Abstract
Atrial fibrillation (AF) is the most common sustained arrhythmia and has significant clinical impact. Over the last decade, our understanding of the genetics of AF has expanded dramatically. After a heritable predisposition for AF was identified, many investigators have in turn identified both common and rare variants associated with AF. Ongoing work is focused on translating these variants into disease pathways and novel therapeutic modalities. In this review, we focus on our understanding of the current concepts behind the genetics of AF and outline a vision for the incorporation of genetic data into clinical practice.
The prevalence of atrial fibrillation (AF) increases with age, affecting approximately 10% of individuals by 80 years of age.1 AF results in increased risk of congestive heart failure, stroke, dementia, and mortality. Although we have a deep understanding of the epidemiology and the many clinical risk factors for AF,2 current treatment options remain limited and are largely focused on controlling the adverse outcomes attributable to the arrhythmia. In recent years, we and others have used genetics in an attempt to further define the molecular pathways underlying AF and to improve patient specific AF risk prediction.3 This review is based on a systematic literature search using PubMed to identify all studies investigating the genetic basis of AF (emphasizing recent publications), with the goal to present the current landscape of AF genetics and its potential clinical applications.
Atrial Fibrillation: A Disease with a Genetic Basis
A genetic basis for AF was initially suggested in the 1940s with the report of three brothers with the arrhythmia.4 While large families with AF remain rare, in the last decade the heritability of AF in the general population has been well described. Investigators from the Framingham Heart Study found that one in three participants with AF had at least one parent who was also affected with AF.5 Individuals with a first-degree relative with AF had about a 40% increased risk even after accounting for established clinical risk factors for AF.6 Similarly, investigators in Iceland demonstrated that risk of AF decreased with increasing genetic distance between relatives.7 For example, if an individual had a brother with AF, their risk was higher than if they had a cousin with AF. Further support of the heritability of AF has arisen from studies of twins,8 chart reviews in cardiology clinics9 and studies of early-onset AF.10, 11
The Role of Rare Genetic Variants in AF
In a seminal study, Yi-Han Chen and colleagues identified a gain of function mutation in KCNQ1 in a large Chinese kindred with AF.12 KCNQ1 encodes the potassium channel α-subunit underlying the slowly repolarizing current, IKs. The association between mutations in KCNQ1 and AF in turn led to the screening of ion channels and wide range of other genes in many subsequent candidate gene studies. In sum, these efforts have identified rare mutations in a variety of genes generally involved in cardiac depolarization and repolarization, such as potassium channels, the cardiac sodium channel, and gap junction proteins.13–15 In addition, mutations in several transcription factors involved in atrial development have also been identified in patients with lone AF.16–18 Yet the influence of these variants is complex: for instance in one example, two different alleles of the same SNP are associated with increased AF risk.19, 20 Supplemental Table S1 provides a comprehensive list of AF associated rare variants or mutations that have been reported to date. Although the importance of these discoveries to our understanding of AF pathogenesis should not be diminished,21 the clinical implications may be limited since individually the discovered mutations appear to account for a small proportion of patients with AF.
Genome Wide Association Studies: Identification of Common Disease Variants
Large families with monogenic AF are rare in the general population, limiting the utility of classical genetic mapping techniques in the study of AF genetics. However in the last decade, technological advances have enabled efficient genome-wide genotyping of hundreds of thousands of single nucleotide polymorphisms (SNPs), allowing genome-wide association studies (GWAS) to be performed which have identified genetic regions associated with a variety of traits and conditions including AF. The first application of GWAS to AF led to the identification of the chromosome 4q25 AF susceptibility locus in an Icelandic population, a finding replicated in two additional cohorts of European and Asian ancestry.22 Since this initial report, chromosome 4q25 has been the most comprehensively studied of the AF risk loci. A full list of GWAS loci associated with AF is provided in Supplemental Table S2.
The AF-associated SNPs at the 4q25 locus are located in an extensive genomic desert that lies about 150 kilobases (kb) from the paired-like homeodomain transcription factor-2 (PITX2), a transcription factor that plays a critical role in cardiac and gastrointestinal rotation during embryogenesis.23 Moreover, Pitx2c has been associated with suppression of sinus node formation in the developing left atrium,24 and Pitx2 knock-out mice were found to have an increased susceptibility to inducible atrial arrhythmias with programmed stimulation.25 A recent report demonstrated that three independent alleles at the 4q25 locus increase AF risk when occurring simultaneously.26 Interestingly, a separate study of common SNPs at the 4q25 locus demonstrated that expression of common SNPs can modify expression of ion channel genes and influence whether carriers of rare polymorphisms at other loci will develop AF.27 Therefore this locus seems to have wide ranging effects on AF susceptibility, and recent investigations suggest another mechanism may involve microRNA regulation of other transcription factors.28
AF-related genetic variants have also been found at chromosome 1q21 intronic to the KCNN3 gene.29 Also known as SK3 or KCa2.3, this gene encodes a calcium-activated, small conductance potassium channel that is highly expressed in electrically excitable cells in the cardiac and neural tissues.30–32 In rabbit models, inhibition of this channel results in inhibition of pacing-induced shortening of pulmonary venous atrial action potential duration,33 and blockade of SKCa in canines resulted in increased atrial action potential duration (APD) heterogeneity and facilitated the development of reentrant arrhythmias.34 These findings make KCNN3 a potential drug target for AF therapy.
In 2012, a GWAS from 16 studies consisting of over 6,000 individuals with AF and more than 50,000 individuals without AF led to the identification of six additional AF-related genetic loci.35 Three of the AF associated genetic variants were located near transcription factors presumably regulating atrial size, structure or development (PITX2, ZFHX3, PRRX1). Another three variants were near ion channels or related proteins that potentially regulate the atrial action potential (KCNN3, HCN4, CAV1/2). In an effort to identify more target loci in AF, Sinner, et al combined genotyping from two ethnic groups with expression quantitative trait loci (eQTL) mapping to identify another 5 loci associated with AF.36 This study also provided functional characterization of the genes implicated at 2 loci, demonstrating an effect on APD in zebrafish knockdown experiments of the genes NEURL and CAND2. With many AF loci now identified, current studies are focusing on the mechanism of action of these gene products and how these genes are ultimately involved in AF pathogenesis.
Clinical Screening for AF Mutations and AF-associated SNPs
Unlike Long QT syndrome in which 3 genes account for more than 60% of the disease, over 30 genes have been implicated in AF through candidate gene studies, and 14 loci through GWAS. This genetic heterogeneity and the low prevalence of mutations in any single gene precludes the clinical utility of genetic testing despite the commercial availability of genetic panels. 37 However it is now possible to rapidly and cost efficiently sequence the exome (protein-coding region of the genome) or even the entire genome. In upcoming years these more comprehensive techniques will likely shift us away from our current focus on a small number of genes or gene panels. Using such an approach we may find that individuals with AF have an increased burden of rare or novel variants throughout the genome that lead to the arrhythmia,21 or that combinations of genotypes place patients at increased risk.26 Such a finding could in turn lead to a reconsideration of the utility of mutation screening in AF.
Applying Genetics to AF Risk Prediction
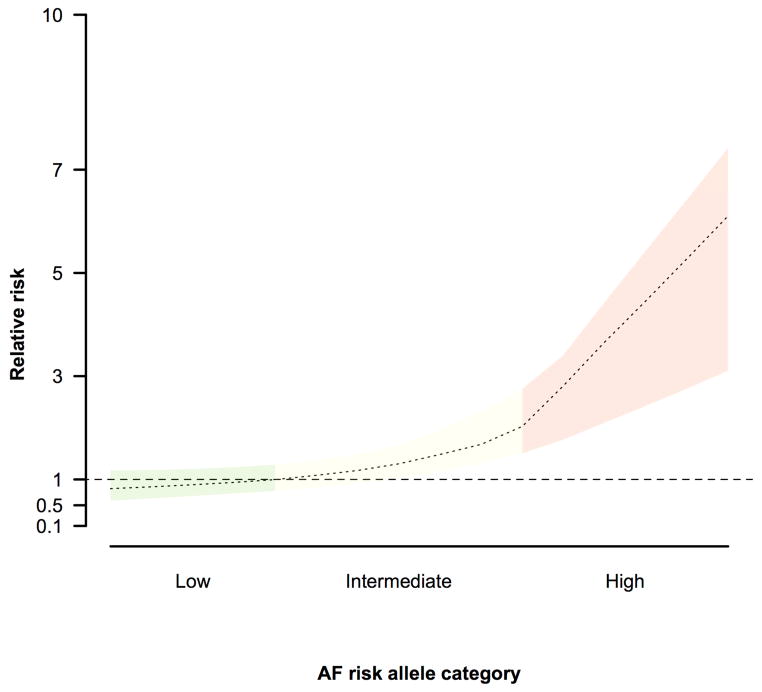
Risk prediction models for AF have been derived using a variety of clinical variables, able to correctly assign higher predicted risks to those that develop AF about 60–70% of the time.38–40 In 2010, we found that adding the top 3 AF SNPs at PITX2, ZFHX3 and KCNN3 offered little improvement to our predictive ability for AF beyond typical clinical risk factors.6 However a separate study demonstrated that the relative risk of developing AF was predicted by the number of risk alleles that a patient harbored (Figure).26, 41 More recently, Everett et al. devised an AF risk prediction algorithm in the Women’s Genome Health Study, which consists of 20,822 women without cardiovascular disease at baseline. Inclusion of a more comprehensive genetic risk score based on 9 SNPs resulted in slight improvements in predictive accuracy beyond established AF risk factors though it did not improve the ability to classify participants into prespecified risk categories.3
Figure.
All common genetic variants can be combined in a genetic risk score for AF. The risk score can then be applied to stratify patients into various categories of risk. One could imagine that patients in the highest risk group would be appropriate for more intensive monitoring for the development of AF.
AF SNPs and Clinical Outcomes: From PVI outcomes to drug response
As noted, the AF risk locus at PITX2/4q25 is by far the most well studied of the risk loci identified to date. Since approximately 20% of individuals of European ancestry carry at least one copy of the top PITX2/4q25 AF risk allele, many recent studies have examined the relation between AF risk SNPs and a variety of clinical outcomes (summarized in Table 1). For instance, recent studies by investigators at Vanderbilt have found that a 4q25 SNP is associated with the response to antiarrhythmic agents and is an independent predictor of AF recurrence after cardioversion.42–44 Body et al and Virani et al have independently demonstrated associations between risk variants on 4q25 and post-operative AF,45, 46 implying a genetic susceptibility to the arrhythmia even in the context of what appears to be a secondary trigger. In addition, multiple reports have found that common AF risk alleles on 4q25 conferred an increased risk of AF recurrence after pulmonary vein isolation procedures.42, 47, 48 However, a separate study in individuals of Asian ancestry found that the presence of the top 4 SNPs associated with AF at the PITX2 and ZFHX3 loci did not predict clinical AF recurrence after ablation, suggesting that earlier positive predictive results may have been either due to a small sample size or genetic differences amongst disparate ethnic groups.49 Therefore defining the clinical niche where the presence of specific genotypes translate into clinical outcome prediction is still a work in progress.
Table 1.
Clinical implications of genetic variation at the 4q25 locus for AF
| Condition | SNPs | Study size | OR | CI | P value | References |
|---|---|---|---|---|---|---|
| Cardioversion success | rs2200733 rs10033464 |
184 | 2.1 | 1.2–3.3 | 8×10−3 | 44 |
|
| ||||||
| AF recurrence post ablation | rs220073 rs10033464 |
200 | 0.76 | 0.6–1.0 | 0.016 | 42 |
| rs2200733 rs10033464 |
195 | 2.0 | 1.0–3.8 | 0.039 | 47 | |
|
| ||||||
| Antiarrhythmic drug response | rs10033464 | 399 | 4.7 | 1.8–12 | 1.3×10−3 | 43 |
|
| ||||||
| Stroke | rs2200733 | 6,222 | 1.3 | 1.2–1.4 | 2.18 × 10−12 | 51 |
| rs1906591 | 4,199 | 1.6 | 1.4–1.9 | 9.2×10−12 | 57 | |
|
| ||||||
| Sudden cardiac death | rs2200733 | 27,629 | 1.3 | 1.1–1.5 | 7.9×10−4 | 58 |
Stroke Risk and AF
Perhaps the best studied of AF outcomes is stroke, and the relation between AF and stroke offers a unique opportunity for the incorporation of genetics in clinical practice. Clinically, current AF related stroke risk is estimated using risk prediction scores such as CHADS2 and CHA2DS2VASc, 50 yet these prediction tools are obviously imperfect. Prior observations suggest a heritable component underlying ischemic stroke, with an estimated 37.9% heritability associated with all ischemic strokes and 32.6% for cardioembolic strokes.51 In line with these observations, AF-associated genetic variants on chromosomes 4q25 and 16q22 have been associated with cardioembolic strokes.51–53 Assessment of multiple independent genetic markers for AF appears to identify individuals at high and low risks of stroke.3, 26 Recently, an AF genetic risk score has been shown to predict a cohort of patients at highest risk for incident AF and stroke, which may become a useful clinical tool to target therapy to patients at highest risk (Figure).54 It is exciting to think that patients at highest risk for stroke may be able to be identified a priori through intensive surveillance of those at highest risk, or perhaps with implantation of long term monitoring devices to identify these patients before a stroke occurs. Future work will need to delineate if AF-associated genetic markers associate with stroke independent of known AF, and if knowing a patient’s genotype will improve risk stratification beyond the CHA2DS2VASc score.
Future studies relating AF variants to clinical outcomes
Current published studies examining the relation between AF-risk variants and clinical outcomes have been limited, typically testing a few SNPs and looking at one or two clinical outcomes of interest. Often these studies have used a limited number of individuals with each outcome (Table 1). With a limited number of individuals with each endpoint and genotype, it is quite possible that the published results illustrate a positive publication bias rather than a true association. Thus, it is not surprising that in most studies the clinical impact of the known AF risk variants has been minimal.
There are now more than a dozen loci that have been found to be associated with AF through GWAS. Additionally, multiple susceptibility signals at a single locus can identify individuals at a marked increased risk for the arrhythmia.26 Therefore it is clear that the next iteration of studies examining AF related outcomes will require testing with a more comprehensive panel of SNPs with a larger number of patients to ensure adequate statistical power. This effort will likely require multi-center collaboration with thousands of individuals to ensure appropriate power. The recent study by Tada, et al can serve as a model for this approach, where testing 12 SNPs in over 27,000 led to the identification of patients at elevated risk for incident AF and stroke.54 Similar methods be used to investigate clinical outcomes that have been tested in smaller patient samples (for instance outcomes after ablation, cardioversion success, or response to antiarrhythmic therapy) to more rigorously test the hypothesis that genotypes may predict clinical outcomes. Such an approach will not only enhance our knowledge of the pathophysiology of AF, but will eventually pave the way for the next frontier of personalized medicine with therapy tailored to improved estimates of a patient’s individual risk.
Conclusions
Unraveling the complex genetics of AF may ultimately allow identification of discrete clinical subtypes and mechanisms for the arrhythmia.55, 56 With our rapidly expanding understanding of the genetic risk factors associated with AF, we hopefully will be able to identify new pathways involved in its pathogenesis, new pharmacologic targets, and patients at risk for specific clinical outcomes. In particular, the intersection of AF with stroke and genetics is particularly promising as it offers the possibility of identifying patients at greatest risk for incident AF and possibly an opportunity to intervene before adverse outcomes occur. We anticipate that within the next few years, much of the data that has been accrued regarding the genetic underpinnings of atrial fibrillation will now be applied to improve clinical risk prediction and used to guide therapies.
Supplementary Material
Summary.
Atrial fibrillation (AF) is heritable and in recent years many genetic loci have been associated with the arrhythmia. Current efforts are directed at determining if AF genetic data can be used to refine clinical risk prediction, predict response to medical or procedural treatments, or help to determine the sequelea of AF such as heart failure and stroke.
Acknowledgments
Dr. Lubitz is supported by an American Heart Association Fellow-to-Faculty award 12FTF11350014. Dr. Ellinor is supported by grants from the National Institutes of Health (1RO1HL092577, R01HL128914, K24HL105780). Dr. Ellinor is also supported by an Established Investigator Award from the American Heart Association (13EIA14220013) and by the Fondation Leducq (14CVD01). Dr. Hucker was supported by Award Number T32HL007208 from the National Heart, Lung, And Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, And Blood Institute or the National Institutes of Health.
Footnotes
Disclosures: Dr. Ellinor is the PI on a grant from Bayer HealthCare to the Broad Institute focused on the genetics and therapeutics of atrial fibrillation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kannel WB, Benjamin EJ. Status of the epidemiology of atrial fibrillation. Med Clin North Am. 2008;92:17–40. ix. doi: 10.1016/j.mcna.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnabel RB, Sullivan LM, Levy D, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J. 2013;34:2243–2251. doi: 10.1093/eurheartj/eht033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff L. Familial auricular fibrillation. N Engl J Med. 1943;229:396–398. [Google Scholar]
- 5.Fox CS, Parise H, D’Agostino RB, Sr, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA : the journal of the American Medical Association. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 6.Lubitz SA, Yin X, Fontes JD, et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. Jama. 2010;304:2263–2269. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnar DO, Holm H, Gudbjartsson DF. Predicting atrial fibrillation. Lancet. 2009;373:1523–1524. doi: 10.1016/S0140-6736(09)60857-6. [DOI] [PubMed] [Google Scholar]
- 8.Christophersen IE, Budtz-Jorgensen E, Olesen MS, Haunso S, Christensen K, Svendsen JH. Familial atrial fibrillation predicts increased risk of mortality: a study in Danish twins. Circ Arrhythm Electrophysiol. 2013;6:10–15. doi: 10.1161/CIRCEP.112.971580. [DOI] [PubMed] [Google Scholar]
- 9.Darbar D, Herron KJ, Ballew JD, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 10.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 11.Oyen N, Ranthe MF, Carstensen L, et al. Familial aggregation of lone atrial fibrillation in young persons. J Am Coll Cardiol. 2012;60:917–921. doi: 10.1016/j.jacc.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 12.Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 13.Wirka RC, Gore S, Van Wagoner DR, et al. A common connexin-40 gene promoter variant affects connexin-40 expression in human atria and is associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2011;4:87–93. doi: 10.1161/CIRCEP.110.959726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olesen MS, Jespersen T, Nielsen JB, et al. Mutations in sodium channel beta-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc Res. 2011;89:786–793. doi: 10.1093/cvr/cvq348. [DOI] [PubMed] [Google Scholar]
- 15.Olesen MS, Refsgaard L, Holst AG, et al. A novel KCND3 gain-of-function mutation associated with early-onset of persistent lone atrial fibrillation. Cardiovascular research. 2013;98:488–495. doi: 10.1093/cvr/cvt028. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Zhang DF, Sun YM, Yang YQ. A novel PITX2c loss-of-function mutation associated with familial atrial fibrillation. European journal of medical genetics. 2014;57:25–31. doi: 10.1016/j.ejmg.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Xu JH, Song HM, et al. Mutational spectrum of the NKX2-5 gene in patients with lone atrial fibrillation. International journal of medical sciences. 2014;11:554–563. doi: 10.7150/ijms.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang JQ, Shen FF, Fang WY, Liu X, Yang YQ. Novel GATA4 mutations in lone atrial fibrillation. Int J Mol Med. 2011;28:1025–1032. doi: 10.3892/ijmm.2011.783. [DOI] [PubMed] [Google Scholar]
- 19.Lubitz SA, Ellinor PT. A common connexion between gap junctions, single nucleotide polymorphisms, and atrial fibrillation? The Canadian journal of cardiology. 2013;29:3–5. doi: 10.1016/j.cjca.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Christophersen IE, Holmegard HN, Jabbari J, et al. Rare variants in GJA5 are associated with early-onset lone atrial fibrillation. The Canadian journal of cardiology. 2013;29:111–116. doi: 10.1016/j.cjca.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Olesen MS, Andreasen L, Jabbari J, et al. Very early-onset lone atrial fibrillation patients have a high prevalence of rare variants in genes previously associated with atrial fibrillation. Heart Rhythm. 2014;11:246–251. doi: 10.1016/j.hrthm.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 23.Mommersteeg MT, Brown NA, Prall OW, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 24.Mommersteeg MT, Hoogaars WM, Prall OW, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubitz SA, Sinner MF, Lunetta KL, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–984. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie MD, Rowan S, Kucera G, et al. Chromosome 4q25 variants are genetic modifiers of rare ion channel mutations associated with familial atrial fibrillation. J Am Coll Cardiol. 2012;60:1173–1181. doi: 10.1016/j.jacc.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Bai Y, Li N, et al. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci U S A. 2014;111:9181–9186. doi: 10.1073/pnas.1405411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohler M, Hirschberg B, Bond CT, et al. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, He Y, Tuteja D, et al. Functional roles of Cav1.3(alpha1D) calcium channels in atria: insights gained from gene-targeted null mutant mice. Circulation. 2005;112:1936–1944. doi: 10.1161/CIRCULATIONAHA.105.540070. [DOI] [PubMed] [Google Scholar]
- 32.Li N, Timofeyev V, Tuteja D, et al. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. J Physiol. 2009;587:1087–1100. doi: 10.1113/jphysiol.2008.167718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozgen N, Dun W, Sosunov EA, et al. Early electrical remodeling in rabbit pulmonary vein results from trafficking of intracellular SK2 channels to membrane sites. Cardiovasc Res. 2007;75:758–769. doi: 10.1016/j.cardiores.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsueh CH, Chang PC, Hsieh YC, Reher T, Chen PS, Lin SF. Proarrhythmic effect of blocking the small conductance calcium activated potassium channel in isolated canine left atrium. Heart Rhythm. 2013;10:891–898. doi: 10.1016/j.hrthm.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinner MF, Tucker NR, Lunetta KL, et al. Integrating Genetic, Transcriptional, and Functional Analyses to Identify Five Novel Genes for Atrial Fibrillation. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.January CT, Wann LS, Alpert JS, et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. J Am Coll Cardiol. 2014 doi: 10.1016/j.jacc.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 38.Benjamin EJ, Levy D, Vaziri SM, D’Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. Jama. 1994;271:840–844. [PubMed] [Google Scholar]
- 39.Psaty BM, Manolio TA, Kuller LH, et al. Incidence of and risk factors for atrial fibrillation in older adults. Circulation. 1997;96:2455–2461. doi: 10.1161/01.cir.96.7.2455. [DOI] [PubMed] [Google Scholar]
- 40.Alonso A, Krijthe BP, Aspelund T, et al. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. Journal of the American Heart Association. 2013;2:e000102. doi: 10.1161/JAHA.112.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubitz SA, Lunetta KL, Lin H, et al. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol. 2014;63:1200–1210. doi: 10.1016/j.jacc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamin Shoemaker M, Muhammad R, Parvez B, et al. Common atrial fibrillation risk alleles at 4q25 predict recurrence after catheter-based atrial fibrillation ablation. Heart Rhythm. 2013;10:394–400. doi: 10.1016/j.hrthm.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parvez B, Vaglio J, Rowan S, et al. Symptomatic response to antiarrhythmic drug therapy is modulated by a common single nucleotide polymorphism in atrial fibrillation. J Am Coll Cardiol. 2012;60:539–545. doi: 10.1016/j.jacc.2012.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parvez B, Benjamin Shoemaker M, Muhammad R, et al. Common genetic polymorphism at 4q25 locus predicts atrial fibrillation recurrence after successful cardioversion. Heart Rhythm. 2013 doi: 10.1016/j.hrthm.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virani SS, Brautbar A, Lee VV, et al. Usefulness of single nucleotide polymorphism in chromosome 4q25 to predict in-hospital and long-term development of atrial fibrillation and survival in patients undergoing coronary artery bypass grafting. The American journal of cardiology. 2011;107:1504–1509. doi: 10.1016/j.amjcard.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Body SC, Collard CD, Shernan SK, et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 48.Shoemaker MB, Bollmann A, Lubitz SA, et al. Common genetic variants and response to atrial fibrillation ablation. Circ Arrhythm Electrophysiol. 2015;8:296–302. doi: 10.1161/CIRCEP.114.001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi EK, Park JH, Lee JY, et al. Korean Atrial Fibrillation (AF) Network: Genetic Variants for AF Do Not Predict Ablation Success. Journal of the American Heart Association. 2015;4:e002046. doi: 10.1161/JAHA.115.002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odum LE, Cochran KA, Aistrope DS, Snella KA. The CHADS(2)versus the new CHA2DS2-VASc scoring systems for guiding antithrombotic treatment of patients with atrial fibrillation: review of the literature and recommendations for use. Pharmacotherapy. 2012;32:285–296. doi: 10.1002/j.1875-9114.2012.01023.x. [DOI] [PubMed] [Google Scholar]
- 51.Gretarsdottir S, Thorleifsson G, Manolescu A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Annals of neurology. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 52.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whang W, Davidson KW, Conen D, Tedrow UB, Everett BM, Albert CM. Global Psychological Distress and Risk of Atrial Fibrillation Among Women: The Women’s Health Study. Journal of the American Heart Association. 2012;1:e001107. doi: 10.1161/JAHA.112.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tada H, Shiffman D, Smith JG, et al. Twelve-Single Nucleotide Polymorphism Genetic Risk Score Identifies Individuals at Increased Risk for Future Atrial Fibrillation and Stroke. Stroke. 2014 doi: 10.1161/STROKEAHA.114.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts JD, Gollob MH. Impact of genetic discoveries on the classification of lone atrial fibrillation. Journal of the American College of Cardiology. 2010;55:705–712. doi: 10.1016/j.jacc.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Roberts JD, Gollob MH. A contemporary review on the genetic basis of atrial fibrillation. Methodist DeBakey cardiovascular journal. 2014;10:18–24. doi: 10.14797/mdcj-10-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemmens R, Buysschaert I, Geelen V, et al. The association of the 4q25 susceptibility variant for atrial fibrillation with stroke is limited to stroke of cardioembolic etiology. Stroke. 2010;41:1850–1857. doi: 10.1161/STROKEAHA.110.587980. [DOI] [PubMed] [Google Scholar]
- 58.Lahtinen AM, Noseworthy PA, Havulinna AS, et al. Common genetic variants associated with sudden cardiac death: the FinSCDgen study. PLoS One. 2012;7:e41675. doi: 10.1371/journal.pone.0041675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.