Abstract
MicroRNAs (miRs) are non-coding gene transcripts abundantly expressed in both the developing and adult mammalian brain. They act as important modulators of complex gene regulatory networks during neuronal development and plasticity. miR-181c is highly abundant in cerebellar cortex and its expression is increased in autism patients as well as in an animal model of autism. To systematically identify putative targets of miR-181c, we repressed this miR in growing cortical neurons and found over 70 differentially expressed target genes using transcriptome profiling. Pathway analysis showed that the miR-181c-modulated genes converge on signaling cascades relevant to neurite and synapse developmental processes. To experimentally examine the significance of these data, we inhibited miR-181c during rat cortical neuronal maturation in vitro; this loss-of miR-181c function resulted in enhanced neurite sprouting and reduced synaptogenesis. Collectively, our findings suggest that miR-181c is a modulator of gene networks associated with cortical neuronal maturation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2179-0) contains supplementary material, which is available to authorized users.
Keywords: Neurodevelopment, Neuronal morphology, Cortex, Non-coding RNA, Post-transcriptional regulation
Introduction
The development of neurons and the establishment of mature synaptic connectivity require an intricate orchestration of defined gene networks. Regulation of neuronal gene network expression is modulated at several levels by a number of different post-transcriptional mechanisms, such as the ubiquitin–proteasome system [1], control of mRNA stability [2], translation initiation [3] and elongation [4], and by non-coding microRNAs (miRs) [5]. miRs coordinately regulate the expression of multiple genes coding for proteins, which often have converging cellular functions [6–8]. The function(s) of individual miRs in neurons are beginning to emerge, and multiple lines of evidence point to the widespread involvement of miRs in various fundamental neurodevelopmental processes (reviewed in [9–11]). Consequently, disturbances in miR expression levels results in aberrant signaling pathways at different levels, leading to impaired intercellular connections and cellular functioning causing various neurodevelopmental disorders. In cell lines derived from autism spectrum disorder (ASD) patients, the levels of miR-181c have been found to be strongly elevated [12]. Moreover, we recently observed that the expression of miR-181c is altered in the amygdala of a valproate rat model of autism, and that miR-181c inhibition affects the growth of amygdala neurons [13]. During retinoic acid (RA)-induced neuronal differentiation of progenitor cells, miR-181c is upregulated, suggesting dynamic changes in the expression of this miR that could be linked to specific neurodevelopmental stages [14]. Furthermore, miR expression profiling showed that miR-181c is widely expressed in the central nervous system, including in cortical regions [15]. Here, we combined neuromorphological studies with whole-genome mRNA expression profiling and bioinformatic analysis to examine the impact of miR-181c repression on growing cortical neurons in vitro. We find a role for this small regulatory RNA in modulating neuronal outgrowth and synaptic formation by acting as a post-transcriptional regulator of genes involved in neuronal growth and maturation.
Materials and methods
Animals
Wistar rats (Harlan Laboratories) embryos were used as a resource for the isolation of primary cortical neurons. Animals were housed 2–3 per cage with ad libitum food and water access with a 12 h light cycle at controlled ambient temperature (21 ± 1 °C). All experiments were performed according to protocols approved by the Committee for Animal Experiments of the Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands.
Cell culture
All media and reagents were purchased from Life Technologies. Primary cortical neurons were isolated from embryonic day 18 (E18) old rat embryos brains. Pregnant rats were anesthetized with isoflurane and killed by cervical dislocation. Cortical region was isolated from E18 brains and placed in ice-cold dissection buffer (HBSS without Ca2+ and Mg2+, 100 U/mL penicillin, 100 μg/mL streptomycin, 1× Glutamax). Cortical tissue was washed two times with the dissection buffer before adding a 0.025 % trypsin in HBSS solution followed by incubation at 37 °C for 15 min. After incubation, the tissue was washed 3 times with the HBSS washing buffer before adding Neurobasal (NB) medium, supplemented with 10 % FBS and 2 mmol/L GlutaMAX. The tissue was titrated several times with a glass Pasteur pipet, this treatment was repeated with a flame thinned tip glass Pasteur pipet to obtain fully dissociated cells. Separated cells were seeded in cell culture plates, which were overnight coated with 0.1 g/L, mol wt 70,000–150,000 poly-d-lysine (PDL) (Sigma Aldrich). For the first 5 h, the cells were cultured in medium containing NB medium with 10 % FBS and 2 mmol/L glutaMAX. Afterwards, the medium was replaced with culturing medium containing NB with the serum free neural supplement B27 and 2 mmol/L glutaMAX. For culturing pure axonal fractions, microfluidic chambers were prepared by firstly ethanol cleaning the chambers. After the chambers were completely dry, they were placed on PDL coated coverglasses (Hecht Assistent). The chambers were subsequently filled with 100 µL of NB medium containing 10 % FBS and 2 mmol/L GlutaMAX. This was done at least 1 day before seeding the cells so that any formed bubbles will be removed. In most cases a seeding density of 100,000 cells per chamber was used. All the neuronal cell preparations were maintained at 37 °C and with 5 % CO2.
Sponge-miR-181c construct
The miR-181c-sponge vector was made by designing DNA oligonucleotides (Sigma Aldrich) as previously described [13, 56, 57]. In short, miR sponge sequences were designed to contain four complementary artificial miR binding sites of the rat miR-181c-3p sequence, each separated by four nucleotides. The miR-181c binding site contains bulged sites that are non-complementary to the miR-181c, to enhance sponge efficacy. Presynthesized DNA oligonucleotides were ligated 3′ to the stop codon of the cDNA encoding eGFP. For cloning miR sponges into the lentiviral pFUGW (Addgene) constructs the restriction sites BsrGI and EcoRI were utilized.
Transfection and virus production
Transfection of cortical neurons was performed using the Screenfect A transfection agent (Genaxxon), following the protocol provided by the manufacturer. In brief, Screenfect A together with either the anti-miR-181c, miR-181c mimic or non-targeting (NT) control were diluted in the provided buffer to achieve a final concentration of 30 nM, respectively. To enable to visualize the transfection efficiency, 20 nM of siGlo red transfection indicator (Dharmacon) was added to the transfection mixture. After 20 min of incubation, the mix was applied to the adherent primary neurons and left to incubate at 37 °C for no longer than 24 h. For lentivirus production, HEK-293T cells were transfected using the CaCl2 method. The viral particles were produced and purified as previously described [58]. The purified viral pellets were resuspended in 100 µL calcium and magnesium free HBSS. For infecting primary neurons, ±0.5 µL/cm2 of concentrated virus was added to the cultures. If a lower infection rate was needed the virus was diluted 10–20 times before addition to the primary neuronal cultures.
RNA isolation, cDNA synthesis and qPCR
Total RNA was isolated from primary cortical neurons using the miRNeasy kit (Qiagen) according to the manufacturer’s protocol. RNA purity was determined using the Nanodrop ND1000 (Thermo Scientific) UV-spectrophotometer. The 260/280 nm ratios were measured and samples with ratios of 2.0 ± 0.05 were considered pure. This was followed by RNA integrity assessment with a 1 % agarose gel imaged with the Gel doc XR system (Bio-Rad). Samples showing clearly visible S28 and S18 ribosomal RNA bands were considered to be intact. cDNA was synthesized from isolated RNA according to the protocol provided with the miScript reverse transcription kit (Qiagen) to reverse transcribe both small RNA and mRNA. Quantitative Real-time PCR (QRT-PCR) detection of mRNA transcripts was performed using the miScript SYBR green PCR kit (Qiagen) for the detection of mature miRs and mRNA according to the manufacturer’s manual. The following gene-specific qPCR primers were used (in 5′ to 3′ direction): β-actin forward, CCAGATCATGTTTGAGACCTTC; β-actin reverse, AGGATCTTCATG-AGGTAGTCTG; U6 forward, GCTTCGGCAGCACATATA; U6 reverse, CGCTTCACGAATTTGCGT; mature rno-miR-181c, AACATTCAACCTGTC-GGTGAGT. The data was normalized to housekeeping genes (β-actin and U6) using the GeNorm software [59].
RNA sequencing
RNA isolated from primary cortical neurons (at 21 DIV) infected either with control or sponge-miR-181c lentiviruses at 8 DIV. RNA quality was assessed using Tapestation analysis, which revealed RNA quality above RIN 8.0 for all samples. RNA samples from the same condition were processed by the HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA). The RNA sequencing was performed in three separate flowcells generating a total of 45–50 million 50 bp reads per sample. RNA sequencing data analysis was carried out with the GeneSifter gene expression analysis suite (Geospiza). Expression data was normalized against the number of mapped reads, and the RPKM threshold was set at 1. Significance was calculated using the Benjamini and Hochberg multiple testing correction and the level of significance was set to 0.05.
miR target prediction
The miRWalk prediction database was used to identify predicted targets of the miRs we found significantly dysregulated due to prenatal VPA exposure. miRWalk combines the prediction data of several databases from which we included TargetScan, miRanda, miRDB and miRWalk prediction lists taking into account a minimal seed length of 7 base pairs and a p value cut-off of 0.05 [60].
Functional classification of miR targets
To detect significantly enriched gene categories in the predicted miR target genes/mRNAs, we used the IPA software package (http://www.ingenuity.com). This software package uses the Ingenuity Knowledge Base, which is based on information from the published literature as well as many other sources, including gene expression and gene annotation databases, to assign genes to different groups and categories of functionally related genes. Each of these categories can be further divided into many subcategories. Ingenuity calculates single p values for the enrichment of each gene category using the right-tailed Fisher’s exact test, and by taking into consideration both the total number of molecules from the analyzed data set and the total number of molecules that is linked to the same gene category according to the Ingenuity Knowledge Base. Furthermore, for each gene category, a multiple testing corrected p value of enrichment, calculated using the Benjamini–Hochberg correction, is provided.
Immunohistochemistry
Cultured neurons were fixed in warm (37 °C) 4 % paraformaldehyde and 16 % sucrose in PBS for 30 min. After washing with PBS (3 × 5 min at RT) cells were incubated for 15 min at RT with 50 mM NH4Cl to quench residual aldehydes. Cells were washed (3 × 5 min at RT), permeabilized with 0.1 % Trixon X-100 for 5 min at RT, incubated with 2 % bovine serum albumin (BSA, ICN Biomedicals Inc) in PBS for 30 min at RT to block non-specific staining, and were incubated with the primary antibodies in 1 % BSA in PBS. Incubation in primary antibodies was done overnight at 4 °C. Subsequently, cells were washed in 2 % BSA in PBS (3 × 5 min) and incubated in species-specific, Alexa-conjugated secondary antibodies in 1 % BSA in PBS for 30–60 min at RT, washed in 2 % BSA in PBS (3 × 5 min), and washed (3 × 5 min) in PBS containing fluorescent DAPI (Sigma, 1:3000). The coverslips were mounted in Prolong Gold (Invitrogen). Primary antibodies included chicken anti-GFP (A10262, Life Technologies, 1:500), rabbit anti-MAP2 (ab32454, Abcam, 1:1000), mouse anti-Tau1 (MAB3420, Millipore, 1:1000). Secondary antibodies included goat anti-rabbit 488 (Life Technologies, 1:500), goat anti-chicken 488 (Life Technologies, 1:500), goat anti-mouse 568 (Life Technologies, 1:500). For visualizing and measuring the axons in the microfluidic chambers, a similar protocol was used. After permeabilizing the neurons with 0.1 % Trixon X-100 and washing with PBS the cells were incubated with 100 nM Acti-stain 488 phalloidin (Cytoskeleton) in PBS for 30 min. After washing the cells were mounted and imaged.
Image analysis
Images of neurons were captured using a Leica TCS SP2 AOBS Confocal Laser Scanning Microscope (CLSM) with 20× magnification for neurite tracing or at 63× magnification for dendritic spine imaging. For outgrowth analysis, the ImageJ plugin NeuronJ was used with which neurons were traced [61]. Neurites were assigned with the correct branch order where neurites originating from the soma were labeled as primary or first branch order and increasing in order values after each successive branch point. Sholl analysis was performed with the concentric circle ImageJ plugin to draw an overlay of circles (20 µm distance step size) onto the images of neurons, followed by manual quantification of the number of intersecting neurites with each ring. Neuronstudio software was used to quantify spine density by determining the amount of spines per micrometer of dendrite followed by spine classification. Spine maturity was automatically assigned based on the length of the spine and the quantified diameter of the spine neck and head, with thin filopodia like spines indicated as immature and mushroom like spines indicated as mature [62].
Results
Inhibition of miR-181c alters the expression of transcripts involved in nervous system development
Due to their relatively small seed regions, miRs have the potential to regulate hundreds of downstream targets, and consequently, control key cellular processes [16]. To investigate which cellular pathways are controlled by miR-181c we used RNA sequencing to examine genome-wide gene expression changes upon inhibition of miR-181c function in primary cortical neurons. To suppress miR-181c, we generated and functionally characterized a competitive ‘miR sponge’ to allow for long-term sequestering of miR-181c in cortical neurons. This lentiviral based construct contains a concatemer of four partially complementary miR-181c binding sites within the 3′ untranslated region (UTR) of a GFP fluorophore (Supplementary Fig. 1A). To test whether the introduction of the miR-181c sponge into cells leads to a decrease in mature miR-181c levels, RNA was isolated from days in vitro (DIV) 9 cortical neurons infected at DIV 1 with the miR-181c sponge or a GFP control (Supplementary Fig. 1B). qPCR analysis revealed that cells expressing the miR-181c sponge exhibited a significant decrease in miR-181c expression levels (mean 1.0 ± 0.0994 control versus 0.3433 ± 0.01252 miR-181c sponge relative miR-181c expression; p = 0.0028) (Supplementary Fig. 1C).
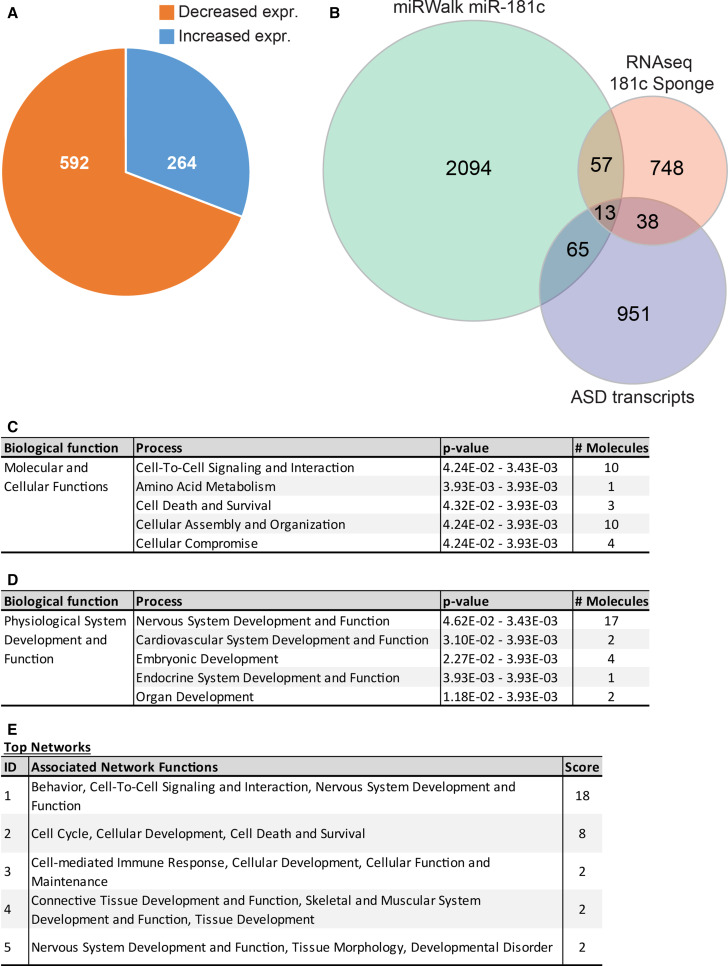
Total RNA was isolated from DIV 21 cortical neurons infected with lentiviral particles encoding the miR-181c sponge or a GFP control, and subjected to RNA deep sequencing [13]. miR-181c inhibition in cortical neurons resulted in significant expression changes of 1596 transcripts, with 880 transcripts decreased and 716 transcripts increased in their expression. A 1.2 fold-change cut-off was applied to remove marginally altered genes. This investigation resulted in the identification of 856 significantly differentially expressed genes, with 592 transcripts decreased and 264 transcripts increased in their expression (Fig. 1a) (Supplementary Table 1). Further analysis using the miRWalk binding site prediction algorithm on these 856 genes showed that 70 transcripts (8.2 %) contained one or more putative miR-181c binding sites within the 3′UTR of these transcripts (Fig. 1b). Of these 70 genes, 16 were increased, while 54 genes were decreased in their expression. Since miR-181c was previously associated with ASD, we cross-compared the miR-181c regulated transcriptome with the set of genes that were found to be dysregulated in cortical material from ASD patients to examine the potential impact this miR may have in modulating genes previously linked with ASD [17–19]. Furthermore, an unbiased comparison suggested that 51 genes within the transcriptomes from postmortem cortical tissue of ASD patients were among the genes altered in their expression by miR-181c suppression. Among these 51 genes, 13 transcripts contained a binding site for miR-181c in their 3′UTR (Fig. 1b).
Fig. 1.
Inhibition of miR-181c function alters the expression of genes functionally involved with neurodevelopmental processes. a Pie diagram depicts the number of genes significantly (p < 0.05) increased (blue) and decreased (orange) in expression by more than 1.2 fold. b Venn diagram shows the overlap between the putative miR-181c targets identified with miRWalk (2229 molecules), the significantly altered transcripts after miR-181c inhibition using the sponge in cortical neurons (786 molecules) and significantly altered transcripts identified in cortical material from ASD patients (1067 molecules). This analysis resulted in the identification of 70 significantly altered transcripts containing a miR-181c binding site and 13 of which previously found to be dysregulated in ASD. c, d Overview of the gene ontology study shows 70 genes containing a miR-181c binding site in their 3′UTR, altered in their levels upon inhibition of miR-181c. c The most significant process in the category molecular and cellular functions is cell-to-cell signaling and interaction. d Within the category physiological system development and function the process nervous system development and function has the highest number of dysregulated genes. e A list of the top five highest scoring gene networks identified by the IPA. Each individual network has functions assigned to them with the highest scoring network involved in behavior, cell-to-cell signaling and interaction, nervous system development and function
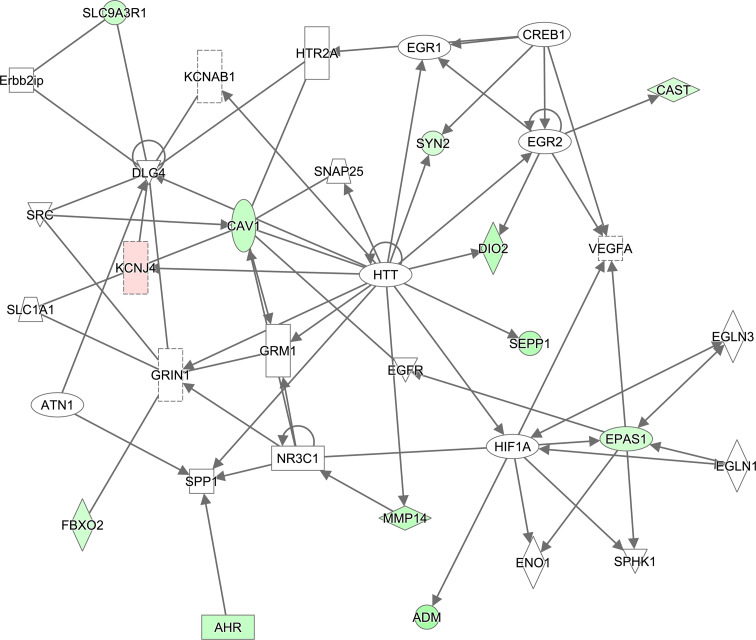
To identify miR-181c regulated gene clusters involved in common gene networks, we performed an Ingenuity pathway analysis (IPA) gene ontology clustering on the 70 genes containing a miR-181c binding site. This study revealed that within the biological function category “molecular and cellular functions” the top five enriched themes are cell-to-cell signaling and interaction, amino acid metabolism, cell death and survival, cellular assembly and organization, and cellular compromise (Fig. 1c). The theme cell-to-cell signaling mainly contains genes involved in functions such as activation of neuronal cells, synaptic transmission, and cell adhesion of neuronal cells. Examples of genes from this group include Apln, Ptger3, Syn2, Cast and Fbxo2. The biological function “physiological system development and function”, nervous system development and function was the theme with the largest number of genes (Fig. 1d). Prominent sub-themes of this category are abnormal morphology of neuronal cells, activation of neurons, and development of cortical or dopaminergic neurons. Genes within these sub-themes are Cast, Cav1, Cyr61, Fgfr3, Lrp4, Pdgfrα, Rhoq and Syn2. Furthermore, using the IPA software to examine the molecular relationship among genes, we identified a number of associated networks (Fig. 1e), with a score of 18, meaning it has a 1E−18 chance that the input genes are found together by random chance, containing genes associated with the functions behavior, cell-to-cell signaling and interaction, and nervous system development (Figs. 1e, 2). This network consists of 35 genes of which 11 showed an increase and one a decrease in their expression upon inhibition of miR-181c function (Fig. 2; Table 1). Within the network, Huntington (Htt) connects with 16 genes, and appears to act as a central regulatory hub. Other central genes within the network, with nine or more connections, are Dlg4, Cav1 and Hif1a. Dlg4 and Cav1 interact primarily with ion channel subunits or genes involved in synaptic vesicle trafficking while the transcription factor Hif1a activates a set of neuronal development genes (Fig. 2). Together, these results suggest that miR-181c inhibition in cortical neurons leads to significant changes in transcripts involved in nervous system development and function.
Fig. 2.
MiR-181c control of gene networks. Highest scoring network identified by IPA with genes involved in behavior, cell-to-cell signaling and interaction, nervous system development and function. Highlighted within the diagram are transcripts that are increased (red) or decreased (green) in their expression by miR-181c inhibition
Table 1.
List of genes functioning in behavior, cell-to-cell signaling and interaction, nervous system development and function
| Gene ID | Other ID | Ratio | Direction | p value | Adj. p value |
|---|---|---|---|---|---|
| Adm | 25026 | 2.58 | Down | 0.0001983 | 5.15E−03 |
| Ahr | 25690 | 1.73 | Down | 4.42E−05 | 1.50E−03 |
| Cast | 25403 | 1.29 | Down | 1.35E−05 | 5.73E−04 |
| Cav1 | 25404 | 1.71 | Down | 0.0002832 | 6.76E−03 |
| Dio2 | 65162 | 1.96 | Down | 1.65E−05 | 6.74E−04 |
| Epas1 | 29452 | 1.44 | Down | 0.0005326 | 1.10E−02 |
| Fbxo2 | 85273 | 1.42 | Down | 6.79E−05 | 2.10E−03 |
| Kcnj4 | 116649 | 1.25 | Up | 1.11E−16 | 4.57E−14 |
| Mmp14 | 81707 | 1.92 | Down | 2.18E−11 | 4.13E−09 |
| Sepp1 | 29360 | 2.13 | Down | 1.77E−09 | 2.31E−07 |
| Slc9a3r1 | 59114 | 1.58 | Down | 5.47E−05 | 1.77E−03 |
| Syn2 | 29179 | 1.21 | Down | 0.0007638 | 1.45E−02 |
MiR-181c controls the growth of neurites and dendritic spines in developing cortical neurons
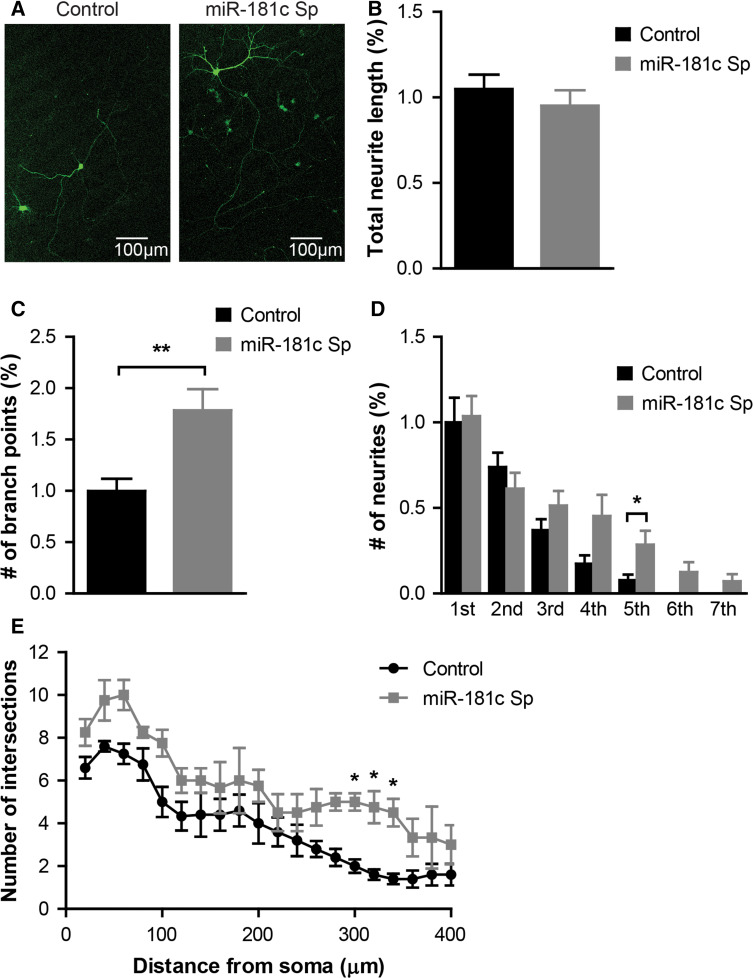
Our transcriptome analysis suggests that miR-181c controls the activity of gene networks strongly associated with neuronal development, supporting previous findings that have implicated aberrant miR-181c expression with the etiology of neurodevelopmental disorders such as ASD [12, 13]. This led us to evaluate whether miR-181c has the capacity to modulate basic neural maturation processes in cultured cortical neurons. Utilizing miR-181c-sponge to knock-down endogenous miR-181c levels, primary rat cortical neurons were infected 1 day after seeding and analyzed at DIV 9 for differences in neurite growth as compared to GFP control infected neurons (Fig. 3a). miR-181c inhibition did not result in changes in total dendritic length (mean 1.05 ± 0.084 control versus 0.95 ± 0.09 miR-181c sponge length in percentage; p = 0.44) (Fig. 3b), whereas it did enhance dendritic branching compared to control infected neurons (mean 1.0 ± 0.118 control versus 1.79 ± 0.206 miR-181c sponge length in percentage; p = 0.0045) (Fig. 3c). Further analysis of the position of the branch points showed that enhanced branching was mainly due to an increase in the number of higher order branches (Fig. 3d). Moreover, Sholl analysis showed the expression of the miR-181c sponge enhanced neurite arborisation revealed by a significantly exacerbated increase in the number of intersecting dendrites at 300 µm distance from the soma compared to control infected cells (Fig. 3e).
Fig. 3.
Inhibition of miR-181c affects outgrowth in cortical neurons. a Representative images of DIV9 neurons infected with control or sponge-miR-181c lentiviruses. b Quantification of total dendritic length. c Graph showing the number of branching points of control and sponge-miR-181c infected neurons. d Quantification of the number of branches per branch order. Data are shown as mean ± SEM; p values are determined by two-tailed unpaired Student’s t test. *p ≤ 0.05; **p ≤ 0.01. e Sholl analysis revealing dendritic arborisation of neurons infected with either control (black line) or sponge-miR-181c (gray line) lentivirus. Data are shown as mean ± SEM; p values are determined by two-way ANOVA followed by Bonferroni multiple comparisons test. *p ≤ 0.05
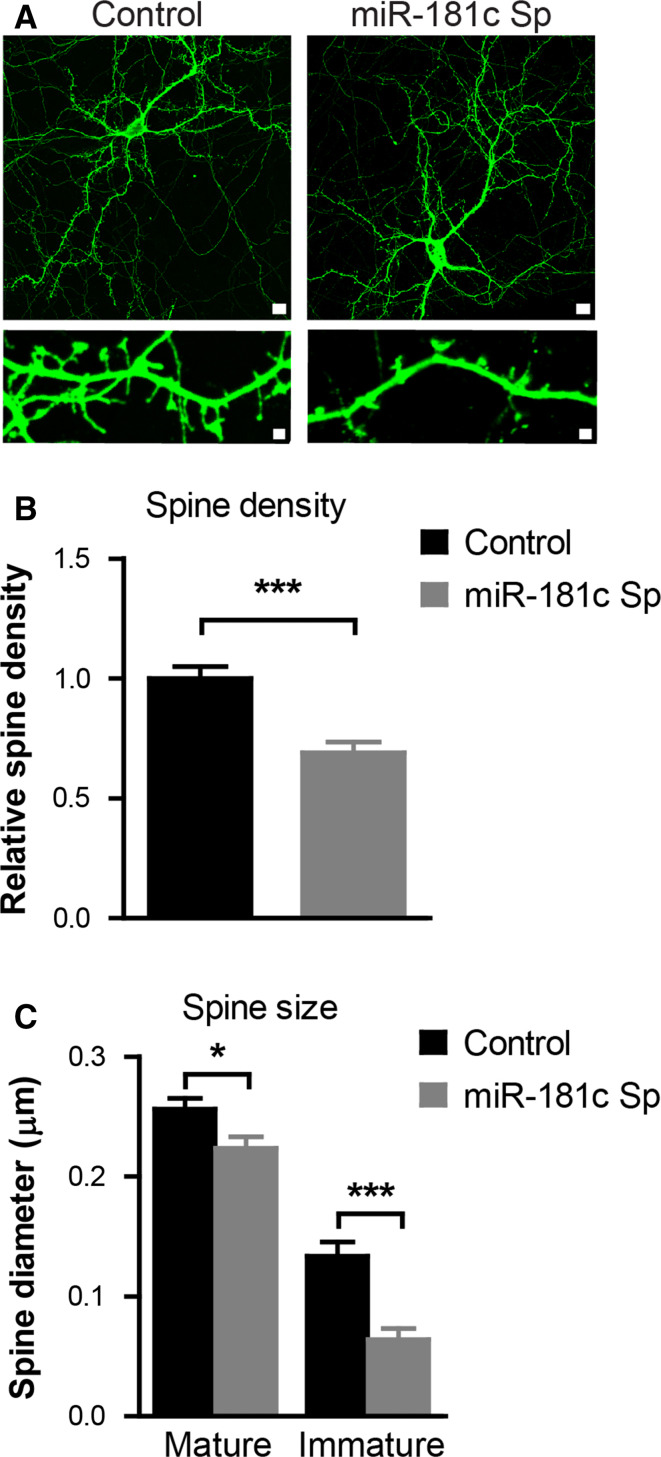
To determine whether miR-181c also affects the process of synapse formation, we measured the effects of selective miR-181c repression on the number and shape of dendritic spines. Accordingly, neurons were infected at DIV 9 with viruses driving the expression of either GFP control or sponge-miR-181c. GFP-positive neurons were studied when they reached maturity at 21 DIV. Analysis of the obtained images revealed that the spine density of sponge-miR-181c infected cells was decreased as compared to control virus infected neurons (mean 1.0 ± 0.04973 control versus 0.69 ± 0.04370 miR-181c sponge relative spine density; p ≤ 0.0001) (Fig. 4a, b). Measurements of spine width revealed that inhibiting miR-181c function results in a significant reduction in the size of both mature and immature synapses (mean 0.2562 ± 0.0091 control versus 0.2236 ± 0.0093 miR-181c mature spine diameter in µm; p = 0.0147 and mean 0.1335 ± 0.0128 control versus 0.06398 ± 0.0094 miR-181c immature spine diameter in µm; p ≤ 0.0001) (Fig. 4c).
Fig. 4.
Inhibition of miR-181c affects spine density and morphology. a Representative images of DIV21 neurons infected with either control or sponge-miR-181c at DIV8. b Quantification of spine density of control and sponge-miR-181c infected neurons. c Graph showing the measurements of spine head diameter of mature (mushroom and stubby) and immature thin spines. Data are shown as mean ± SEM; p values are determined by two-tailed unpaired Student’s t test. *p ≤ 0.05 and ***p ≤ 0.0001
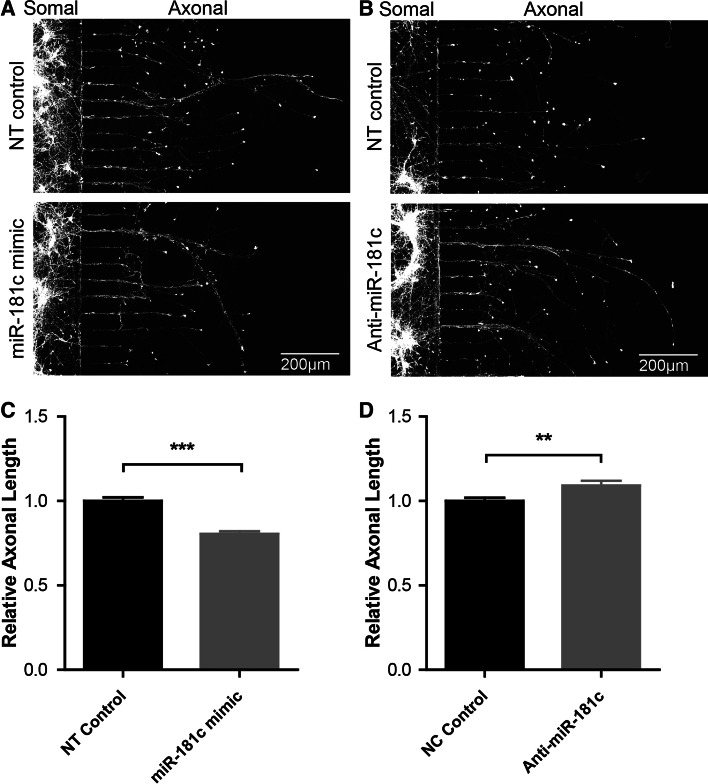
Previously, it was shown that miRs can be transported to distal parts of the axon and are important for axon development and function [20–24]. To investigate whether miR-181c has the capacity to regulate the growth of axons from young cortical neurons we made use of microfluidic chambers, which allowed us to selectively study axons in compartmentalized isolation from the soma and dendrites (Supplementary Fig. 2A, B) [25]. For modulating miR-181c in this setup we made use of miR-181c mimics and locked nucleic acid (LNA) anti-miRs to examine how changes in miR-181c levels result in altered axonal outgrowth. We opted for the use of small RNA molecules since they have the capacity to rapidly modulate miR levels (as opposed to the miR sponges which act slowly), and because LNAs are readily transfected into primary neurons grown in compartmentalized culture systems (Supplementary Fig. 1D). qPCR analysis revealed that transfection of neurons with the miR-181c mimic lead to a significant increase of mature miR-181c (66 % increase; p = 0.0028) (Supplementary Fig. 1E). Conversely, transfection with an anti-miR-181c resulted in a significant decrease of miR-181c compared to control transfected neurons (Supplementary Fig. 1F). For the axon outgrowth experiments these synthetic RNAs were transfected into DIV 3 cortical neurons grown in microfluidic chambers and subsequently analyzed at DIV 6 (Fig. 5a, b). Compared to a non-targeting (NT) control, miR-181c overexpressing neurons exhibited an attenuated axon growth (mean 20 % decrease; p ≤ 0.0001) (Fig. 5d), whereas inhibiting miR-181c modestly but significantly increased axon outgrowth (mean 9 % increase; p = 0.0054) (Fig. 5e). These findings suggest that modulation of miR-181c expression levels in cortical neurons results in altered axonal growth.
Fig. 5.
miR-181c attenuates axonal growth in cortical neurons. a Representative images of DIV 6 cortical neurons grown in microfluidic chambers transfected with NT control or miR-181c mimic. b Representative images of DIV 6 cortical neurons transfected with NT control or anti-miR-181c inhibitor. c Relative axonal length of neurons overexpressing miR-181c compared to the control. d Relative axon length of neurons transfected with the miR-181c inhibitor compared to the control. Data are shown as mean ± SEM; p values are determined by two-tailed unpaired Student’s t test. **p ≤ 0.001 and ***p ≤ 0.0001
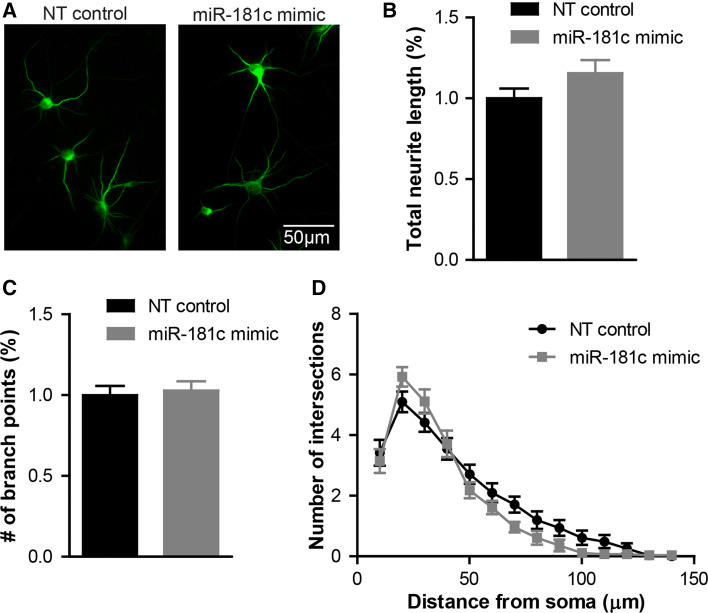
Given that miR-181c is overexpressed in some ASD cells, we performed a miR-181c gain-of-function assay by transfecting DIV 3 cortical neurons with a miR-181c mimic. Three days after transfection, neurons overexpressing this miR exhibited only a minimal change in dendrite length, branching and complexity compared to cells transfected with NT control oligonucleotides (Fig. 6a–d). This finding suggests that miR-181c overexpression does not significantly alter dendrite formation during this growth period. Moreover, to examine the impact of miR-181c overexpression in dendritic spine maturation, we investigated spine formation in DIV16 primary cortical neurons transfected with miR-181c mimic at DIV3. Since most neurons exhibited a necrotic phenotype during the time when spine maturation occurs, we concluded that miR-181c overexpression in cortical neurons within an extended period of time results in cell death (Supplementary Fig. 3).
Fig. 6.
miR-181c overexpression has minimal impact on dendrite outgrowth. a Representative images of DIV 6 cortical neurons transfected at DIV 3 with NT control or miR-181c mimic and stained with the dendrite marker MAP2. Quantification of b total dendritic length and c total number of branching points of NT control (black bars) or miR-181c mimic (gray bars) transfected cells. Sholl analysis of neurons transfected with either NT control (black line) or miR-181c mimic (gray line). Data are shown as mean ± SEM
Discussion
Subtle changes in neurite outgrowth, synaptic development and function, and neural network formation underlie the etiology of neurodevelopmental disorders and cognitive dysfunction [26–30]. Here, we provide accumulating evidence for a critical role for miR-181c in neurite growth and synaptic development. Using transcriptome profiling, we show that inhibition of miR-181c during development of cortical cells altered the expression of a large set of transcripts. Gene network analysis points towards a significant enrichment of genes involved in neural development and synaptic function. Subsequent neuromorphological analysis of cells, in which miR-181c was knocked-down, revealed a significant increase in dendritic branching and decreased dendritic spine size. Furthermore, cortical cells lacking miR-181c displayed longer axons, while increasing miR-181c expression results in shorter axons. Conversely, short-term overexpression of miR-181c in young cortical neurons had negligible impact on dendritic outgrowth and branching. A possibility for a lack of a clear dendrite outgrowth phenotype during overexpression could be that endogenous abundance of miR-181c impeded any additional gain-of-function by the introduction of exogenous miR-181c. Moreover, the transient miR-181c overexpression was conducted between DIV 3 and DIV 6, a time period in which this miR may not have critical importance for neuronal outgrowth and branching.
Overexpression of miR-181c for an extended period of time resulted in premature cell death of cortical neurons as illustrated by condensed nuclei and diminished PSD-95 staining. This observation suggests that long-term overexpression of miR-181c resulted in either apoptosis or necrosis in these cells. Examining the extant literature we encountered recent studies that implicated miR-181 family members in the induction of apoptosis. Multiple members of the Bcl-2 family were found to be regulated by miR-181 family members (including miR-181c) and specific overexpression of miR-181a increased apoptosis in primary astrocyte cultures [31]. Moreover, miR-181 was shown to regulate members of the heat shock protein family resulting in aggravated cell injury during cellular stress, while increasing the expression of miR-181a-b exacerbated cerebral ischemia [32]. However, of important note, we did not see increased cell death compared to control when we transfected neurons with the miR-181c mimic for 3 days.
Collectively, the results of our study suggest that miR-181c has the capacity to regulate the expression of hundreds of downstream transcripts, indicating a vast regulatory potential for this miR during neuronal development [16]. Following long-term inhibition of miR-181c function in cortical neurons, whole transcriptome analysis resulted in the identification of 856 genes significantly altered in their expression by 1.2-fold or more. More than half of the genes were decreased in their expression. Surprisingly, most of the significantly altered genes lack putative miR-181c regulation sites, suggesting that a large number of these genes are possibly regulated by miR-181c in a non-canonical manner. It has recently become evident that miRs can indeed control gene expression in an indirect manner [33, 34]. Thus, within our identified gene set miR-181c may control a number of genes in this mode of action. Furthermore, miR-181c could indirectly induce expression changes of transcripts that are downstream of direct targets. It is therefore important to study miR regulation using high-throughput experimental setups together with prediction algorithms to chart global gene network changes.
IPA gene ontology clustering analysis of the genes significantly induced by the introduction of the miR-181c sponge in cortical neurons resulted in the identification of gene clusters involved in neuronal development. In particular, eight genes significantly altered by the miR-181c sponge play important roles in the development of neurons, namely Cast, Cav1, Cyr61, Fgfr3, Lrp4, Pdgfra, Rhoq and Syn2 [35–40]. Furthermore, an enrichment of genes involved in the activation of neurons and regulation of synaptic transmission were found, namely Apln, Ptger3, Syn2, Cast and Fbxo2 [41–43]. Together, these results suggest that miR-181c controls, through direct or indirect interactions, several gene clusters, which modulate neuronal developmental processes and synaptic function.
Due to the high degree of sequence identity between the various miR-181 family members (miR-181a–d) it is possible that the sponge used in this study reduced the expression of other miR-181 family members besides miR-181c. Indeed, introduction of a miR-181a LNA inhibitor into neuronal cell cultures has led to decreased expression of all family members [44]. Unfortunately, the currently available tools do not allow a more specific manipulation of the expression of highly conserved miR family members, such as the miR-181 family members. The extent of the sequence identity among the miR-181 family members further suggests that the members regulate similar gene networks. This is exemplified by the finding that when we compare predicted targets for the various miR-181 family members we find extensive overlap between the putative targets. This could be indicative that the miR-181 family could cooperatively control similar targets. miR-181 family members sharing the same seed sequence have actually been found to be dysregulated in neurodevelopmental disorders or control neurodevelopmental processes. For example, miR-181a, miR-181a* and miR-181b were significantly elevated in ASD patients, similar to miR-181c. Furthermore, two independent studies have shown increased levels of miR-181b in gray matter of the dorsolateral prefrontal cortex and temporal cortex of schizophrenia patients [45, 46]. Finally, miR-181a acts as a negative regulator of AMPA-R surface expression by direct targeting of GluA2, resulting in reduced spine development and miniature excitatory postsynaptic current (mEPSC) frequency in hippocampal neurons [44]. Thus, the miR-181 family controls overlapping downstream target genes, and potentially modulate the expression of similar cellular pathways. Multiple miRs from a single family sharing sequence similarities can indeed cooperatively control cellular processes [47, 48]. For example, miR-135a/b control chronic stress resilience through regulation of overlapping genes within the serotonergic system [49], while miR-48, mir-84, and mir-241 family members with sequence similarity to let-7 cooperatively control critical developmental timing events in Caenorhabditis elegans [50].
Our data indicate that miR-181c inhibition dysregulates the levels of a number of neurodevelopmentally relevant genes, implying that disruption of miR-181c activity during cortical neuron development may result in altered neuronal morphologies. We showed that miR-181c inhibition leads to increased dendritic branching while increasing axon length and the number and size dendritic spines were decreased. In a recent study, we showed that increased levels of miR-30d and miR-181c were found in the amygdala of a VPA-induced rat model for autism [13]. Additional studies showed that inhibition of miR-181c in neurons isolated from the amygdala resulted in a decrease in neurite growth and branching and fewer dendritic spines. The observed differences in the effects of miR-181c on neurite growth could be attributed to spatiotemporally divergent gene expression in amygdala cells compared to cortical cells. Alternatively, the primary cell cultures used in those studies contain a variety of cell types, thus the cellular composition of amygdala cultures could differ from those isolated from the cortex. This underlines the importance for investigating miR regulation in a multitude of genetic and cellular contexts.
Previously, the involvement of the non-coding transcriptome, including miRs, has been studied in the context of neurodevelopmental disorders such as ASD, schizophrenia and intellectual disability disorders (ID) [12, 51–55]. Expression profiling of cell lines derived from ASD patients has revealed 12 differentially expressed miRs with miR-181c being the second most significantly elevated miR [12]. This suggests the possibility that altered miR expression could underlie some of the expression changes found in ASD. Indeed, when we compared transcriptome changes identified in the cortex of ASD patients we found a considerable overlap with the miR-181c transcriptome changes. This opens up the possibility that miR-181c and its family members are partially responsible for the mRNA expression changes in ASD patients, which may be contributing to disease susceptibility and/or progression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Figure 1: MiR-181c transduction efficiency into primary cortical neurons. A. Schematic overview of the lentiviral miR-181c-sponge construct designed to continuously and specifically sequester miR-181c. A concatemer of four complementary miR-181c binding sites is placed within the 3′UTR of the eGFP gene. As example, one binding site of the sponge with the mature miR-181c is shown, the central mismatch is included to enhance efficiency [56,63,64]. B. Micrographs of primary cortical neurons infected with the miR-181c sponge lentivirus. The left image shows cell nuclei visualized with DAPI (bleu), the middle panel shows GFP from the miR-181c sponge (green) and on the right is the DAPI and GFP overlay. C. Relative expression of miR-181c in DIV 9-old primary cortical neurons expressing the GFP control or miR-181c sponge construct normalized to control. D. Representative images of primary cortical neurons transfected with siGLO transfection indicator. The micrographs from left to right show the nuclear DAPI staining (blue), siGLO (red) and a DAPI with siGLO overlay. E. Relative expression of miR-181c in DIV 6 primary cortical neurons transfected with NT control or miR-181c mimic, levels were normalized to control. F. Relative expression of miR-181c in DIV 6 primary cortical neurons transfected with NT control or anti-miR-181c, levels were normalized to control. Data are shown as mean ± SEM; p values are determined by two-tailed unpaired Student’s t test. *p ≤ 0.01 and ** p ≤ 0.001 (JPEG 1349 kb)
Supplement Figure 2: Growing pure axonal fractions in microfluidic chambers. A. Schematic representation of a microfluidic chamber with the soma side (gray) and the axon side (green) connected with microgrooves in the middle. Neurons are shown in red, which grow their axons through the microgrooves to the axonal side. B. An example immunostaining of cortical neurons cultured in microfluidic devices. The DIC image shows the microgrooves of the microfluidic chamber, MAP2 highlights the dendrites in green and Tau1 for visualizing axons in red. The image on the bottom right corner shows an overlay of DIC, MAP2 and Tau1 (JPEG 403 kb)
Supplement Figure 3: Mature cortical neurons transduced with miR-181c mimic have a necrotic phenotype. Representative micrographs of DIV16 primary cortical neurons transfected with 30 nM NT control (upper panels) or miR-181c mimic (lower panels) and 20 nM SiGlo fluorescent transfection indicator at DIV 3. The panels depict micrographs of DIV16 neurons stained with DAPI (blue), PSD-95 (green) and the SiGlo transfection indicator (red). The necrotic phenotype (condensed nuclei) in miR-181c transfected neurons was observed for around 90% of the cells; this experiment was repeated three times, with similar outcomes (JPEG 853 kb)
Supplement Table 1: List of genes functioning in behavior, cell-to-cell signaling and interaction, nervous system development and function (PDF 100 kb)
References
- 1.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin–proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smart F, Aschrafi A, Atkins A, Owens GC, Pilotte J, et al. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J Neurochem. 2007;101:1367–1379. doi: 10.1111/j.1471-4159.2007.04521.x. [DOI] [PubMed] [Google Scholar]
- 3.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 4.Huang F, Chotiner JK, Steward O. The mRNA for elongation factor 1alpha is localized in dendrites and translated in response to treatments that induce long-term depression. J Neurosci. 2005;25:7199–7209. doi: 10.1523/JNEUROSCI.1779-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aschrafi A, Kar AN, Natera-Naranjo O, Macgibeny MA, Gioio AE, et al. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci. 2012;69:4017–4027. doi: 10.1007/s00018-012-1064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 7.Barca-Mayo O, De Pietri Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell Mol Life Sci. 2014;71:2975–2995. doi: 10.1007/s00018-014-1576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bicker S, Lackinger M, Weiss K, Schratt G. MicroRNA-132, -134, and -138: a microRNA troika rules in neuronal dendrites. Cell Mol Life Sci. 2014;71:3987–4005. doi: 10.1007/s00018-014-1671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, et al. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer AN, Bellon A, Baudet ML. microRNAs in axon guidance. Front Cell Neurosci. 2014;8:78. doi: 10.3389/fncel.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380:85–97. doi: 10.1016/j.brainres.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 13.Olde Loohuis NF, Kole K, Glennon JC, Karel P, Van der Borg G, et al. Elevated microRNA-181c and microRNA-30d levels in the enlarged amygdala of the valproic acid rat model of autism. Neurobiol Dis. 2015;80:42–53. doi: 10.1016/j.nbd.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith B, Treadwell J, Zhang D, Ly D, McKinnell I, et al. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS One. 2010;5:e11109. doi: 10.1371/journal.pone.0011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, et al. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 17.Chow ML, Pramparo T, Winn ME, Barnes CC, Li HR, et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet. 2012;8:e1002592. doi: 10.1371/journal.pgen.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki Y, Gross C, Xing L, Goshima Y, Bassell GJ. Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev Neurobiol. 2014;74:397–406. doi: 10.1002/dneu.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock ML, Preitner N, Quan J, Flanagan JG. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J Neurosci. 2014;34:66–78. doi: 10.1523/JNEUROSCI.3371-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, et al. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci. 2012 doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- 24.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, et al. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Microfluidic culture platform for neuroscience research. Nat Protoc. 2006;1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 26.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher D, Voronova A, Zander MA, Cancino GI, Bramall A, et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev Cell. 2015;32:31–42. doi: 10.1016/j.devcel.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 29.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, et al. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006;9:1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 31.Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12:213–219. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang YB, Lu Y, Yue S, Xu LJ, Xiong XX, et al. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012;45:555–563. doi: 10.1016/j.nbd.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang WX, Wilfred BR, Xie K, Jennings MH, Hu YH, et al. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol. 2010;7:373–380. doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Head BP, Peart JN, Panneerselvam M, Yokoyama T, Pearn ML, et al. Loss of caveolin-1 accelerates neurodegeneration and aging. PLoS One. 2010;5:e15697. doi: 10.1371/journal.pone.0015697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Head BP, Hu Y, Finley JC, Saldana MD, Bonds JA, et al. Neuron-targeted caveolin-1 protein enhances signaling and promotes arborization of primary neurons. J Biol Chem. 2011;286:33310–33321. doi: 10.1074/jbc.M111.255976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik AR, Urbanska M, Gozdz A, Swiech LJ, Nagalski A, et al. Cyr61, a matricellular protein, is needed for dendritic arborization of hippocampal neurons. J Biol Chem. 2013;288:8544–8559. doi: 10.1074/jbc.M112.411629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y, Tian QB, Endo S, Suzuki T. A role for LRP4 in neuronal cell viability is related to apoE-binding. Brain Res. 2007;1177:19–28. doi: 10.1016/j.brainres.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 39.Yumoto N, Kim N, Burden SJ. Lrp4 is a retrograde signal for presynaptic differentiation at neuromuscular synapses. Nature. 2012;489:438–442. doi: 10.1038/nature11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei Y, Wang Z, Zhang L, Zhang Y, Li X, et al. Regulation of neuroblastoma differentiation by forkhead transcription factors FOXO1/3/4 through the receptor tyrosine kinase PDGFRA. Proc Natl Acad Sci USA. 2012;109:4898–4903. doi: 10.1073/pnas.1119535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feliciano P, Andrade R, Bykhovskaia M. Synapsin II and Rab3a cooperate in the regulation of epileptic and synaptic activity in the CA1 region of the hippocampus. J Neurosci. 2013;33:18319–18330. doi: 10.1523/JNEUROSCI.5293-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takao-Rikitsu E, Mochida S, Inoue E, Deguchi-Tawarada M, Inoue M, et al. Physical and functional interaction of the active zone proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J Cell Biol. 2004;164:301–311. doi: 10.1083/jcb.200307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuen EY, Wei J, Liu W, Zhong P, Li X, et al. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron. 2012;73:962–977. doi: 10.1016/j.neuron.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saba R, Storchel PH, Aksoy-Aksel A, Kepura F, Lippi G, et al. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32:619–632. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 46.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamanu TK, Radovanovic A, Archer JA, Bajic VB. Exploration of miRNA families for hypotheses generation. Sci Rep. 2013;3:2940. doi: 10.1038/srep02940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med. 2014;24:105–112. doi: 10.1016/j.tcm.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Issler O, Haramati S, Paul ED, Maeno H, Navon I, et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 50.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans . Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willemsen MH, Valles A, Kirkels LA, Mastebroek M, Olde Loohuis N, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 53.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 56.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14:1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 2011;14:1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- 58.Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 61.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Figure 1: MiR-181c transduction efficiency into primary cortical neurons. A. Schematic overview of the lentiviral miR-181c-sponge construct designed to continuously and specifically sequester miR-181c. A concatemer of four complementary miR-181c binding sites is placed within the 3′UTR of the eGFP gene. As example, one binding site of the sponge with the mature miR-181c is shown, the central mismatch is included to enhance efficiency [56,63,64]. B. Micrographs of primary cortical neurons infected with the miR-181c sponge lentivirus. The left image shows cell nuclei visualized with DAPI (bleu), the middle panel shows GFP from the miR-181c sponge (green) and on the right is the DAPI and GFP overlay. C. Relative expression of miR-181c in DIV 9-old primary cortical neurons expressing the GFP control or miR-181c sponge construct normalized to control. D. Representative images of primary cortical neurons transfected with siGLO transfection indicator. The micrographs from left to right show the nuclear DAPI staining (blue), siGLO (red) and a DAPI with siGLO overlay. E. Relative expression of miR-181c in DIV 6 primary cortical neurons transfected with NT control or miR-181c mimic, levels were normalized to control. F. Relative expression of miR-181c in DIV 6 primary cortical neurons transfected with NT control or anti-miR-181c, levels were normalized to control. Data are shown as mean ± SEM; p values are determined by two-tailed unpaired Student’s t test. *p ≤ 0.01 and ** p ≤ 0.001 (JPEG 1349 kb)
Supplement Figure 2: Growing pure axonal fractions in microfluidic chambers. A. Schematic representation of a microfluidic chamber with the soma side (gray) and the axon side (green) connected with microgrooves in the middle. Neurons are shown in red, which grow their axons through the microgrooves to the axonal side. B. An example immunostaining of cortical neurons cultured in microfluidic devices. The DIC image shows the microgrooves of the microfluidic chamber, MAP2 highlights the dendrites in green and Tau1 for visualizing axons in red. The image on the bottom right corner shows an overlay of DIC, MAP2 and Tau1 (JPEG 403 kb)
Supplement Figure 3: Mature cortical neurons transduced with miR-181c mimic have a necrotic phenotype. Representative micrographs of DIV16 primary cortical neurons transfected with 30 nM NT control (upper panels) or miR-181c mimic (lower panels) and 20 nM SiGlo fluorescent transfection indicator at DIV 3. The panels depict micrographs of DIV16 neurons stained with DAPI (blue), PSD-95 (green) and the SiGlo transfection indicator (red). The necrotic phenotype (condensed nuclei) in miR-181c transfected neurons was observed for around 90% of the cells; this experiment was repeated three times, with similar outcomes (JPEG 853 kb)
Supplement Table 1: List of genes functioning in behavior, cell-to-cell signaling and interaction, nervous system development and function (PDF 100 kb)