation (Fig. 2C), cell cycle progression (Table 1) and anchorage-independent growth (Fig. 2D) in MCF-7L and T47D TamR cells, but not in MCF-7L or T47D parental cells. Re-introduction of recombinant human InsR (hIR) in shIR#2 and shIR#6 transduced MCF-7L TamR cells rescued insulin-mediated signaling (Fig. S1A) and cell proliferation (Fig. S1B).
Table 1.
Distribution of cell cycle phases after overnight insulin treatment in respective population of cell lines
| A | ||||
|---|---|---|---|---|
| Cell lines | Insulin (nM) | Phases (%) | ||
| G0/G1 | S | G2/M | ||
| MCF-7L shScrb | 0 | 47.8 ± 7.8 | 27.4 ± 1.2 | 9.28 ± 3.8 |
| 1 | 49.4 ± 4.1 | 29.0 ± 1.9 | 14.6 ± 2.9 | |
| 10 | 45.5 ± 3.0 | 35.1 ± 1.5 | 11.9 ± 5.1 | |
|
| ||||
| MCF-7L shIR#2 | 0 | 51.2 ± 6.6 | 27.8 ± 2.8 | 9.9 ± 5.7 |
| 1 | 53.9 ± 3.7 | 27.2 ± 2.6 | 12.4 ± 1.6 | |
| 10 | 49.4 ± 7.3 | 35.7 ± 5.1 | 6.71 ± 4.0 | |
|
| ||||
| MCF-7L shIR#6 | 0 | 49.6 ± 8.2 | 30.9 ± 9.7 | 11.6 ± 3.8 |
| 1 | 52.1 ± 7.3 | 28.2 ± 2.0 | 11.7 ± 3.2 | |
| 10 | 44.4 ± 5.5 | 38.1 ± 4.1 | 7.7 ± 7.0 | |
|
| ||||
| MCF-7L TamR shScrb | 0 | 55.3 ± 1.9 | 24.5 ± 2.4 | 14.4 ± 1.1 |
| 1 | 39.0 ± 15.3 | 34.2 ± 1.4 | 11.9 ± 4.6 | |
| 10 | 41.4 ± 1.7 | 35.6 ± 2.7 | 13.8 ± 4.6 | |
|
| ||||
| MCF-7L TamR shIR#2 | 0 | 39.8 ± 1.6 | 17.8 ± 1.4 | 26.2 ± 3.8 |
| 1 | 39.8 ± 3.5 | 16.3 ± 1.6*** | 28.5 ± 6.6 | |
| 10 | 38.0 ± 3.5 | 22.1 ± 3.1*** | 22.0 ± 5.3 | |
|
| ||||
| MCF-7L TamR shIR#6 | 0 | 45.7 ± 3.0 | 20.3 ± 4.3 | 19.2 ± 3.8 |
| 1 | 43.7 ± 5.9 | 19.7 ± 2.7*** | 21.2 ± 2.2 | |
| 10 | 44.6 ± 5.6 | 20.7 ± 0.1*** | 19.7 ± 5.8 | |
| B | ||||
|---|---|---|---|---|
| Cell lines | Insulin (nM) | Phases (%) | ||
| G0/G1 | S | G2/M | ||
| 0 | 53.0 ± 5.0 | 24.8 ± 2.5 | 11.3 ± 5.4 | |
| 1 | 53.8 ± 6.8 | 25.6 ± 1.4 | 10.7 ± 6.1 | |
| 10 | 41.9 ± 8.2 | 33.3 ± 2.2 | 13.1 ± 0.8 | |
|
| ||||
| T47D shIR#2 | 0 | 50.5 ± 2.4 | 26.9 ± 0.7 | 11.0 ± 1.4 |
| 1 | 52.4 ± 8.6 | 25.0 ± 3.7 | 12.1 ± 0.8 | |
| 10 | 4.4 ± 8.7 | 33.8 ± 5.5 | 13.4 ± 1.2 | |
|
| ||||
| T47D shIR#6 | 0 | 52.2 ± 8.1 | 27.1 ± 6.2 | 7.5 ± 1.3 |
| 1 | 49.2 ± 5.9 | 28.1 ± 6.0 | 10.9 ± 0.5 | |
| 10 | 35.9 ± 8.1 | 33.6 ± 1.3 | 10.8 ± 7.0 | |
|
| ||||
| T47D TamR pLKO.1 | 0 | 60.3 ± 6.5 | 21.0 ± 5.1 | 13.0 ± 1.3 |
| 1 | 55.2 ± 4.2 | 27.9 ± 6.4 | 10.7 ± 3.9 | |
| 10 | 42.0 ± 2.8 | 36.8 ± 1.6 | 10.7 ± 2.6 | |
|
| ||||
| T47D TamR shIR#2 | 0 | 63.9 ± 3.6 | 20.3 ± 1.7 | 8.9 ± 0.2 |
| 1 | 52.5 ± 8.6 | 19.8 ± 2.0ns | 17.7 ± 5.8 | |
| 10 | 49.0 ± 7.1 | 25.8 ± 1.0* | 14.1 ± 3.4 | |
|
| ||||
| T47D TamR shIR#6 | 0 | 54.8 ± 9.9 | 19.5 ± 0.7 | 12.8 ± 2.6 |
| 1 | 54.7 ± 8.5 | 18.6 ± 1.4* | 15.3 ± 4.9 | |
| 10 | 48.6 ± 5.0 | 22.8 ± 1.9*** | 14.8 ± 1.5 | |
Mean ± SD (n = 3)
not significant;
p<0.05;
p<0.001
shIRs versus control of relative treatment groups
In contrast, InsR overexpression in both MCF-7L and TamR cells showed an up-regulation of InsR protein level and also insulin-mediated phosphorylation of IRS (pY-20), AKT, P70S6K and MAPK (Fig. S2A). However, only MCF-7L TamR cells showed a significant increase in cell proliferation (Fig. S2B).
S961 inhibited insulin-regulated PI3K/MAPK signaling and growth in TamR cells, but not in parental cells
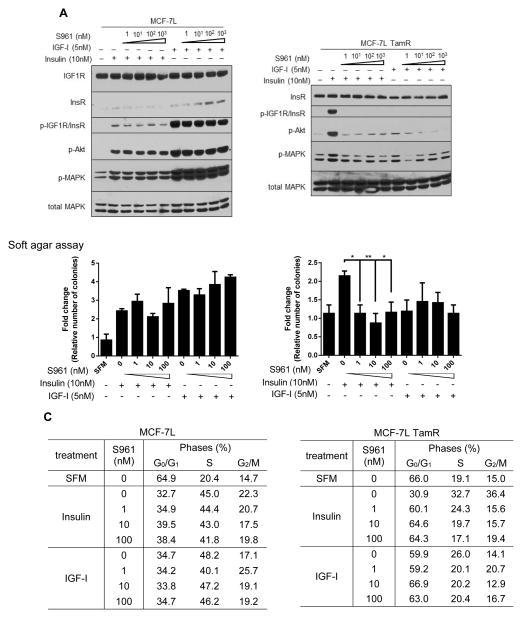
S961, a small peptide was synthesized and shown to be a competitive antagonist with a slightly higher affinity to InsR than insulin and partial agonist effects at lower concentrations (27). S961 has very low affinity for IGF1R. To examine the effect of S961 in endocrine resistance cells, cells were pre-treated with increasing concentrations of S961 before exposing cells to either IGF-I or insulin. S961 did not inhibit IGF-I or insulin-regulated signaling in MCF-7L and T47D parental cells even at high concentration as shown by IGF1R/InsR, AKT and MAPK phosphorylation (Fig. 3A, S3A). Similarly, S961 did not inhibit insulin or IGF-I stimulated cell cycle progression or anchorage-independent growth in parental cells (Fig. 3B, 3C, S4A and Table S1). Even though agonist effects of S961 have been reported, we did not observe S961-induced signaling or cell proliferation in these breast cancer cells (Fig. 5B).
Figure 3. S961 inhibited insulin-stimulated PI3K/MAPK signaling and growth in MCF-7L TamR cells, but not in parental cells.
(A) MCF-7L and TamR cells were serum starved overnight and pre-treated with varying concentrations of S961 for 30 minutes before treating the cells with either 10nM insulin or 5nM IGF-I for 10 minutes. Whole cell lysates were separated by SDS-PAGE and immunoblotted for indicated antibodies. (B) Anchorage-independent growth assay was carried out on MCF-7L and TamR cells treated with varying concentrations of S961 and either 10nM insulin or 5nM IGF-I. Treatments were spiked in after 7 days. Colonies formed were counted 14 days later. Two-way ANOVA with Bonferroni comparison was performed to compare between treated and untreated group. *, p<0.05; **, p<0.01. (C) Cell cycle analysis was performed using flow cytometry. MCF-7L and TamR cells were plated, serum starved for 8 hours before treating with varying concentrations of S961 and either 10nM insulin or 5nM IGF-I overnight.
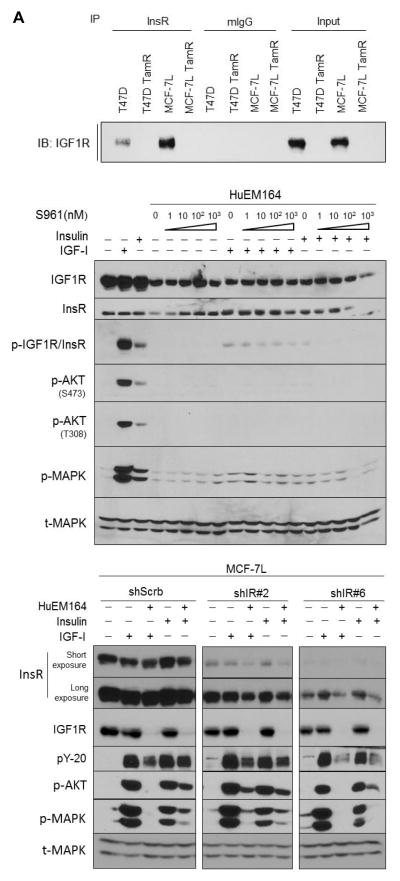
Figure 5. Inhibition of InsR via S961 and shIR was not effective in parental cells due to the presence of IGF1R/InsR hybrid receptors.
The presence of IGF1R/InsR hybrid receptors was identified via immunoprecipitation (IP). (A) Whole cell lysates were collected in full media and immunoprecipitated with either anti-InsR antibody or mouse IgG overnight. IP was then resolved with SDS-PAGE and subjected for IGF1R immunoblotting. (B) MCF-7L parental cells were serum starved overnight, pre-treated with 20 μg/mL HuEM164 for 3 hours and varying concentrations of S961 for 30 minutes before exposing toeither 10nM insulin or 5nM IGF-I for 10 minutes. Whole cell lysates were collected, separated by SDS-PAGE and subjected for immunoblotting analyses. (C) Lentiviral transduced MCF-7L cells were serum starved overnight and pre-treated with 20 μg/mL HuEM164 overnight before exposing to either 10nM insulin or 5nM IGF-I for 15 minutes. Whole cell lysates were collected, separated by SDS-PAGE and subjected for indicated immunoblotting analyses.
In contrast, S961 blocked insulin-regulated signaling even at concentrations as low as 1 nM concentration in TamR cells (Fig. 3A and S3B). Similar sensitivity was reflected in anchorage-independent growth assay (Fig. 3B and S4B) and cell cycle analysis (Fig. 3C and Table S1), where 1 nM concentration of S961 fully diminished insulin-stimulated colony formation growth and S-phase induction, respectively in MCF-7L TamR cells. A higher S961 concentration was needed to fully block insulin-stimulated growth in T47D TamR cells. Since TamR cells lack IGF1R, they do not respond to IGF-I stimulation.
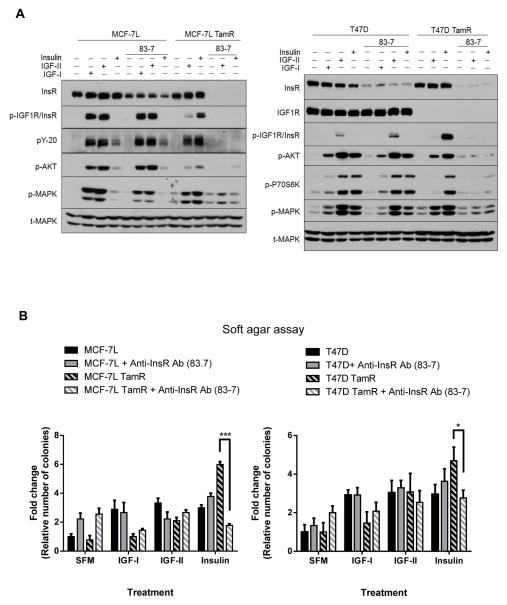
83-7 mAb down-regulates InsR, thus inhibiting insulin-stimulated signaling and growth in TamR cells
Monoclonal antibody clone 83-7 (83-7 mAb) binds alpha-subunit of InsR allosterically without interfering insulin binding (28) and is specific for InsR binding although its functional roles in cells are not well studied. The antibody has been reported to stimulate lipogenesis, inhibit lipolysis, and activate receptor kinase by cross-linking receptor molecules (29).
To explore the effect of 83-7 mAb in cancer cells, we pre-treated 83-7 mAb overnight before treating with IGF-I, IGF-II, or insulin in MCF-7L, T47D and TamR cells. As shown in Figure 4A, 83-7 mAb did not induce receptor phosphorylation. In contrast, 83-7 mAb down-regulated InsR and effectively blocked IGF-II and insulin-stimulated IGF1R/InsR, IRS, AKT and MAPK phosphorylation (Fig. 4A). To study the biological effects of InsR, anchorage independent growth assays showed that 83-7 mAb effectively inhibited insulin-stimulated colony formation (Fig. 4B). Although 83-7mAb caused some InsR down-regulation in the parental cells, there was little to minimal inhibitory effect of 83-7 mAb on MCF-7L and T47D parental cells in terms of IGF-I, IGF-II or even insulin-stimulated signaling and anchorage-independent growth (Fig. 4).
Figure 4. Monoclonal antibody clone 83-7 against insulin receptor (83-7) downregulated insulin receptor, inhibiting insulin-stimulated signaling and growth in TamR cells.
(A) MCF-7L and T47D cells were serum starved and pre-treated with 2 μg/mL of 83-7 overnight before treating with either 5nM IGF-I, 10nM IGF-II or 10nM insulin for 15 minutes. Whole cell lysate were collected, separated by SDS-PAGE and subjected for indicated immunoblotting analyses. Anchorage-independent growth assay was carried out on (B) MCF-7L and T47D cells treated with 2 μg/mL of 83-7 without or with either 10nM IGF-I or 10nM insulin. Colonies formed were counted 14 days for MCF-7Ls and 21 days for T47D cells after first plated. Two-way ANOVA with Bonferroni comparison was performed to identify significance among untreated vs. treated groups. *, p<0.05; **, p<0.01.
Inhibition of InsR was not effective in parental breast cancer cells due to the presence of IGF1R/InsR hybrid receptors
To further explore why InsR inhibition was not effective in MCF-7L and T47D parental cells, we used immunoprecipitation and immunoblotting to examine the ability of these cells to express IGF1R/InsR hybrid receptors. Co-immunoprecipitation studies showed that IGF1R/InsR hybrid receptors are present in MCF-7L and T47D parental cells but not in TamR cells because of their downregulation of IGF1R (Fig. 5A). As previously shown, S961 was not effective in parental cells. To inhibit IGF1R and hybrid receptors, we used HuEM164 (also known as AVE1642), an anti-IGF1R mAb shown to specifically bind IGF1R and result in its down-regulation (30). In the parental cells, insulin signaling was more completely extinguished by the use of both IGF1R mAb and S961 (Fig. 5B and S5).
Similarly, when shIR transduced parental cells was treated with HuEM164, a synergistic inhibitory effect was achieved, as measured by IGF1R/InsR, IRS, AKT and MAPK phosphorylation. Unlike S961, down-regulation of InsR by HuEM164 however was able to only partially block insulin-mediated signaling in parental cells (Fig. 5C and S6). The most complete inhibition of ligand signaling was achieved by the combination of shIR#6 and HuEM164.
Discussion
While the insulin/InsR signaling system is responsible for glucose homeostasis; it is also a cellular growth factor. Metabolic syndrome associated with obesity and type II diabetes, both states of relative insulin resistance resulting in hyperinsulinemia are associated with cancer risk (31). Breast cancer patients who have these conditions are more likely to suffer metastatic disease, disease recurrence, and mortality (32, 33). When a number of anti-IGF1R mAbs trials failed to show benefits in cancer patients, one mechanistic explanation is the presence of InsR acting as a compensatory pathway to IGF1R inhibition and IGF1R loss (12). Our endocrine resistant model showed greater sensitivity towards insulin when IGF1R expression level is lacking (Fig. 1). Other studies have also shown similar pattern in prostate cancer and pancreatic neuroendocrine tumors in vivo, where InsR induces mitogenic activities and compensates for IGF1R inhibition or resistance to anti-IGF1R (34, 35).
InsR exists in two isoforms: InsR-A and InsR-B due to alternative splicing of exon 11, differing by 12 amino acids. Previous studies have shown that InsR-B tends to be a metabolic receptor expressed in adult muscle, liver, and fat. InsR-B binds only insulin at physiological concentrations. In contrast, InsR-A, a predominant isoform during fetal development is commonpressed in cancer and binds with high affinity to insulin and IGF-II (36, 37). Up-regulation of InsR-A has been reported in breast, ovarian, lung, colon cell lines and/or human tumors and is thought to mediate tumorigenesis and survivor in response to insulin and IGF-II (38–42). However, our endocrine-resistant model did not show an increase in InsR-A/InsR-B ratio of mRNA level compared to parental cells (data not shown). To date, antibodies have not been developed that can distinguish between levels of InsR isoforms, thus the exact protein expression of isoforms is uncertain in cells. In tumors, the data regarding the role of InsR-A are derived from mRNA levels detected by PCR. Additional study is needed to determine if InsR-B has an important role in cancer biology.
In this study, we blocked InsR function by three different techniques: 1.) genetic knockdown of InsR using lentiviral shRNA, 2.) competitively blocked of insulin binding to its receptor by S961 and 3.) down-regulation of InsR without affecting insulin binding by a mAb. These different techniques showed consistent results, inhibiting InsR was effective in the inhibition of insulin-regulated signaling and growth in TamR breast cancer cells, but not in parental cells. These data show that insulin signaling is important in endocrine resistant cells, but less relevant to parental MCF-7L and T47D cells. The presence of IGF1R or hybrid receptors and little of holo-InsR make insulin only a weak mitogen in parental cells. Similar functions of insulin receptor have been described in an ER-negative model of mouse breast cancer (43). In this system, InsR suppression was necessary to inhibit murine breast cancers in both normal and hyper-insulinemic hosts. Importantly, we have shown that inhibitors, in addition to genetic knockdown techniques, can function to disrupt cell growth but only in a subset of breast cancers with diminished IGF1R levels.
The presence of IGF1R/InsR hybrid receptors allows cancer cells to expand their ligand binding capacity (Fig. 5A). Insulin can still signal through the other available IGF1R heterodimer upon InsR inhibition; likewise IGF-I may signal through InsR in a hybrid confirmation suggesting single target inhibition of IGF1R or InsR is not sufficient to suppress hybrid receptor signaling. However in TamR cells, where there is little IGF1R, InsR becomes the predominant receptor driving insulin- (and IGF-II) stimulated growth. Thus, InsR is an important target in TamR cells. Unfortunately, there is no reliable method to quantify the level of hybrid receptors in cells or patient tumors making it difficult to predict response to antibody-based therapy on only examining levels of receptor expression. However, the use of broader range of receptor biochemical inhibition such a TKI may provide a better therapeutic advantage.
The major concern about targeting InsR is the resulting disruption of glucose homeostasis in normal tissues. Hyperglycemia can be managed by metformin, a commonly used drug for type 2 diabetes that reduces hepatic gluconeogenesis, circulating insulin level, and stimulates glucose uptake in muscle independent of insulin (44, 45). However the ability of metformin to directly affect cancer cell biology outside of modulating serum insulin levels is not understood. While there are preclinical data suggesting that metformin has little effect in models of non-diabetic rodent models (46), the clinical benefits of metformin in non-diabetic women with breast cancer awaits reporting of an adjuvant clinical trial where women were assigned to receive metformin or placebo for five years after surgical therapy for breast cancer (NCT01101438).
Thus, direct targeting of InsR would be a preferable strategy. Here we show that monoclonal antibodies and a competitive peptide inhibitor have active against InsR, but there could have significant metabolic effects in vivo. As noted the two isoforms of InsR provide a theoretical strategy to only inhibit the cancer associated function of InsR signaling. If InsR-A specific agents could be developed, then this would not perturb the metabolic functions of InsR-B in normal tissues. Furthermore, disruption of estrogen receptor signaling in breast cancer has been an extremely effective therapy despite the presence of this receptor in many normal tissues. InsR inhibition might be tolerated with appropriate strategies to manage glucose homeostasis.
In conclusion, we highlighted the role of InsR in breast cancer biology, especially in the context of resistance to endocrine therapies in ER+ breast cancer cells. Thus InsR should not be neglected as a cancer target. Whether InsR inhibition could overcome primary (de novo) endocrine resistance or treat secondary (acquired) resistanceis not certain. Combination use of metformin may address hyperglycemia condition resulted from InsR inhibition, however additional efforts at developing InsR-A blocking agents are warranted.
Materials and Methods
Cell lines and culture
MCF-7L and T47D are human ER-positive breast cancer cell lines. MCF-7L (parental cell line) was kindly provided by C. Kent Osborne (Baylor College of Medicine, Houston, TX) and maintained in improved MEM Richter’s modification medium (zinc option) supplemented with 5% FBS, and 11.25 nM insulin. MCF-7L karyotyping and gene expression profiling have shown that these cells are consistent with the originally described cell line (data not shown). T47D (parental cell line) was obtained from ATCC and maintained in MEM supplemented with 5% FBS, 1X nonessential amino acids and 6 ng/mL insulin. MCF-7L TamR and T47D TamR cells were generated as described (13). Cells lines were confirmed to be mycoplasma negative and STR profiling is performed annually on these cells (data not shown). MCF-7L TamR cells were maintained in phenol-red free IMEM (zinc option) supplemented with 11.25 nM insulin, 5% charcoal/dextran-treated FBS and 100 nM 4-OH tamoxifen; while T47D TamR cells were maintained in phenol-red free IMEM supplemented with 6 ng/mL insulin, 1X nonessential amino acids, 5% charcoal/dextran-treated FBS and 100 nM 4-OH tamoxifen. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2. All growth media were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin and purchased from Gibco®.
Reagents
IGF-I, IGF-II and insulin were purchased from Gemini and Eli Lily, respectively. Puromycin solution was purchased from Sigma-Aldrich. Geneticin (G418 sulfate) solution was purchased from Life Technologies. Humanized anti-IGF1R monoclonal antibody HuEM164 was generously provided by Immunogen Inc. Anti-InsR monoclonal antibody, alpha subunit clone 83-7 was purchased from EMD Millipore. S961 peptide was generously provided by Novo Nordisk, Denmark (27).
Antibodies
Antibodies for total IGF1R (#3027), phosphorylated IGF1R tyrosine 1135 (#3918), phosphorylated AKT serine 473 (#9271) and threonine 308 (#9275), total p44/42 MAPK (#9102), phosphorylated p44/42 MAPK (#4376), phosphorylated p70 S6 kinase (#9205) used in immunoblotting were purchased from Cell Signaling Technology. Normal mouse IgG (sc-2025), InsR antibody for immunoprecipitation (sc-57342) and InsR antibody for immunoblotting (sc-711) were purchased from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated anti-phosphotyrosine (pY-20) was purchased from BD Transduction Lab. (#610012). Anti-rabbit horseradish peroxidase-conjugated secondary antibody was purchased from Pierce.
Generation of stable InsR knockdown with shRNA
Lentiviral pLKO.1 vectors encoding either InsR mRNA specific short hairpin RNA (shRNA) sequences or vector controls were purchased from Open Biosystems through BioMedical Genomics Center at the University of Minnesota, Minneapolis, MN. Two different constructs of shRNA against InsR are described as shIR#2 and shIR#6. Their full sequences are respectively CCGGCACTGATTACTTGCTGCTCTTCTCGAGAAGAGCAGCAAGTAATCAGTGTTTTT and CCGGCTCAGGATTCTCACGACTCTACTCGAGTAGAGTCGTGAGAATCCTGAGTTTTT. Lentivirus production was carried out in HEK293T packaging cells and the viral particles were used to transduce MCF-7L and T47D cells. Final concentration of 10 μg/mL polybrene was used to increase transduction efficiency. Cells underwent puromycin selection and were maintained in 2 μg/mL for MCF-7L and MCF-7L TamR; 1.5 μg/mL for T47D parental cells and 15 μg/mL for T47D TamR.
Overexpression of insulin receptor
pEGFP-N2 vector containing full length human InsR cDNA was obtained from Addgene (#22286) (47) and transiently transfected into MCF-7L and MCF-7L TamR using Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. Cells were selected and maintained in 500 μg/mL G418 for a week before re-plating for immunoblotting analysis or monolayer growth assay.
Immunoblotting analysis
Cells were plated at a density of 3 × 105 cells in 60 mm diameter dishes and allow to equilibrate overnight. Full medium was replaced with serum-free medium (SFM) for 24 hours. Cells were then treated, washed twice with ice-cold PBS and lysed with lysis buffer of 50 mM Tris-Cl (pH 7.4), 1% Nonidet P-40, 2 mM EDTA (pH 8.0), 100 mM NaCl, 10 mM sodium orthovanadate, and with complete proteases inhibitor cocktail (Roche Diagnostics). Lysates were centrifuged at 12,000 g for 30 minutes at 4°C. Protein concentrations were measured using bicinchoninic acid protein assay reagent kit (Pierce). Whole cell lysates (50 μg) were boiled in 5X Laemmli loading buffer, separated by 8% SDS-PAGE, transferred to PVDF membrane and immunoblotted according to manufacturer guidelines. For Immunoprecipitation (IP), whole-cell lysates were incubated with either anti-InsR antibody or mIgG overnight at 4°C. Protein A/G PLUS-Agarose bead slurry was added into the samples and incubated for 4 hours at 4°C. Beads were washed with lysis buffer 5 times and boiled in 5X Laemmli loading buffer. Samples were resolved by 8% SDS-PAGE, transferred to PVDF membrane and immunoblotted.
Reverse transcription and quantitative real-time PCR
Cells were seeded at a density of 2 × 105 cells in 6-well plates in growth media until reaching 80% confluent. Cellular RNA was isolated using TriPure Reagent according to the manufacturer (Roche). For RNA quality verification, a ratio of 260 nm to 280 nm was determined. A total of 1 μg of RNA was reverse transcribed using qScript cDNA synthesis kit (Quanta Biosciences), and quantitative PCR was performed using the University SYBR Green Kit according to the manufacturer’s protocol (Roche) on an Eppendorf Mastercycler Realplex machine. The relative abundance of InsR mRNA was calculated using cycle threshold values that were derived from a standard curve and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) level as an internal control. The forward and reverse primers are as followed: InsR 5′-CAACGTGGTTTTCGTCCCC-3′ and 5′-AGATGACCAGCGACTCCTTG-3′; GAPDH 5′-TGAAGGTCGGAGTCAACGATTTGGT-3′ and 5′-GAAGATGGTGATGGGATTTC-3′
Monolayer growth assay
Cells were plated at a density of 15,000 cells per well in 24-well plates and allowed to attach overnight. Full media were replaced and starved with SFM for 24 hours. After 5 or 6 days of treatment, growth was assessed via MTT assay. Each well was added 60 μL of 5 mg/mL thiazolyl blue tetrazolium bromide solution (MTT) from Sigma-Aldrich in SFM for 4 hours at 37°C in dark. Media were aspirated and purple formazan crystals were lyzed with 500 μL of solubilization solution (95% dimethylsulfoxide and 5% Improved MEM). Absorbance was measured with a plate reader at 570 nm using a 650 nm differential filter to access growth.
Anchorage-independent growth assay (soft agar)
A 1-mL layer of 0.8% SeaPlaque-agarose (bioWhittaker) in 1% FBS-containing growth media was solidified into each well of a 6-well plate. The bottom layer was overlaid with 1mL of 0.5% top agar mixture for 15,000 cells per well with appropriate treatment. All plates were incubated at 37°C. The number of colony formation was assessed on a light microscope with an ocular grid. Only colonies exceeding two thirds of grid square were scored. Five random fields were counted per well.
Cell cycle analysis
Cells were plated at density of 8 × 105 cells in 60 mm dishes, starved with SFM for 24 hours and treated overnight. Cells were trypsinized, washed twice with ice-cold PBS containing 1% bovine serum albumin (BSA) and re-suspended with staining buffer (PBS containing 0.1 mg/mL propidium iodide, 0.5% triton X-100, 16 μg/mL RNase, 1% BSA) for 2 hours. Cell cycle analyses were performed for DNA content using BD Accuri C6 flow cytometry. Single cells were gated and 10,000 events were collected. The proportion of cells in G0G1, S and G2/M phases was quantified using FlowJo software.
Supplementary Material
Acknowledgments
Financial support:
NIH/NCI 2P30-CA077598, NIH/NCI P50CA116201 (Ingle), Komen for the Cure SAC110039
We would like to acknowledge the assistance of the University Flow Cytometry Resource at the University of Minnesota; Novo Nordisk for supplying S961 peptides. Funding was provided by NIH/NCI 2P30-CA077598 (JYC, KL, DY), NIH/NCI P50CA116201 (DY), and Komen for the Cure SAC110039 (DY).
Footnotes
Disclosure of any potential conflicts of interest:
The authors declare no conflicts of interest.
References
- 1.Nadji M, Gomez-Fernandez C, Ganjei-Azar P, Morales AR. Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol. 2005;123:21–7. doi: 10.1309/4wv79n2ghj3x1841. [DOI] [PubMed] [Google Scholar]
- 2.Macaskill EJ, Renshaw L, Dixon JM. Neoadjuvant use of hormonal therapy in elderly patients with early or locally advanced hormone receptor-positive breast cancer. Oncologist. 2006;11:1081–8. doi: 10.1634/theoncologist.11-10-1081. [DOI] [PubMed] [Google Scholar]
- 3.Paridaens RJ, Dirix LY, Beex LV, Nooij M, Cameron DA, Cufer T, et al. Phase III study comparing exemestane with tamoxifen as first-line hormonal treatment of metastatic breast cancer in postmenopausal women: the European Organisation for Research and Treatment of Cancer Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:4883–90. doi: 10.1200/JCO.2007.14.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocrine-related cancer. 2004;11:643–58. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- 5.Giuliano M, Schifp R, Osborne CK, Trivedi MV. Biological mechanisms and clinical implications of endocrine resistance in breast cancer. Breast. 2011;20(Suppl 3):S42–9. doi: 10.1016/S0960-9776(11)70293-4. [DOI] [PubMed] [Google Scholar]
- 6.Baselga J, Campone M, Piccart M, Burris HA, Rugo HS, Sahmoud T, et al. Everolimus in Postmenopausal Hormone-Receptor–Positive Advanced Breast Cancer. New England Journal of Medicine. 2011;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garber K. The cancer drug that almost wasn’t. Science. 2014;345:865–7. doi: 10.1126/science.345.6199.865. [DOI] [PubMed] [Google Scholar]
- 8.Fagan DH, Yee D. Crosstalk between IGF1R and estrogen receptor signaling in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:423–9. doi: 10.1007/s10911-008-9098-0. [DOI] [PubMed] [Google Scholar]
- 9.Arpino G, De Angelis C, Giuliano M, Giordano A, Falato C, De Laurentiis M, et al. Molecular mechanism and clinical implications of endocrine therapy resistance in breast cancer. Oncology. 2009;77(Suppl 1):23–37. doi: 10.1159/000258493. [DOI] [PubMed] [Google Scholar]
- 10.Lee AV, Weng CN, Jackson JG, Yee D. Activation of estrogen receptor-mediated gene transcription by IGF-I in human breast cancer cells. J Endocrinol. 1997;152:39–47. doi: 10.1677/joe.0.1520039. [DOI] [PubMed] [Google Scholar]
- 11.Becker MA, Ibrahim YH, Cui X, Lee AV, Yee D. The IGF pathway regulates ERalpha through a S6K1-dependent mechanism in breast cancer cells. Mol Endocrinol. 2011;25:516–28. doi: 10.1210/me.2010-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yee D. Insulin-like growth factor receptor inhibitors: baby or the bathwater? J Natl Cancer Inst. 2012;104:975–81. doi: 10.1093/jnci/djs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagan DH, Uselman RR, Sachdev D, Yee D. Acquired resistance to tamoxifen is associated with loss of the type I insulin-like growth factor receptor: implications for breast cancer treatment. Cancer Res. 2012;72:3372–80. doi: 10.1158/0008-5472.CAN-12-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drury SC, Detre S, Leary A, Salter J, Reis-Filho J, Barbashina V, et al. Changes in breast cancer biomarkers in the IGF1R/PI3K pathway in recurrent breast cancer after tamoxifen treatment. Endocr Relat Cancer. 2011;18:565–77. doi: 10.1530/ERC-10-0046. [DOI] [PubMed] [Google Scholar]
- 15.Arnedos M, Drury S, Afentakis M, A’Hern R, Hills M, Salter J, et al. Biomarker changes associated with the development of resistance to aromatase inhibitors (AIs) in estrogen receptor-positive breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2014;25:605–10. doi: 10.1093/annonc/mdt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HX, Sharon E. IGF-1R as an anti-cancer target--trials and tribulations. Chinese journal of cancer. 2013;32:242–52. doi: 10.5732/cjc.012.10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ullrich A, Gray A, Tam AW, Yang-Feng T, Tsubokawa M, Collins C, et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. Embo J. 1986;5:2503–12. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fulzele K, DiGirolamo DJ, Liu Z, Xu J, Messina JL, Clemens TL. Disruption of the insulin-like growth factor type 1 receptor in osteoblasts enhances insulin signaling and action. The Journal of biological chemistry. 2007;282:25649–58. doi: 10.1074/jbc.M700651200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Pelzer AM, Kiang DT, Yee D. Down-regulation of type I insulin-like growth factor receptor increases sensitivity of breast cancer cells to insulin. Cancer Res. 2007;67:391–7. doi: 10.1158/0008-5472.CAN-06-1712. [DOI] [PubMed] [Google Scholar]
- 20.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 21.Milazzo G, Giorgino F, Damante G, Sung C, Stampfer MR, Vigneri R, et al. Insulin receptor expression and function in human breast cancer cell lines. Cancer Res. 1992;52:3924–30. [PubMed] [Google Scholar]
- 22.Zhang H, Fagan DH, Zeng X, Freeman KT, Sachdev D, Yee D. Inhibition of cancer cell proliferation and metastasis by insulin receptor downregulation. Oncogene. 2010;29:2517–27. doi: 10.1038/onc.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belfiore A, Malaguarnera R. Insulin receptor and cancer. Endocrine-related cancer. 2011;18:R125–47. doi: 10.1530/ERC-11-0074. [DOI] [PubMed] [Google Scholar]
- 24.Papa V, Pezzino V, Costantino A, Belfiore A, Giuffrida D, Frittitta L, et al. Elevated insulin receptor content in human breast cancer. J Clin Invest. 1990;86:1503–10. doi: 10.1172/JCI114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puzanov I, Lindsay CR, Goff L, Sosman J, Gilbert J, Berlin J, et al. A phase I study of continuous oral dosing of OSI-906, a dual inhibitor of insulin-like growth factor-1 and insulin receptors, in patients with advanced solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:701–11. doi: 10.1158/1078-0432.CCR-14-0303. [DOI] [PubMed] [Google Scholar]
- 26.Jones RL, Kim ES, Nava-Parada P, Alam S, Johnson FM, Stephens AW, et al. Phase I study of intermittent oral dosing of the insulin-like growth factor-1 and insulin receptors inhibitor OSI-906 in patients with advanced solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:693–700. doi: 10.1158/1078-0432.CCR-14-0265. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen L, Hansen BF, Jensen P, Pedersen TA, Vestergaard K, Schaffer L, et al. Agonism and antagonism at the insulin receptor. PLoS One. 2012;7:e51972. doi: 10.1371/journal.pone.0051972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soos MA, Siddle K, Baron MD, Heward JM, Luzio JP, Bellatin J, et al. Monoclonal antibodies reacting with multiple epitopes on the human insulin receptor. The Biochemical journal. 1986;235:199–208. doi: 10.1042/bj2350199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien RM, Soos MA, Siddle K. Monoclonal antibodies to the insulin receptor stimulate the intrinsic tyrosine kinase activity by cross-linking receptor molecules. The EMBO journal. 1987;6:4003–10. doi: 10.1002/j.1460-2075.1987.tb02743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Sachdev D, Wang C, Hubel A, Gaillard-Kelly M, Yee D. Detection and downregulation of type I IGF receptor expression by antibody-conjugated quantum dots in breast cancer cells. Breast Cancer Res Treat. 2009;114:277–85. doi: 10.1007/s10549-008-0014-5. [DOI] [PubMed] [Google Scholar]
- 31.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, et al. Diabetes and cancer: a consensus report. CA: a cancer journal for clinicians. 2010;60:207–21. doi: 10.3322/caac.20078. [DOI] [PubMed] [Google Scholar]
- 32.Josefson D. High insulin levels linked to deaths from breast cancer. BMJ: British Medical Journal. 2000;320:1496. [PMC free article] [PubMed] [Google Scholar]
- 33.Rose DP, Haffner SM, Baillargeon J. Adiposity, the metabolic syndrome, and breast cancer in African-American and white American women. Endocrine reviews. 2007;28:763–77. doi: 10.1210/er.2006-0019. [DOI] [PubMed] [Google Scholar]
- 34.Weinstein D, Sarfstein R, Laron Z, Werner H. Insulin receptor compensates for IGF1R inhibition and directly induces mitogenic activity in prostate cancer cells. Endocrine Connections. 2014;3:24–35. doi: 10.1530/EC-13-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulanet DB, Ludwig DL, Kahn CR, Hanahan D. Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy. Proceedings of the National Academy of Sciences. 2010;107:10791–8. doi: 10.1073/pnas.0914076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frasca F, Pandini G, Scalia P, Sciacca L, Mineo R, Costantino A, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;19:3278–88. doi: 10.1128/mcb.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moller DE, Yokota A, Caro JF, Flier JS. Tissue-specific expression of two alternatively spliced insulin receptor mRNAs in man. Molecular endocrinology (Baltimore, Md) 1989;3:1263–9. doi: 10.1210/mend-3-8-1263. [DOI] [PubMed] [Google Scholar]
- 38.Jones HE, Gee JMW, Barrow D, Tonge D, Holloway B, Nicholson RI. Inhibition of insulin receptor isoform-A signalling restores sensitivity to gefitinib in previously de novo resistant colon cancer cells. Br J Cancer. 2006;95:172–80. doi: 10.1038/sj.bjc.6603237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, Zhu W, Streicher K, Morehouse C, Brohawn P, Ge X, et al. Increased IR-A/IR-B ratio in non-small cell lung cancers associates with lower epithelial-mesenchymal transition signature and longer survival in squamous cell lung carcinoma. BMC cancer. 2014;14:131. doi: 10.1186/1471-2407-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrington SC, Weroha SJ, Reynolds C, Suman VJ, Lingle WL, Haluska P. Quantifying insulin receptor isoform expression in FFPE breast tumors. Growth hormone & IGF research: official journal of the Growth Hormone Research Society and the International IGF Research Society. 2012;22:108–15. doi: 10.1016/j.ghir.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalli KR, Falowo OI, Bale LK, Zschunke MA, Roche PC, Conover CA. Functional insulin receptors on human epithelial ovarian carcinoma cells: implications for IGF-II mitogenic signaling. Endocrinology. 2002;143:3259–67. doi: 10.1210/en.2001-211408. [DOI] [PubMed] [Google Scholar]
- 42.Belfiore A, Frasca F, Pandini G, Sciacca L, Vigneri R. Insulin Receptor Isoforms and Insulin Receptor/Insulin-Like Growth Factor Receptor Hybrids in Physiology and Disease. Endocrine reviews. 2009;30:586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- 43.Rostoker R, Abelson S, Bitton-Worms K, Genkin I, Ben-Shmuel S, Dakwar M, et al. Highly specific role of the insulin receptor in breast cancer progression. Endocr Relat Cancer. 2015;22:145–57. doi: 10.1530/ERC-14-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowling RJO, Niraula S, Stambolic V, Goodwin PJ. Metformin in cancer: translational challenges. Journal of Molecular Endocrinology. 2012;48:R31–R43. doi: 10.1530/JME-12-0007. [DOI] [PubMed] [Google Scholar]
- 45.Jaques G, Kiefer P, Rotsch M, Hennig C, Goke R, Richter G, et al. Production of insulin-like growth factor binding proteins by small-cell lung cancer cell lines. Exp Cell Res. 1989;184:396–406. doi: 10.1016/0014-4827(89)90339-x. [DOI] [PubMed] [Google Scholar]
- 46.Thompson MD, Grubbs CJ, Bode AM, Reid JM, McGovern R, Bernard PS, et al. Lack of effect of metformin on mammary carcinogenesis in nondiabetic rat and mouse models. Cancer prevention research (Philadelphia, Pa) 2015;8:231–9. doi: 10.1158/1940-6207.CAPR-14-0181-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramos RR, Swanson AJ, Bass J. Calreticulin and Hsp90 stabilize the human insulin receptor and promote its mobility in the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10470–5. doi: 10.1073/pnas.0701114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.