Abstract
Schizophrenia is a chronic deliberating neuropsychiatric disorder that affects about 1% of the population. Dystrobrevin-binding protein 1 (DTNBP1 or dysbindin) is one of Research Domain Constructs (RDoC) associated with cognition and is significantly reduced in the brain of schizophrenia patients. To further understand the molecular underpinnings of pathogenesis of schizophrenia, we have performed microarray analyses of the hippocampi from dysbindin knockout mice, and found that genes involved in lipogenic pathway are suppressed. Moreover, we uncovered that maturation of a master transcriptional regulator for lipid synthesis, sterol regulatory element binding protein-1 (SREBP1) is induced by neuronal activity, and is required for the induction of the immediate early gene protein ARC (activity-regulated cytoskeleton-associated protein), necessary for synaptic plasticity and memory. We revealed that nuclear SREBP1 is dramatically reduced in dysbindin-1 knockout mice and postmortem brain tissues from human schizophrenia patients. Furthermore, activity-dependent maturation of SREBP1 as well as ARC expression were attenuated in the dysbindin-1 knockout mice, and these deficits were restored by the atypical antipsychotic drug, clozapine. Together, results indicate an important role of dysbindin-1 in neuronal activity induced SREBP1 and ARC, which could be related to cognitive deficits in schizophrenia.
Introduction
Schizophrenia is a major health burden for patients, families and public health systems. It is characterized by positive symptoms such as hallucinations and negative symptoms including flattening of affect and cognitive deficits, that are more resistant to treatment. Recent work has identified potential genetic risk factors of the disorder, as well as more proximate synaptic and circuit mechanisms that may underlie specific clinical endophenotypes [1-5]. One of the potential genes involved in schizophrenia is dystrobrevin binding protein-1 or dysbindin-1. This is a coiled-coil domain containing protein and is one of eight subunits of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Single-nucleotide polymorphism of the dysbindin gene is associated with higher risk of schizophrenia and postmortem brains of schizophrenia patients show decreased expression of dysbindin protein [6-8]. Dysbindin-1 is involved in the regulation of vesicle formation and synaptic release as a partner of BLOC-1 [9,10]. Analysis of dysbindin-1 mutant mice reveals increase in dopamine turnover [11,12], disruption of SNARE complex [9], disruption of dopamine receptor trafficking [13] and NMDA receptor surface expression [14]. Recently, dysbindin-1 has been identified as a protein associated with cognition [15,16] using the Research Domain Criteria (RDoC) defined by National Institute of Mental Health (NIMH) to associate discoveries from neuroscience research and clinical symptoms.
Lipids form cellular membranes, neuroactive hormones and intracellular signaling molecules in the brain, and are involved in protein modifications of many molecules including G-proteins [17-20]. A normal flux of cholesterol and other lipid species is dependent on a metabolic web of enzymes that synthesize and break down lipids. The transcription of these enzymes is controlled by a small family of Sterol Regulatory Element Binding Proteins (SREBPs) [21], which regulate the expression of genes involved in biosynthesis of fatty acids, cholesterol, triglycerides and phospholipids. SREBPs are synthesized and reside in the endoplasmic reticulum (ER) as precursor full length proteins (fSREBPs) complexed with SREBP cleavage-activating protein (SCAP) and insulin-induced gene (Insig) [21]. In response to signals such as lipid deprivation, full-length fSREBPs are transported to the Golgi apparatus, where they are cleaved sequentially by two proteases, site 1 protease (S1P) and site 2 protease (S2P). The newly-generated N-terminal fragments (nSREBPs) are translocated into the nucleus, and bind to sterol regulatory elements (SRE) in promoter regions to stimulate the transcription of lipid-related genes [21,22] including fatty acid synthase (FASN), acetyl-CoA carboxylase, low-density lipoprotein receptor, and 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGCR), stearoyl-CoA desaturase and Glycerol-3-phosphate acyltransferase [23,24]. There are three well-characterized SREBP isoforms in mammals. SREBP1A and C are alternatively spliced isoforms from the same gene on chromosome 17p11.2 while SREBP2 is from a gene on 22q13 [25]. SREBP1 markedly increases the expression of genes producing both fatty acids and cholesterol, while SREBP2 causes preferential induction of genes related to cholesterol biosynthesis.
SREBPs respond to signaling pathways linked to energy balance and growth, and have a key central role in increasing lipid synthesis in many organs, yet their role in the brain has not been well established. Here we show that expression of SREBP1 is significantly reduced in the dysbindin-1 knockout mouse model of schizophrenia (Sandy mice), and postmortem brain tissue from patients with schizophrenia. Moreover, we demonstrate that maturation of neuronal SREBP is regulated in an activity-dependent manner and is required for induction of an immediate early gene, activity-regulated cytoskeleton-associated protein (ARC or Arg3.1) that is important in memory and cognition [26,27]. Furthermore, we found that activation of SREBP1 and induction of ARC were attenuated in dysbindin-1 knockout mice, whereas the atypical antipsychotic drug clozapine restored levels of nSREBP1 and ARC. Together, results point to a link between neuronal activity, lipid biosynthesis and cognition, which may be disrupted in schizophrenia.
Materials and Methods
Cells
PC12 cells were obtained from the American Type Culture Collection (Manassas, VA). HT22, murine hippocampal culture cells were a generous gift from Dr. David Schubert at the Salk Institute. PC12 cells were maintained in a humid atmosphere containing 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium supplemented with 5% horse serum, 5% calf serum, L-glutamine (300 μg/ml), penicillin (100 U/ml), and streptomycin (100 μg/ml). One day after plating, PC12 cells were differentiated in Dulbecco’s modified Eagle’s medium supplemented with 1% horse serum, 1% Pen-Strep and 100ng / mL nerve growth factor (NGF).
Generation of Constitutively active SREBP (nSREBP1)
Human SREBP1A (GenBank ID: U00968) cDNAs encoding the two nuclear mature forms (1-490) were generated from construct pSREBP1A (ATCC 79811) and cloned into pCMV-Myc (Clontech) between SalI and NotI sites. The N-terminal fragment of SREBP1A bypasses the requirement of protease-mediated activation process of full length SREBP1, and can be overexpressed in the nucleus as an active transcription factor.
Overexpression of plasmids in PC12 cells
PC12 cells were plated at a density of 5 × 105 cells per well in six-well plates. Two or three days after plating, cells were transfected with plasmids for nSREBP-myc via Lipofectamine 2000 (Life Technologies, Grand Island, NY), according to the manufacturer’s instructions [28].
Immunofluorescence staining
PC12 cells were seeded on glass coverslips and transfected with full-length SREBP-myc plasmid. After 24 hours, cells were depolarized with a buffer containing 50 mM KCl. They were fixed with 4% PFA, washed, permeabilized and blocked with 1% BSA, 2% normal goat serum in PBS. Cells were then incubated with a mouse anti-Myc epitope primary antibody and visualized with an appropriate Alexa Fluor 596 secondary antibody (Invitrogen, Grand Island, NY). Confocal microscopy was performed using a 63X oil immersion objective in a Leica DMI6000 microscope (Leica Microsystems, Buffalo Grove, IL)
Animals and housing
Mice were maintained on a 12:12 light:dark cycle at 22±2°C, with ad libitum access to water and food. They were weaned at 4 weeks postnatal and housed 4 per cage, randomly assigned to one of two conditions, standard-housing (SH) or environmentally enriched (EE) conditions. SH mice were housed in open top standard mouse cages (34 × 16 × 16 cm) with basic nesting materials. EE animals were housed in larger cages (80 × 60 × 60 cm) containing toys made of various materials, wooden blocks, climbing platforms, plastic tubes and small houses that were rearranged and renewed twice a week to stimulate the animals’ exploratory behavior. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Microarray analyses
Four homozygous dysbindin −/− (Sandy) mice and four wild type mice were deeply anaesthetized and killed by decapitation. The brains were harvested and the right hippocampus dissected out and stored in RNA Later (Ambion, Austin, TX, USA). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Extracted RNA was purified using the RNeasy® Kit (Qiagen, Hilden, Germany). RNA was extracted from the hippocampus and submitted to the BFIG Core Facility Lab (Department of Paediatrics, National University of Singapore). The quality of RNA was analyzed using an Agilent 2100 Bio-analyzer (Agilent, Santa Clara, CA). cRNA was generated and labeled using the one-cycle target labeling method and hybridized to Affymetrix Mouse Genome 430 2.0 microarrays (Affymetrix, Santa Clara, CA), using standard Affymetrix protocols. A total of eight microarrays were used for the two groups of animals - four for the homozygous dysbindin −/− right hippocampi, and four for the wild type right hippocampi. Initial image analysis of the microarray chips was performed with Affymetrix GCOS 1.2 software. The data were exported into GeneSpring v11 (Agilent) software for analysis using parametric test based on cross gene error model (PCGEM). T-test was used to identify differentially expressed genes between the two groups. P < 0.05 was considered significant.
Ingenuity Pathway Analyses of Gene Networks
Gene identifiers and corresponding expression values of differentially expressed genes between dysbindin−/− and wild type mice with more than 1.5 fold change were uploaded into the Ingenuity Pathway Analysis (IPA) application (Ingenuity® Systems, www.ingenuity.com). Each identifier mapped to its corresponding object in Ingenuity's Knowledge Base, and was overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. Network Eligible genes are differentially expressed genes that have at least one other molecule in the Knowledge Base that interacts with it to form a network.
Ex vivo brain slices and Western blots
The frontal lobes of mice were freshly dissected out and cut at 0.4 mm intervals in the sagittal and coronal plane using a McIlwain tissue chopper. The slices were dispersed in oxygenated artificial cerebrospinal fluid buffer (ACSF, 113mM NaCl, 4.5mM KCl, 1mM MgCl2, 25mM NaHCO3, 1mM NaH2PO4, 25mM glucose, 2mM CaCl2), incubated with 25 mM TEA (Sigma, St. Louis, MO) for 15 min, and washed with ice cold ACSF. Tissues were homogenized in lysis buffer and subjected to SDS-PAGE and immunoblots as previously described [29-31]. Commercial phospho-specific antibodies for proteins of interest included SREBP and ARC, with GAPDH as a loading control (SREBP1 and ARC antibodies were obtained from Santa Cruz Biotechnology, Santa Cruz, CA and GAPDH from Cell Signaling, Danvers, MA).
Analyses of human postmortem tissues from subjects with schizophrenia
Postmortem hippocampi were studied from 12 non-psychiatric controls (age 79.6 years [SD=11.2], 50% female, postmortem interval 11.8 hours [SD=6.6]) and 13 schizophrenia cases (age 78.6 years [SD=9.0], 54% female, postmortem interval 12.2 hours [SD=4.5]) participating in a longitudinal study of prospectively diagnosed subjects approved by an Institutional Review Board (IRB) and conducted by the Schizophrenia Research Center at the University of Pennsylvania [32]. Psychiatric subjects met DSM-IV (American Psychiatric Association, Washington, D.C.) diagnostic criteria for schizophrenia determined in consensus conference after review of medical records, direct clinical assessments of patient and interviews with caregivers. Control and psychiatric subjects were well matched for sex, age and postmortem interval. Neuropathological examination revealed no degenerative processes in control or psychiatric cases, i.e. normal densities of amyloid plaques, neurofibrillary tangles and Lewy bodies with no infarcts or gross cell loss.
Formalin-fixed, paraffin tissue blocks of the body of the hippocampus were sectioned at 6 μm and mounted on APES-coated slides. Immunohistochemistry and microscopic image analyses were conducted as previously described.[33,6,34]. Briefly, tissue sections were dewaxed in xylene and rehydrated in descending alcohols. Endogenous peroxidase activity was quenched in 5% H2O2 -methanol for 30 minutes. Sections were rinsed in distilled water, followed by boiling in 1mM EDTA (pH 8.0) for 15 minutes for antigen retrieval. They were rinsed in water, washed in 0.1 M Tris buffer with 0.01% Triton X-100 (TTB), blocked in 10% normal horse serum for 45 minutes and incubated in primary antibody overnight at 4°C. Primary antibody was directed against SREBP1 (Santa Cruz 13551, Mouse 1:50). The specificity of the antibody was confirmed by Western blots. The following day, sections were incubated for 1 hour at room temperature in biotinylated anti-mouse secondary antibody, washed in TTB and incubated in an avidin-biotin complex for 1 hour. Finally, sections were washed in TTB and reacted with a 0.05% diaminobenzidine (DAB) – 0.03% hydrogen peroxide solution for 17 minutes. Immunoreaction signal was enhanced by adding NiSO4 (0.25% final dilution) to the DAB solution. Sections were rinsed in water, dehydrated in ascending alcohols, cleared in xylenes and coverslipped under Cytoseal 60 (Fisher Scientific).
Quantification of SREBP1 immunoreactivity in cell nuclei in the CA1/subiculum subregion of the hippocampus was conducted by semi-automated image analysis of net optical density (OD), defined as the OD of a region of interest (nuclei) minus the OD of the background (i.e., neighboring white matter). OD analysis was performed on high resolution, gray-scale photomontages made on a motorized microscope stage with Image-Pro Plus software (Media Cybernetics: Silver Springs, MD, USA). Nuclear regions of interest were algorithmically determined using OD, size and shape filters. Quantitative comparisons were performed only on sections photographed at the same light intensity. Operators were blinded to any identifying information throughout data accrual and analysis.
ATP measurement
ATP levels in brain tissues were measured using ATP colorimetric/fluorometric assay kit from BioVision (San Francisco, CA) according to the manufacturer’s instruction.
Data analysis
All quantitative data were present as mean ± S.E., if they were derived from at least 3 experiments. For comparison of multiple groups, the data were analyzed by one-way analysis of variance (ANOVA) followed by a Tukey post hoc test. Student’s t-test was employed for directly testing the difference between two sets of independent samples. All statistical analyses were performed using GraphPad Prism (GraphPad Software, San Diego, CA). p value less than 0.05 was considered significant.
Results
1. Microarray analyses of dysbindin knockout (Sandy mice)
Based on Research Domain Criteria (RDoC) [15,16], dysbindin is linked to cognition, which may be affected in various neuropsychiatric disorders such as schizophrenia. Moreover, polymorphism in the dysbindin-1 gene has been identified as one of the risk factors associated with schizophrenia, and its expression level is significantly reduced in the hippocampus of subjects with schizophrenia [35,36]. We therefore carried out microarray analyses on the hippocampi of dysbindin-1 knockout (dys1−/−) versus wildtype mice, to determine potential genes that might be involved in schizophrenia.
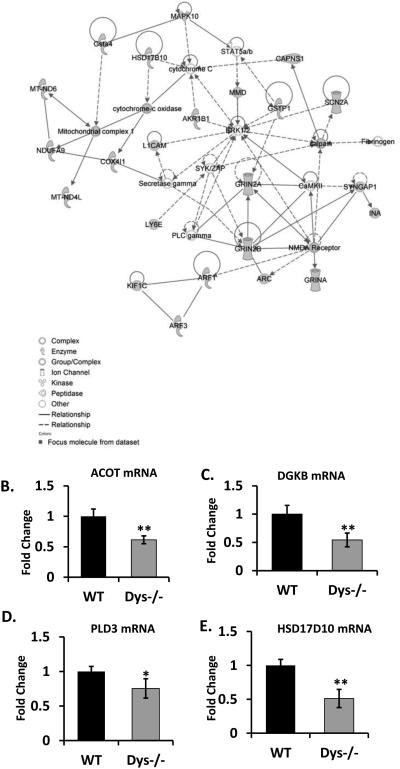
A total of 53 genes were up-regulated and 165 genes were down-regulated in dys1−/− mice hippocampus (P<0.05 and at least 1.5 fold change). These included genes associated with neuronal activity, such as the NMDA receptor subunit 2A and 2B (Figure 1A and Supplemental Table 1) and those related to lipids, including acyl-CoA thioesterase (ACOT), diacylglycerol kinase (DGKB), phospholipase D (PLD3), hydroxysteroid (17-beta) dehydrogenase 10 (HSD17D10). The changes in expression of several of genes were confirmed by real time qPCR (Figure 1B-E). Based on the above results, we hypothesized that absence of dysbindin-1 may affect neuronal activity-induced lipid signaling with consequent effect on cognition, and proceeded to explore a link between neuronal activity and a master regulator of lipid biosynthesis, SREBP1 [21].
Figure 1. Lipid pathway is suppressed in dysbindin −/− mice.
(A) Ingenuity Systems analysis of genes identified by microarray, with fold change of greater than 1.5. (B-E) The expression levels of ACOT, DGKB, PLD3 and HSD17D10 mRNA were examined by quantitative RT-PCR (n=5). The representative data is shown from 3 or 4 independent experiments.
2. In vitro studies on the relation between depolarization and SREBP1 in PC12 cells
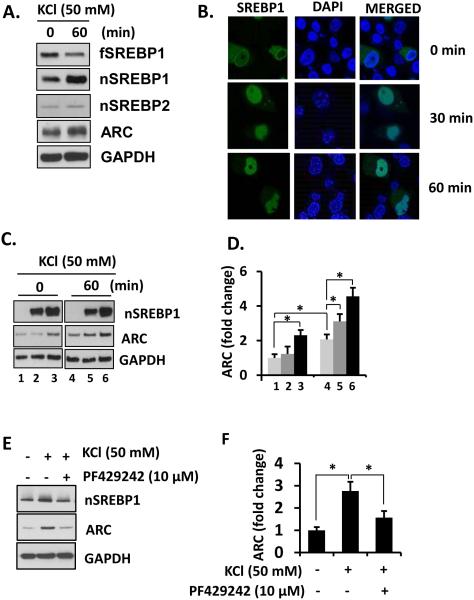
Differentiated PC12 cells were treated with 50 mM KCl for 60 min to induce depolarization and expression levels of SREBPs determined. Treatment with 50 mM KCl for 60 min resulted in cleavage of full length SREBP1 (fSREBP1) to produce nSREBP1 while there was no change in nSREBP2 (Figure 2A). Moreover, ARC (activity-regulated cytoskeleton-associated protein, or Arg3.1) expression was increased upon KCl treatment. We performed immunofluorescence staining on differentiated PC12 cells overexpressing full-length SREBP1 plasmid that was conjugated with green fluorescent protein (GFP) on the N-terminal to enable detection of both the unprocessed and cleaved forms of SREBP1. SREBP1 was predominantly located in the cytosol of PC12 cells before treatment with KCl, but after KCl-induced depolarization, SREBP1 was mostly found in the nucleus. This indicated that full length SREBP is cleaved and its N-terminal fragment which contains the SRE binding domain is translocated to the nucleus (Figure 2B).
Figure 2. SREBP1 maturation is sensitive to neural activation.
(A) Differentiated PC12 cells were treated with 50 mM KCl for 60 min and the lysates were analyzed by immunoblotting and (B) confocal microscopy. (C) Differentiated PC12 cells were transfected with different amount of nSREBP-myc and depolarized for 60 min. Cell lysates were subjected to immunoblotting. (D) Arc expression was quantified and normalized by GAPDH levels. Student's t-tests were performed (* p < 0.05). (E) A blockade of SREBP processing inhibits Arc activation in differentiated PC12 cells. (F) Arc expression was quantified and normalized by GAPDH levels. Student's t-tests were performed (* p < 0.05). The representative data is shown from 3 or 4 independent experiments.
Microarray analysis of altered genes in the dys1−/− mouse hippocampus indicated reduction in expression of the cytoskeleton related protein, ARC, which plays a critical role in cytoskeleton remodeling necessary for consolidation of memory. Hence, we sought to elucidate a possible relationship between SREBP1 and ARC. PC12 cells were transfected with the nuclear form of SREBP1 tagged with myc (nSREBP1-myc, amino acid 1-490), which can bypass the cleavage process for maturation and activation of SREBP, and expressed in the nucleus as an active transcription factor. Overexpression of nSREBP1 induced the expression of ARC, similar to the effect of 50 mM KCl depolarization (Figure 2C, lane 3 vs 4). The expression of ARC in nSREBP1 overexpressing cells was further increased by KCl depolarization (Figure 2C lanes 4-6).
To confirm that SREBP1 drives the expression of ARC, cells were pretreated with PF-429242 (10 μM) to block site 1 protease (S1P)-mediated cleavage and activation of fSREBP1 for 30 min prior to depolarization. PF-429242 not only blocked an activity dependent generation of nSREBP1 but also induction of ARC (Figure 2E and F). Together, results indicate that cellular depolarization induces maturation of SREBP1 which is necessary for induction of ARC.
3. In vitro and in vivo studies on the relation between neuronal activity and SREBP in cultured neurons, brain slices, and NMDA hypomorph mice
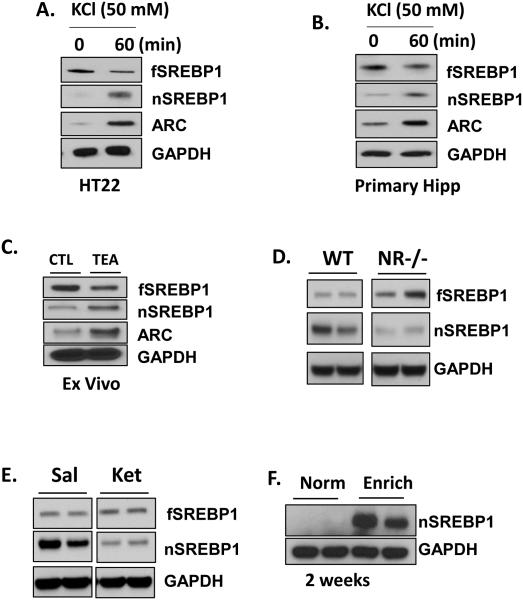
Since PC12 cells are a type of neuroendocrine cells, we sought to verify the relation between depolarization and SREBP1 in neurons, using immortalized mouse hippocampal cell line, mouse primary cultured hippocampal neurons, mouse hippocampal slices and behaving mice in vivo. As with PC12 cells, KCl depolarization induced cleavage of fSREBP1 in differentiated HT22 cells (immortalized mouse hippocampal cell line) as well as mouse primary hippocampal neurons (Figure 3A and B).
Figure 3. Maturation of nSREBP1 is NMDA-dependent in vivo.
HT22 cells (A) or primary hippocampal neurons (B) were treated with 50 mM KCl for 60 min and the lysates were analyzed by immunoblotting. (C) Hippocampal ex vivo preparation from WT mouse (8 week) was treated with 25 mM TEA (LTP inducer) for 15 min and then subjected to immunoblotting (n=4). (D) NMDA hypomorph mice brains were harvested and analyzed for an expression of SREBPs by immunoblotting. (E) A ketamine (300 mg/Kg) was injected intraperitoneally and mice were sacrificed in 24 hours. Mice hippocampi were dissected out and lysates were subjected for immunoblotting (n=5). (F) Mice (4 week, n=5-7) were placed under both standard or environment-enrichment conditions, and brains were harvested for immunoblotting. The representative data is shown from 3 or 4 independent experiments.
To confirm the effect of neuronal activity on SREBP1 processing in an ex vivo model, we determined possible effects of tetraethylammonium chloride (TEA), a potassium channel blocker and inducer of neuronal activity on SREBP1 activation and ARC in mouse hippocampus slices [37,29-31]. Fifteen-minute treatment with 25 mM TEA resulted in increased nSREBP1, indicating that processing of SREBP1 is enhanced by TEA (Figure 3C). As expected from findings in PC12 cells, the same treatment also resulted in increased expression of ARC (Figure 3C).
Since NMDA receptor function is closely related to ARC, and NMDA activity is down regulated in the dys1−/− mouse model of schizophrenia [38-40,6], we determined whether SREBP1 expression could be altered in NMDA receptor1 (NR1) hypomorph mice. Significant reduction in nSREBP1 was found in the brains of these mice (Figure 3D). Similarly, administration of a NMDA receptor antagonist, ketamine in wildtype mice led a reduction in nSREBP1 (Figure 3E). Together, results indicate that NMDA receptor activation is associated with nSREBP1 expression.
To demonstrate a possible link between an enriched environment and SREBP1 processing in behaving animals, we employed an environmental enrichment paradigm, which is a combination of complex inanimate and social stimulation that has been shown to increase hippocampal neurogenesis, enhance synaptic plasticity and improve memory [41-43]. Four week-old wild type B57/C6 mice were exposed to an enriched environment for 2 weeks and expression levels of nSREBP1 in the hippocampus determined. Mice in standard condition had low levels of nSREBP1, while those exposed to environmental enrichment showed significantly greater in nSREBP1 expression (Figure 3F). Together, the above results indicate that neuronal depolarization induces maturation of SREBP1 and induction of the plasticity related protein ARC, which could a cellular substrate for improved cognition in an enriched environment.
4. In vitro and in vivo studies demonstrating disruption in the relation between neuronal activity, SREBP and ARC in dysbindin-1 knockout mice
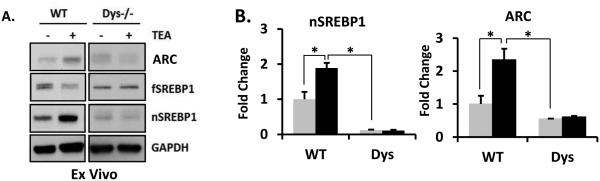
In view of changes in lipid related genes found in our microarray analyses of dysbindin-1 knockout mice, we sought to establish whether neuronal activity induced-increase in SREBP1 and ARC might be disrupted in these mice. The N-terminal fragment of active nSREBP1 and ARC was significantly down-regulated (Figure 4A and B) in brains of dys1−/− mice. Moreover, TEA induced nSREBP1 and ARC were reduced in brain slices prepared from these mice (Figure 4A and B).
Figure 4. SREBP1 maturation and expression of ARC was inhibited in Dys−/− mice.
(A) Hippocampal ex vivo preparation from WT and Dys1−/− mouse (8 week, n=5-7) was treated with 25 mM TEA for 15 min and then subjected to immunoblotting. (B) The expression levels of ARC and nSREBP1 were quantified and normalized by GAPDH levels. Student's t-tests were performed (* p < 0.05). The representative data is shown from 4 independent experiments.
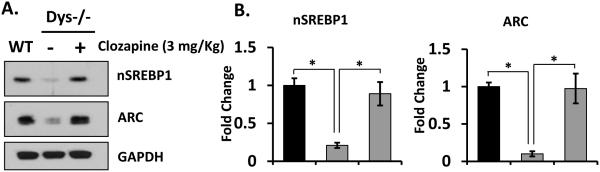
Since many antipsychotic medication, especially atypical antipsychotics drugs have a lipogenic effect and induce weight gain [44-46], we postulated that atypical antipsychotics such as clozapine may increase maturation of SREBP. Mice were intraperitoneally injected with 3 mg/kg of clozapine, and SREBP and ARC expression measured. Administration of clozapine increased the brain levels of nSREBP1 and ARC in dys1−/− mice (Figure 5A and B). Importantly, we confirmed that nSREBP1 expression was downregulated in postmortem brain tissue from schizophrenia patients (Figure 6). The brains of these patients have earlier been shown to have reduced levels of dysbindin-1 [35,36].
Figure 5. Clozapine restores SREBP1 and ARC deficit in Dys−/− mice brain.
(A) Dys1−/− mice (8 week, n=5-7) were administered with clozapine (3mg/kg) and sacrificed 3 hours later. Mice brains were harvested and analyzed by immunoblotting. (B) The expression levels of ARC and nSREBP1 were quantified and normalized by GAPDH levels. Student's t-tests were performed (* p < 0.05). The representative data is shown from 4 independent experiments.
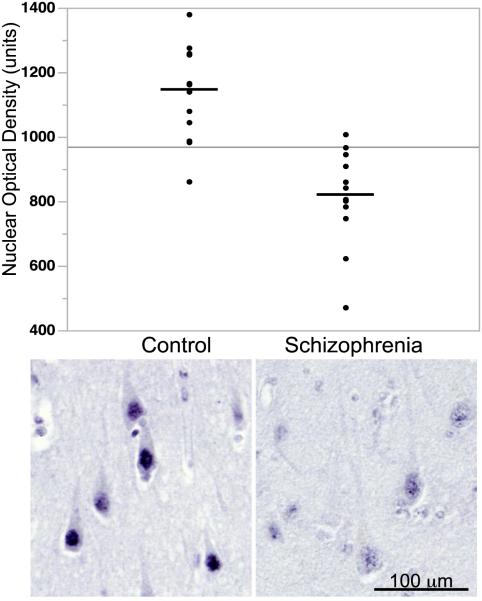
Figure 6. nSREBP1 expression is reduced in postmortem tissues from schizophrenia patients.
Immunohistochemistry was performed from 12 non-psychiatric controls and 13 schizophrenia cases. Measurement of SREBP immunoreactivity in cell nuclei in the CA1/subiculum subregion of the hippocampus was conducted by semi-automated image analysis for net optical density (OD), defined as the OD of a region of interest (nuclei) minus the OD of the background. The representative image was shown here. Semi-quantitative measurement of nuclear optical density of SREBP1 found highly significant decreases in schizophrenia (t=5.4, df=23, p<0.0001).
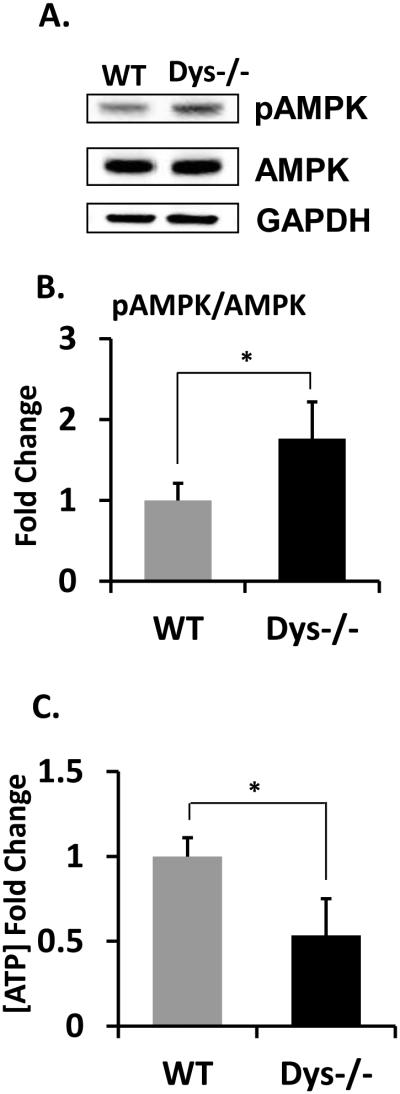
Finally, we examined a possible link between SREBP1 and ATP levels. The SREBP pathway plays a crucial role in fundamental aspects of cell biology inducing lipid biosynthesis and energy production. We detected that phosphorylated adenosine monophosphate activated kinase (AMPK), which is well-characterized cellular energy sensor (Fig 7A and B) is increased in dys1−/− mice brain. We further showed that ATP levels were reduced in the brain of dys1−/− mice (Fig 7C). Taken together, the above results indicate that neuronal depolarization induced maturation of SREBP1 and expression of ARC are affected in dysbindin-1 knockout mice and human cases of schizophrenia, which could be a molecular underpinnings for cognitive deficits in this disease.
Figure 7. AMPK activity is enhanced and ATP level is deficient in Dys1−/− mice brain.
(A) Expression levels of pAMPK were measured by immunoblotting and (B) quantified and normalized by total AMPK levels. The representative data is shown from 4 independent experiments. (C) ATP levels in hippocampi were measured as described in material methods (N=6). Student's t-tests were performed (* p < 0.05).
Discussion
In the present study, we have performed unbiased microarray and pathway analyses, and found altered expression of many lipid-related genes in dys1−/− mice. Based on these analyses, we further identified that a master transcriptional regulator of lipid biosynthesis, SREBP1 is significantly reduced in dys1−/− mice and postmortem brain tissues from patients with schizophrenia. We also found that maturation of SREBP1 in neurons is modulated in an activity dependent manner that can be triggered by depolarization or environmental enrichment. We further identified that maturation of SREBP1 is necessary for induction of an immediate early gene, ARC expression that plays a critical role in learning and memory. Furthermore, we found loss of activity-dependent induction of SREBP1 processing as well as ARC induction in brain slices from dys1−/− mice. This deficit was restored by the antipsychotic drug, clozapine, which is known to induce lipid synthesis [47,48]. Together results indicate thatSREBP1 is dynamically regulated by neuronal activity, and dysregulation may contribute to cognitive deficits in schizophrenia.
We first carried out comprehensive gene expression profiling on the hippocampi of dysbindin-1 knockout (Sandy) mice, to determine possible changes in gene expression in these mice. Based on the microarray and qPCR analyses, we confirmed a decrease in mRNA expression of ACOT, DGKB, PLD3 and HSD17D10. These genes play an essential role maintaining lipid homeostasis. ACOT catalyzes the hydrolysis of acyl-CoAs to free fatty acids and coenzyme A [49]. DGKB and PLD3 modulate glycerophospholipid and phospholipid production respectively [50,51] and HSD17B10 is involved in regulating fat and steroid hormone production including estrogen [52,53]. Even though we did not test their expression levels by qPCR, there are several other genes, thath are downregulated in the dysbindin-1 −/− mice, which could have an effect on lipid balance. For example, the expression of glucose phosphate isomerase 1 (GPI1) is reduced and its suppression is known to reduce lipid synthesis [54]. Based on these results, we hypothesized that the absence of dysbindin-1 may affect neuronal activity induced lipid signaling, with consequent effect on synaptic plasticity and cognitive function; and proceeded to explore a link between neuronal activity and a master regulator of lipid related gene expression, SREBP1 [21].
A change in energy balance such as lipid deprivation rapidly stimulates SREBP maturation in peripheral tissues, but it is not well understood how SREBPs are regulated in the CNS. Studies have reported that expression of SREBP1 in the brain is affected more by age than by metabolic changes, unlike peripheral tissues [55], and our previous studies have shown high basal expression of SREBP1 and SREBP2 in neurons of the CNS [56,57]. In this study, we provide evidence that neuronal SREBP is dynamically and rapidly modulated by neuronal activity. Importantly, neuronal activity or NMDA receptor activity leads to SREBP1 activation, which facilitates the expression of an immediate early gene, ARC. ARC gene expression is activated by hippocampal activity [58,26,27]. Conversely, reduced NMDA function may contribute to an inhibition of SREBP1 processing, leading to decreased expression of nSREBP1. In fact, when an NMDA antagonist, ketamine was administered to mice, we found a dramatic reduction in nSREBP1. Results are consistent with those of a previous study, which showed that NMDA treatment induces accumulation of nSREBPs [59]. We also demonstrated that mice exposed to an enriched environment showed a dramatic increase in nSREBP1. Environmental enrichment involves the generation of novelty and complexity in housing conditions, which facilitates enhanced sensory and cognitive stimulation as well as physical activity. In animal models, environmental enrichment has been found to have beneficial effects, including enhancing synaptic plasticity and cognition [42,43].
Reduction of dysbindin-1 expression is a very common finding in immunostained sections of the hippocampus in patients with schizophrenia [6]. In this context, we found that dys1 −/− mice lack activity dependent-maturation of SREBP1 as well as ARC induction, suggesting that lack of neuronal SREBP1 regulation may contribute to cognitive deficits in this disease. In fact, it has been shown that polymorphism of SREBPs is associated with an increased incidence of schizophrenia [60,61], and it interesting that many antipsychotic drugs are known to cause an increase in SREBP activity, lipid synthesis and obesity [62,47]. Consistently, our data demonstrated that reduced expression of nSREBP1 and ARC in dys1−/− was restored by an atypical antipsychotic drug, clozapine. It is therefore tempting to speculate that the lipogenic effect of antipsychotic drugs may have a role in their efficacy. A recent lipidomic study has shown that drug naïve first episode schizophrenia patients have reduced levels of n-3 polyunsaturated fatty acid including Docosahexaenoic acid, which was corrected after antipsychotic medication [63]. It is postulated that SREBP-induced lipid biosynthesis may be needed for the cell membrane or production of lipid mediators, necessary for neurite outgrowth and synaptic plasticity.
Brain lipid metabolism is crucial for normal neural physiology and brain function, considering the fundamental roles of lipids in membrane molecular composition, cellular energy homeostasis, and signal transduction [64]. In fact, our data suggest that disrupted lipid pathway leads to a reduction of cellular energy production levels reflected by increased pAMPK and decreased ATP (Fig 7). It is well known that ATP plays a crucial role in different aspects of neuronal function as well as neurodevelopment [65,66]. Moreover, evidence has demonstrated that diverse lipids exert multifaceted influences on axonal myelination, neuronal differentiation, neurite outgrowth, formation of synaptic vesicles and synaptic plasticity [17,20,18,19]. Disruption of lipid homeostasis in nervous systems has been causally linked to the onset and progress of many neurological disorders such as Huntington’s disease and Alzheimer’s disease [67,68]. Interestingly, polymorphisms in SREBPs have been associated with increased incidence for schizophrenia [60]. The present results add to these findings, and showed that SREBP1 maturation is positively regulated by neuronal activation, linked to canonical plasticity pathways involving ARC, and is disrupted in the dys1 −/− mouse model of schizophrenia. Together, results suggest that SREBP1 may play a dynamic role in enhancing synaptic plasticity, and its augmentation may be a potential therapeutic strategy for neurocognitive disorders such as schizophrenia.
Supplementary Material
Acknowledgement
We thank Ms. Hui-Jen Lye and Ms. Sau-Yeen Loke for help with microarray analyses. This work was supported by the National Research Foundation, National Medical Research Council and National University Health System, Singapore (WYO) and by HD026979, MH079614, and DK084336 (SFK).
Reference List
- 1.Carlson GC, Talbot K, Halene TB, Gandal MJ, Kazi HA, Schlosser L, Phung QH, Gur RE, Arnold SE, Siegel SJ. Dysbindin-1 mutant mice implicate reduced fast-phasic inhibition as a final common disease mechanism in schizophrenia. ProcNatlAcadSciUSA. 2011;108(43):E962–E970. doi: 10.1073/pnas.1109625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cellular and molecular neurobiology. 2006;26(4-6):365–384. doi: 10.1007/s10571-006-9062-8. doi:10.1007/s10571-006-9062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg SC. Negative and deficit symptoms in schizophrenia do respond to neuroleptics. SchizophrBull. 1985;11(3):453–456. doi: 10.1093/schbul/11.3.453. [DOI] [PubMed] [Google Scholar]
- 4.Hattori S, Murotani T, Matsuzaki S, Ishizuka T, Kumamoto N, Takeda M, Tohyama M, Yamatodani A, Kunugi H, Hashimoto R. Behavioral abnormalities and dopamine reductions in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia. Biochemical and biophysical research communications. 2008;373(2):298–302. doi: 10.1016/j.bbrc.2008.06.016. doi:10.1016/j.bbrc.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Kantrowitz JT, Javitt DC. N-methyl-d-aspartate (NMDA) receptor dysfunction or dysregulation: the final common pathway on the road to schizophrenia? Brain ResBull. 2010;83(3-4):108–121. doi: 10.1016/j.brainresbull.2010.04.006. doi:S0361-9230(10)00087-0 [pii];10.1016/j.brainresbull.2010.04.006 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. The Journal of clinical investigation. 2004;113(9):1353–1363. doi: 10.1172/JCI20425. doi:10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang J, LeGros RP, Louneva N, Yeh L, Cohen JW, Hahn CG, Blake DJ, Arnold SE, Talbot K. Dysbindin-1 in dorsolateral prefrontal cortex of schizophrenia cases is reduced in an isoform-specific manner unrelated to dysbindin-1 mRNA expression. Hum Mol Genet. 2009;18(20):3851–3863. doi: 10.1093/hmg/ddp329. doi:10.1093/hmg/ddp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talbot K, Louneva N, Cohen JW, Kazi H, Blake DJ, Arnold SE. Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS One. 2011;6(3):e16886. doi: 10.1371/journal.pone.0016886. doi:10.1371/journal.pone.0016886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, Malvar JS, de Vellis J, Sabatti C, Dell'Angelica EC. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Molecular psychiatry. 2010;15(2):115–204. doi: 10.1038/mp.2009.58. 115. doi:10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickman DK, Davis GW. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science. 2009;326(5956):1127–1130. doi: 10.1126/science.1179685. doi:10.1126/science.1179685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murotani T, Ishizuka T, Hattori S, Hashimoto R, Matsuzaki S, Yamatodani A. High dopamine turnover in the brains of Sandy mice. Neuroscience letters. 2007;421(1):47–51. doi: 10.1016/j.neulet.2007.05.019. doi:10.1016/j.neulet.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Chen XW, Feng YQ, Hao CJ, Guo XL, He X, Zhou ZY, Guo N, Huang HP, Xiong W, Zheng H, Zuo PL, Zhang CX, Li W, Zhou Z. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J Cell Biol. 2008;181(5):791–801. doi: 10.1083/jcb.200711021. doi:10.1083/jcb.200711021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Y, Yang F, Papaleo F, Wang HX, Gao WJ, Weinberger DR, Lu B. Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(46):19593–19598. doi: 10.1073/pnas.0904289106. doi:10.1073/pnas.0904289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang TT, Yang F, Chen BS, Lu Y, Ji Y, Roche KW, Lu B. Dysbindin regulates hippocampal LTP by controlling NMDA receptor surface expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21395–21400. doi: 10.1073/pnas.0910499106. doi:10.1073/pnas.0910499106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. doi:10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. The American journal of psychiatry. 2010;167(7):748–751. doi: 10.1176/appi.ajp.2010.09091379. doi:10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 17.Bazan NG. Synaptic signaling by lipids in the life and death of neurons. MolNeurobiol. 2005;31(1-3):219–230. doi: 10.1385/MN:31:1-3:219. [DOI] [PubMed] [Google Scholar]
- 18.Davletov B, Montecucco C. Lipid function at synapses. CurrOpinNeurobiol. 2010;20(5):543–549. doi: 10.1016/j.conb.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Rohrbough J, Broadie K. Lipid regulation of the synaptic vesicle cycle. NatRevNeurosci. 2005;6(2):139–150. doi: 10.1038/nrn1608. [DOI] [PubMed] [Google Scholar]
- 20.Soldan MM, Pirko I. Biogenesis and significance of central nervous system myelin. SeminNeurol. 2012;32(1):9–14. doi: 10.1055/s-0032-1306381. [DOI] [PubMed] [Google Scholar]
- 21.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16(4):414–419. doi: 10.1016/j.cmet.2012.09.002. doi:10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77(1):53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 23.Espenshade PJ. SREBPs: sterol-regulated transcription factors. J Cell Sci. 2006;119:973–976. doi: 10.1242/jcs.02866. Pt 6. doi:10.1242/jcs02866. [DOI] [PubMed] [Google Scholar]
- 24.Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001;40(6):439–452. doi: 10.1016/s0163-7827(01)00010-8. [DOI] [PubMed] [Google Scholar]
- 25.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. doi:10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28(46):11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. doi:10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52(3):403–407. doi: 10.1016/j.neuron.2006.10.016. doi:10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Bang S, Steenstra C, Kim SF. Striatum specific protein, Rhes regulates AKT pathway. Neurosci Lett. 2012;521(2):142–147. doi: 10.1016/j.neulet.2012.05.073. doi:10.1016/j.neulet.2012.05.073S0304-3940(12)00756-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold SE, Lucki I, Brookshire BR, Carlson GC, Browne CA, Kazi H, Bang S, Choi BR, Chen Y, McMullen MF, Kim SF. High fat diet produces brain insulin resistance, synaptodendritic abnormalities and altered behavior in mice. NeurobiolDis. 2014;67:79–87. doi: 10.1016/j.nbd.2014.03.011. doi:S0969-9961(14)00072-2 [pii];10.1016/j.nbd.2014.03.011 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bang S, Steenstra C, Kim SF. Striatum specific protein, Rhes regulates AKT pathway. NeurosciLett. 2012;521(2):142–147. doi: 10.1016/j.neulet.2012.05.073. doi:S0304-3940(12)00756-2 [pii];10.1016/j.neulet.2012.05.073 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SF, Huang AS, Snowman AM, Teuscher C, Snyder SH. From the Cover: Antipsychotic drug-induced weight gain mediated by histamine H1 receptor-linked activation of hypothalamic AMP-kinase. ProcNatlAcadSciUSA. 2007;104(9):3456–3459. doi: 10.1073/pnas.0611417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold SE, Gur RE, Shapiro RM, Fisher KR, Moberg PJ, Gibney MR, Gur RC, Blackwell P, Trojanowski JQ. Prospective clinicopathologic studies of schizophrenia: accrual and assessment of patients. Am J Psychiatry. 1995;152(5):731–737. doi: 10.1176/ajp.152.5.731. [DOI] [PubMed] [Google Scholar]
- 33.Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, Bakshi K, Kamins J, Borgmann-Winter KE, Siegel SJ, Gallop RJ, Arnold SE. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12(7):824–828. doi: 10.1038/nm1418. doi:nm1418 [pii]10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 34.Talbot K, Wang HY, Kazi H, Han LY, Bakshi KP, Stucky A, Fuino RL, Kawaguchi KR, Samoyedny AJ, Wilson RS, Arvanitakis Z, Schneider JA, Wolf BA, Bennett DA, Trojanowski JQ, Arnold SE. Demonstrated brain insulin resistance in Alzheimer's disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. The Journal of clinical investigation. 2012;122(4):1316–1338. doi: 10.1172/JCI59903. doi:10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson MA, Sillitoe RV, Blake DJ. Schizophrenia genetics: dysbindin under the microscope. Trends in neurosciences. 2004;27(9):516–519. doi: 10.1016/j.tins.2004.06.004. doi:10.1016/j.tins.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Talbot K. The sandy (sdy) mouse: a dysbindin-1 mutant relevant to schizophrenia research. 2009;179:87–94. doi: 10.1016/S0079-6123(09)17910-4. doi:10.1016/s0079-6123(09)17910-4. [DOI] [PubMed] [Google Scholar]
- 37. PLACE HOLDER DELETE.
- 38.Jeans A, Malins R, Padamsey Z, Reinhart M, Emptage N. Increased expression of dysbindin-1A leads to a selective deficit in NMDA receptor signaling in the hippocampus. Neuropharmacology. 2011;61(8):1345–1353. doi: 10.1016/j.neuropharm.2011.08.007. doi:10.1016/j.neuropharm.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Karlsgodt KH, Robleto K, Trantham-Davidson H, Jairl C, Cannon TD, Lavin A, Jentsch JD. Reduced dysbindin expression mediates N-methyl-D-aspartate receptor hypofunction and impaired working memory performance. Biological psychiatry. 2011;69(1):28–34. doi: 10.1016/j.biopsych.2010.09.012. doi:10.1016/j.biopsych.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glen WB, Jr., Horowitz B, Carlson GC, Cannon TD, Talbot K, Jentsch JD, Lavin A. Dysbindin-1 loss compromises NMDAR-dependent synaptic plasticity and contextual fear conditioning. Hippocampus. 2014;24(2):204–213. doi: 10.1002/hipo.22215. doi:10.1002/hipo.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sztainberg Y, Chen A. An environmental enrichment model for mice. Nature protocols. 2010;5(9):1535–1539. doi: 10.1038/nprot.2010.114. doi:10.1038/nprot.2010.114. [DOI] [PubMed] [Google Scholar]
- 42.Kondo M, Gray LJ, Pelka GJ, Christodoulou J, Tam PP, Hannan AJ. Environmental enrichment ameliorates a motor coordination deficit in a mouse model of Rett syndrome--Mecp2 gene dosage effects and BDNF expression. EurJNeurosci. 2008;27(12):3342–3350. doi: 10.1111/j.1460-9568.2008.06305.x. [DOI] [PubMed] [Google Scholar]
- 43.Nag N, Moriuchi JM, Peitzman CG, Ward BC, Kolodny NH, Berger-Sweeney JE. Environmental enrichment alters locomotor behaviour and ventricular volume in Mecp2 1lox mice. BehavBrain Res. 2009;196(1):44–48. doi: 10.1016/j.bbr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Canfran-Duque A, Casado ME, Pastor O, Sanchez-Wandelmer J, de la Pena G, Lerma M, Mariscal P, Bracher F, Lasuncion MA, Busto R. Atypical antipsychotics alter cholesterol and fatty acid metabolism in vitro. Journal of lipid research. 2013;54(2):310–324. doi: 10.1194/jlr.M026948. doi:10.1194/jlr.M026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauressergues E, Staels B, Valeille K, Majd Z, Hum DW, Duriez P, Cussac D. Antipsychotic drug action on SREBPs-related lipogenesis and cholesterogenesis in primary rat hepatocytes. Naunyn Schmiedebergs Arch Pharmacol. 2010;381(5):427–439. doi: 10.1007/s00210-010-0499-4. doi:10.1007/s00210-010-0499-4. [DOI] [PubMed] [Google Scholar]
- 46.Kristiana I, Sharpe LJ, Catts VS, Lutze-Mann LH, Brown AJ. Antipsychotic drugs upregulate lipogenic gene expression by disrupting intracellular trafficking of lipoprotein-derived cholesterol. PharmacogenomicsJ. 2010;10(5):396–407. doi: 10.1038/tpj.2009.62. [DOI] [PubMed] [Google Scholar]
- 47.Raeder MB, Ferno J, Vik-Mo AO, Steen VM. SREBP activation by antipsychotic- and antidepressant-drugs in cultured human liver cells: relevance for metabolic side-effects? MolCell Biochem. 2006;289(1-2):167–173. doi: 10.1007/s11010-006-9160-4. [DOI] [PubMed] [Google Scholar]
- 48.Steiner J, Martins-de-Souza D, Schiltz K, Sarnyai Z, Westphal S, Isermann B, Dobrowolny H, Turck CW, Bogerts B, Bernstein HG, Horvath TL, Schild L, Keilhoff G. Clozapine promotes glycolysis and myelin lipid synthesis in cultured oligodendrocytes. Front Cell Neurosci. 2014;8:384. doi: 10.3389/fncel.2014.00384. doi:10.3389/fncel.2014.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunt MC, Siponen MI, Alexson SE. The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochimica et biophysica acta. 2012;1822(9):1397–1410. doi: 10.1016/j.bbadis.2012.03.009. doi:10.1016/j.bbadis.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Holmsen H, Hindenes JO, Fukami M. Glycerophospholipid metabolism: back to the future. Thromb Res. 1992;67(3):313–323. doi: 10.1016/0049-3848(92)90006-v. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cellular and molecular life sciences : CMLS. 2005;62(19-20):2305–2316. doi: 10.1007/s00018-005-5195-z. doi:10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcus PI, Talalay P. Induction and purification of alpha- and beta-hydroxysteroid dehydrogenases. The Journal of biological chemistry. 1956;218(2):661–674. [PubMed] [Google Scholar]
- 53.Labrie F, Luu-The V, Lin SX, Labrie C, Simard J, Breton R, Belanger A. The key role of 17 beta-hydroxysteroid dehydrogenases in sex steroid biology. Steroids. 1997;62(1):148–158. doi: 10.1016/s0039-128x(96)00174-2. [DOI] [PubMed] [Google Scholar]
- 54.Haller JF, Krawczyk SA, Gostilovitch L, Corkey BE, Zoeller RA. Glucose-6-phosphate isomerase deficiency results in mTOR activation, failed translocation of lipin 1alpha to the nucleus and hypersensitivity to glucose: Implications for the inherited glycolytic disease. Biochimica et biophysica acta. 2011;1812(11):1393–1402. doi: 10.1016/j.bbadis.2011.07.007. doi:10.1016/j.bbadis.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okamoto K, Kakuma T, Fukuchi S, Masaki T, Sakata T, Yoshimatsu H. Sterol regulatory element binding protein (SREBP)-1 expression in brain is affected by age but not by hormones or metabolic changes. Brain Res. 2006;1081(1):19–27. doi: 10.1016/j.brainres.2006.01.081. [DOI] [PubMed] [Google Scholar]
- 56.Kim JH, Ong WY. Localization of the transcription factor, sterol regulatory element binding protein-2 (SREBP-2) in the normal rat brain and changes after kainate-induced excitotoxic injury. JChemNeuroanat. 2009;37(2):71–77. doi: 10.1016/j.jchemneu.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 57.Ong WY, Hu CY, Soh YP, Lim TM, Pentchev PG, Patel SC. Neuronal localization of sterol regulatory element binding protein-1 in the rodent and primate brain: a light and electron microscopic immunocytochemical study. Neuroscience. 2000;97(1):143–153. doi: 10.1016/s0306-4522(00)00031-2. [DOI] [PubMed] [Google Scholar]
- 58.Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends in neurosciences. 2011;34(11):591–598. doi: 10.1016/j.tins.2011.08.007. doi:10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taghibiglou C, Martin HG, Lai TW, Cho T, Prasad S, Kojic L, Lu J, Liu Y, Lo E, Zhang S, Wu JZ, Li YP, Wen YH, Imm JH, Cynader MS, Wang YT. Role of NMDA receptor-dependent activation of SREBP1 in excitotoxic and ischemic neuronal injuries. NatMed. 2009;15(12):1399–1406. doi: 10.1038/nm.2064. [DOI] [PubMed] [Google Scholar]
- 60.Le HS, Muhleisen TW, Djurovic S, Ferno J, Ouriaghi Z, Mattheisen M, Vasilescu C, Raeder MB, Hansen T, Strohmaier J, Georgi A, Brockschmidt FF, Melle I, Nenadic I, Sauer H, Rietschel M, Nothen MM, Werge T, Andreassen OA, Cichon S, Steen VM. Polymorphisms in SREBF1 and SREBF2, two antipsychotic-activated transcription factors controlling cellular lipogenesis, are associated with schizophrenia in German and Scandinavian samples. MolPsychiatry. 2010;15(5):463–472. doi: 10.1038/mp.2008.110. doi:mp2008110 [pii];10.1038/mp.2008.110 [doi] [DOI] [PubMed] [Google Scholar]
- 61.Le HS, Theisen FM, Haberhausen M, Raeder MB, Ferno J, Gebhardt S, Hinney A, Remschmidt H, Krieg JC, Mehler-Wex C, Nothen MM, Hebebrand J, Steen VM. Association between the insulin-induced gene 2 (INSIG2) and weight gain in a German sample of antipsychotic-treated schizophrenic patients: perturbation of SREBP-controlled lipogenesis in drug-related metabolic adverse effects? MolPsychiatry. 2009;14(3):308–317. doi: 10.1038/sj.mp.4002133. [DOI] [PubMed] [Google Scholar]
- 62.Ferno J, Raeder MB, Vik-Mo AO, Skrede S, Glambek M, Tronstad KJ, Breilid H, Lovlie R, Berge RK, Stansberg C, Steen VM. Antipsychotic drugs activate SREBP-regulated expression of lipid biosynthetic genes in cultured human glioma cells: a novel mechanism of action? The pharmacogenomics journal. 2005;5(5):298–304. doi: 10.1038/sj.tpj.6500323. doi:10.1038/sj.tpj.6500323. [DOI] [PubMed] [Google Scholar]
- 63.McEvoy J, Baillie RA, Zhu H, Buckley P, Keshavan MS, Nasrallah HA, Dougherty GG, Yao JK, Kaddurah-Daouk R. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PLoS One. 2013;8(7):e68717. doi: 10.1371/journal.pone.0068717. doi:10.1371/journal.pone.0068717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ledesma MD, Martin MG, Dotti CG. Lipid changes in the aged brain: effect on synaptic function and neuronal survival. ProgLipid Res. 2012;51(1):23–35. doi: 10.1016/j.plipres.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Krugel U. Purinergic receptors in psychiatric disorders. Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2015.10.032. doi:10.1016/j.neuropharm.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 66.LaSalle JM, Powell WT, Yasui DH. Epigenetic layers and players underlying neurodevelopment. Trends in neurosciences. 2013;36(8):460–470. doi: 10.1016/j.tins.2013.05.001. doi:10.1016/j.tins.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spell C, Kolsch H, Lutjohann D, Kerksiek A, Hentschel F, Damian M, von BK, Rao ML, Maier W, Heun R. SREBP-1a polymorphism influences the risk of Alzheimer's disease in carriers of the ApoE4 allele. DementGeriatrCogn Disord. 2004;18(3-4):245–249. doi: 10.1159/000080023. [DOI] [PubMed] [Google Scholar]
- 68.Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, Strand A, Tarditi A, Woodman B, Racchi M, Mariotti C, Di DS, Corsini A, Bates G, Pruss R, Olson JM, Sipione S, Tartari M, Cattaneo E. Dysfunction of the cholesterol biosynthetic pathway in Huntington's disease. JNeurosci. 2005;25(43):9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.