Abstract
All mucins are highly O-glycosylated by variable glycans depending on species, histo-blood group and organ. This makes the intestinal main mucin MUC2 non-degradable by the host digestive system, but well by both commensal and pathogenic bacteria. The MUC2 glycans are important for selection of the commensal bacteria and act as a nutritional source for the bacteria; this also helps the host to recover some of the energy spent on constantly renewing the protective mucus layer.
Glycosylation is the most diverse and common post-translational modification of cell surfaces and secreted proteins. N-glycosylation is most well studied and predictable, whereas O-glycosylation is more diverse and less well understood. O-glycosylation is also often called mucin-type glycosylation as it is typical for mucins that often have more than 80% of the mass as O-glycans. This review will discuss the mucin-type O-glycosylation and especially the O-glycosylation of human and mice intestinal mucin MUC2 in relation to bacteria and disease.
Keywords: Mucus, Glycosyltransferase, Glycosylation, Colon, Small Intestine, Proteomics, Mass Spectrometry
O-glycosylation
The two hydroxyl-amino acids, serine and threonine, are the attachment sites for the mucin O-glycans. The initiation of O-glycosylation is made by one of the twenty peptidyl-GalNAc transferases in the human genome that utilize UDP-GalNAc to add GalNAc in an alfa-configuration [1]. Each of these 20 transferases have their specificities when it comes to peptide or glycopeptide sequence. Some of these enzymes are more general in their preferences of glycosylation site whereas others are more specific [1]. O-glycosylation is normally initiated in the Golgi apparatus and requires that the Ser/Thr are exposed on the folded protein. The amino acid sequence to be glycosylated cannot be well-folded and typically the amino acid Pro is found close to the attachment site. The 20 different transferase specificities and the restriction to exposed and less folded regions make O-glycosylation prediction from only the amino acid sequence unreliable even if such tools have been developed [2]. The initiating GalNAc is extended by the addition of Gal, GlcNAc or GalNAc on the 3- and/or 6-hydroxyl groups giving primarily the Core1, 2, 3, and 4 structures [3–6]. These are further extended by Gal and GlcNAc and often terminated by sialic acid, sulfate group, GalNAc and/or Fuc. The expressed glycosyltransferases in the specific cell will determine the O-glycan outcome and thus the same protein sequence can have highly different glycans. In addition to the transferases present, the transferase localization within the Golgi stacks will affect the resulting glycosylation [7]. In the secretory pathway of the cell there is a declining pH gradient from the endoplasmic reticulum to the trans-Golgi network that is important for the glycosyltransferase localization. Neutralization of this gradient moves the transferases forward and results in less glycosylated proteins [8].
Mucins
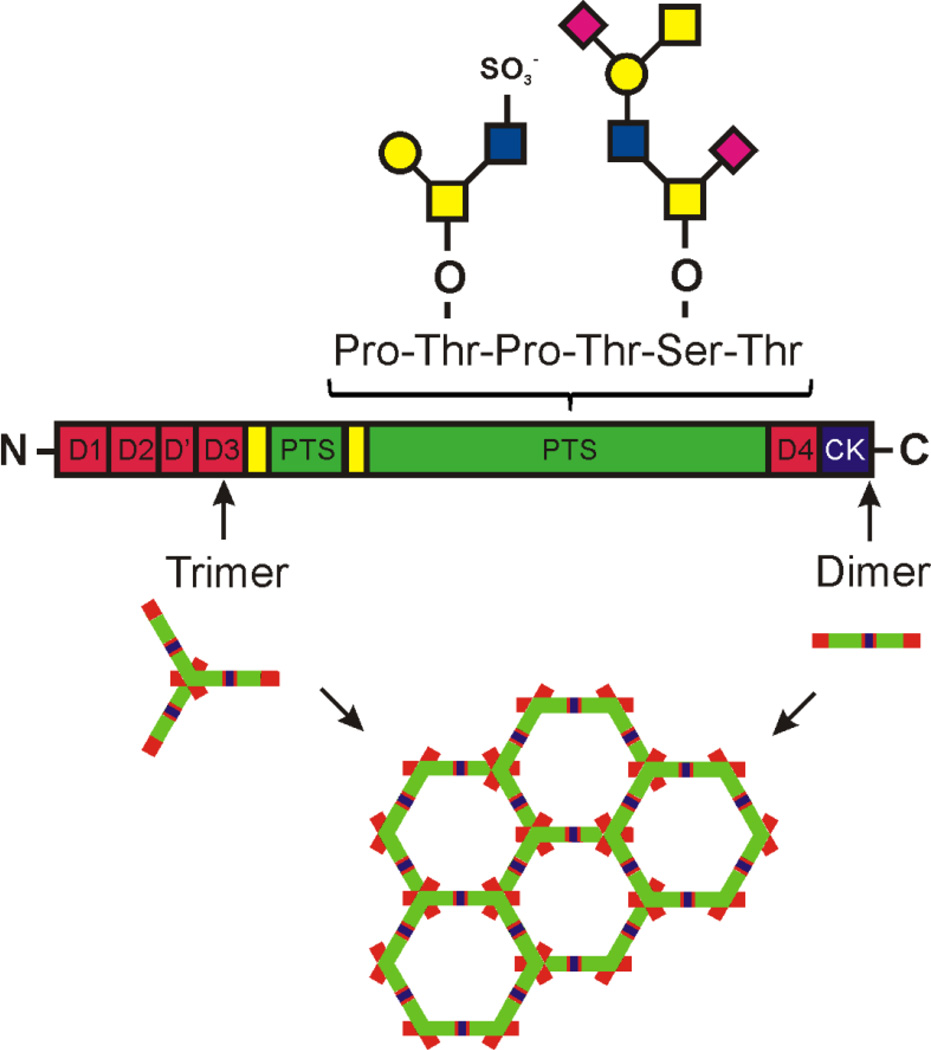
Mucins are characterized by long extended, rod-like glycopeptide domains that are called mucin domains [6,9,10]. These are densely O-glycosylated making them look like a bottle brush with a central protein core and the glycans pointing out in all directions as bristles. The protein cores of the mucins domains are characterized by abundant Ser, Thr and Pro and are called PTS domains for the single letter code of these amino acids [11]. Originally these sequences were called VNTR for varying number of tandem repeats [12]. However, quite few mucins do not show any tandem repeats and do not vary in length. Instead all these sequences are characterized by long stretches of Thr and/or Ser (typically >40% of all amino acids) and Pro (typically >5%). In fact, one can use these criteria in search algorithms for mucin genes as there is no sequence conservation even between closely related species [11,13]. It seems as if only the number of glycan attachment sites and the total length of these domains have been conserved during evolution. These PTS sequences are often long (> 2,000 amino acids) and limited to one single exon. However, there are a few exceptions, where each PTS repeat is encoded in one single exon thus allowing the mucin domain length to vary by alternative splicing [11,14].
All mucins have one or several PTS domains that are either localized N-terminally, as for the type 1 membrane-bound mucins, or centrally, as for the gel-forming mucins [6,10,12,15]. The MUC7 mucin is more or less only a PTS domain [6]. There are two types of transmembrane mucins, the SEA and NIDO-AMOP-VWD types [11]. The SEA mucins all have a SEA domain just outside of the cell membrane. This domain is autocatalytically cleaved during biosynthesis by the folding energy, but remains held together as one molecule at the cell surface [16]. This interaction is strong and can hold the highly O-glycosylated extracellular mucin domain with a mass >2 MDa anchored to the cell membrane, but at the same time act as a breaking point when the physical stress on the surface is too strong threatening to rupture the cell membrane [17]. The second group, the NIDO-AMOP-vWD mucins, has only one member among mammals, the MUC4 mucin. This mucin has its three typical domains next to the large PTS domain at the extracellular cell surface. MUC4 is just as the SEA mucins cleaved during biosynthesis [18]. Most transmembrane mucins are found in the gastrointestinal tract, MUC1, MUC3, MUC4, MUC12, MUC13, MUC16, and MUC17. The function of the transmembrane mucins is, except for their role in cancer [12], less well explored. These mucins are typically found on the apical side of polarized epithelial cells and extend far out from the cell membrane [15,19]. They all appeared during vertebrate evolution, in contrast to the gel-forming mucins that appeared early during metazoan evolution [11]. In addition to the mucins, there are other important glycoproteins in the gastrointestinal tract, for example DMBT1 with its scavenger receptor repeats [20].
The gel-forming mucins are characterized by a long N-terminus built by three vWD assemblies, one or several PTS domains interrupted by CysD domains, and a C-terminus ending by a cysteine knot (Fig. 1) [21–23]. Humans have four expressed gel-forming mucins, MUC2, MUC5AC, MUC5B and MUC6 whereas frogs and fish have many more [11]. All these mucins form large polymers held together by the cysteine-knot two-and-two in the C-terminus and in the N-terminus two-and-two in MUC5B or three-and-three in MUC2 [23–26]. In the gastrointestinal tract, the MUC5AC and MUC6 mucins are found in the stomach suface cells and glands, respectively, whereas the rest of the intestine has MUC2 as its typical mucin [6,23]. Due to the type of MUC2 oligomerization this mucin generates large net-like sheets that when layered on top of each other form efficient size-exclusion filters. The MUC2 mucin is produced by goblet cells where MUC2 is densely packed due to the calcium and low pH in the secretory vesicles. This packing is required for well-organized release that allows the >1,000-fold expansion upon secretion of MUC2 [26]. To allow this expansion, sufficient levels of bicarbonate are required to remove calcium and increase the pH [27]. Most important for this expansion is the highly O-glycosylated mucin domains that are attracting water and by this generate physical forces that are pulling out and expanding the folded MUC2 into flat sheets [26,28].
Fig. 1. The MUC2 mucin structure with examples of O-glycans attached to the PTS sequences.
The MUC2 mucin forms large net-like sheets by N-terminal trimerization and C-terminal dimerization.
The MUC2 mucin is building the intestinal mucus skeleton. In the small intestine the mucus is less dense and after bacterial colonization non-attached to the epithelium [29]. In the large intestine, the mucus is organized into two layers, an inner attached dense mucus that is essentially free of bacteria and an outer non-attached less dense mucus that allow bacteria to enter and thrive [30]. The inner mucus layer of distal colon is renewed every hour and the conversion to the outer layer is dependent on host-controlled proteolysis that allows expansion of MUC2 without a directly dissolving the polymeric nature of the mucin [31,32]. The properties of the inner colon mucus layer are dependent on the commensal bacteria and when normally developed it acts as a physical filter separating bacteria as well as bacteria-sized beads [33,34]. The function of this inner mucus layer is fully relaying on the integrity of the MUC2 polymeric network.
Mucin glycans and their role as nutrients for the commensal bacteria
The O-glycan repertoire on the MUC2 mucin are different between species and also show individual variation as the ABH(O), Lewis, and Secretor histoblood group systems [35]. The human small intestinal MUC2 is fucosylated in Secretor positive individuals by the FUT2 enzyme, forming an epitope that is further extended dependent on the individual AB blood group status [36]. Human colon shows less variability between individuals and the distal colon MUC2 glycans are relatively homogenous among humans and rich in the Sda-Cad epitope [36–39]. The human colon glycans are characterized by NeuAc attached to the C-6 of the peptide-bound GalNAc. In addition, sulfation of Gal is prominent as well as acetylation on the sialic acids [40].
The species differences between mice and humans are reflected at the Core structure level as for example humans are dominated by Core3, whereas mice have mainly Core1 and Core2 structures [39,41]. As only inbred specific strains of mice have been studied, we do not really know the extent of blood group variability in mice living free in nature. The C57BL/6 mice has been mostly studied and these have little fucosylation present in the normal small intestine, but more fucosylated compouds are found in colon [41].
Although there are specific features depending on species, all studied mucins have a very diverse glycan repertoire with up to a hundred different structures on one mucin from one source. In fact it has been calculated that all epitopes cannot be found on one and the same Muc2 mucin domain [42]. The high number of glycan structure is partly explained by the presence of biosynthetic glycan intermediates and each organ typically has a limited number of fully ‘completed’ products. Comparing O-glycans with glycosphingolipids from the same tissue show that in the latter case only certain specific intermediates are present [43]. This argues for differences in the glycosylation processes for these two types of glycoconjugates. The high variability and the presence of all intermediate glycans on mucins is highly functional when assuming that a major function of the mucin is to bind many different bacteria that carry surface adhesins. In this way the mucins can select certain bacteria or trap and move these away from the host.
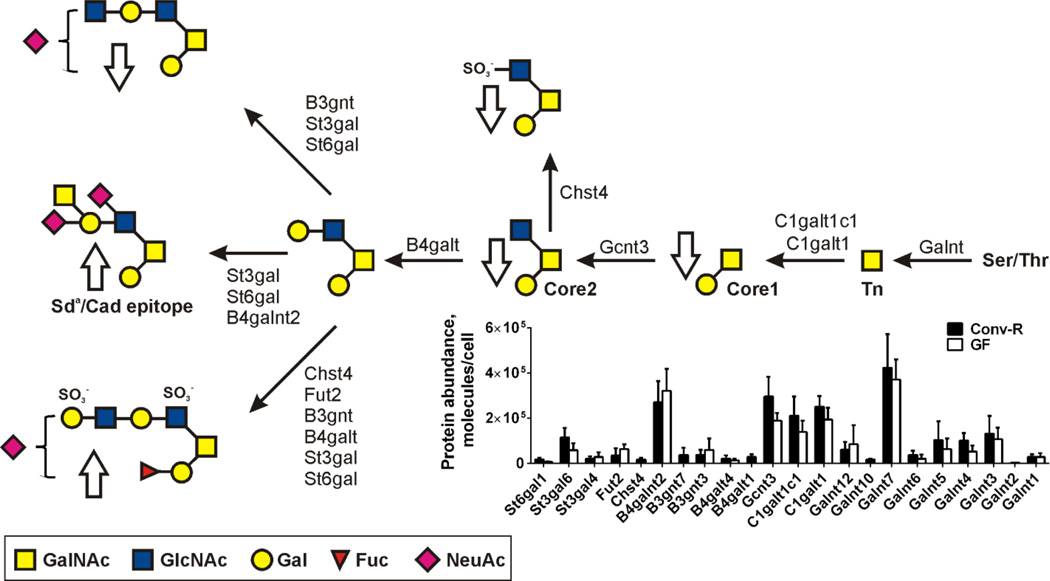
The mucin O-glycans are closely related to the intestinal commensal bacteria [44]. Our recent studies show that the Muc2 glycosylation of mice grown up under germ-free (GF) conditions is different than when colonized at birth e.g. conventionally raised (Conv-R) (Fig. 2) [45]. The levels of most glycosyltransferases are increased by the presence of microbiota, whereas a few are decreased (Fig. 2). The resulting glycan structures are the result of the combined actions of all the transferases and even if for example the B4galnt2 enzyme responsible for the formation of the Sda/Cad epitope is decreased by microbiota, the levels of glycans with this structure are increased likely due to more precursors available. Overall, the Conv-R mice have more extended structures and higher fucosylation. Similar glycans as in the Conv-R mice are obtained if GF mice are colonized by normal mouse microbiota, but it takes up to three weeks until this is accomplished [45]. There is nothing known about how the bacteria can signal this at a distance as well as the host control circuits responsible for these glycosylation alterations.
Fig. 2. Conventionalization of germ-free mice alters glycosyltransferase levels and glycans.
Major glycans found on Muc2 in mouse colon and arrows showing increased/decreased levels upon conventionalization of germ-free (GF) mice. The relative molecular abundances of the glycosyltransferases in conventionally raised (Conv-R) and germ-free (GF) mice. Results from Holmen et al. [41] and Johansson et al. [45]. C1galt1c1 is the cosmic chaperone [76].
The mucin and especially MUC2 O-glycans are probably very important for the selection of the typical bacterial composition found in each species [44]. This has been elegantly illustrated by John Rawls observing that zebrafish and mice have different normal microbiota [46]. After the germ-free zebrafish and mice were cross-colonized with the other species’ microbiota the original zebrafish and mice characteristic microbiota was selected out from the donor bacteria. This illustrate that the host can select its microbiota. Comparing the commensal bacteria between mice and humans show differences that further supports the idea that the host can influence the selection of its bacteria [47,48]. There are probably several mechanisms by which the host can influence or select its microbiota, like the small intestinal repertoire of antimicrobial peptides/proteins [49,50]. Of course, other reasons like food or environment will also influence the bacterial composition [33,51]. However, the mucin O-glycan repertoire is probably influential as the glycans can act as attachment sites for the numerous bacterial adhesins. This is probably especially important for bacteria residing on the interphase between the inner and outer mucus layers as the mucin glycans are essentially intact at this site [31,52].
Of further importance is also the capacity of bacteria to degrade the mucin glycans provided by the host [53,54]. As the glycans varies between species, the bacteria needs to carry a repertoire of enzymes adapted to its specific host. Typical for the commensal bacteria are their rich repertoire of glycan degrading enzymes. It has been estimated that up to 40% of the bacterial genomes are encoding these types of enzymes [53] The released monosaccharides are used by the bacteria providing the enzymes or other members of the microbiota. The importance of the host mucin glycans, polysaccharides and other saccharides degraded by the bacteria is difficult to quantify. In any case, the host mucin glycans are important for the host microbiota as illustrated by mice that partially lack Core1 glycans in the intestine [55]. These mice have higher levels of Bacteroidetes and lower level of Fermicutes than wild-type animals. There is a large interest in the human intestinal microbiota in relation to human health and disease and it has become common to colonize GF mice with human microbiota to study their effects on the host [56,57]. These mice have been called ‘humanized mice’, but these studies have not yet been considering the influence of mouse mucin glycans on the bacteria typical for humans.
The commensal colonic bacteria can efficiently utilize the monosaccharides released by the bacterial hydrolases and transported into the bacteria by specific transporters [53]. Many bacteria carry large operons including sensory molecules, hydrolases and transporters [58]. Not all bacteria have all necessary enzymes for cleaving specific sugar linkages and it is common that several bacteria collaborate in a community. The bacteria metabolize the monosaccharides to smaller molecules, most commonly to the short fatty acids acetate, propionate and butyrate [53]. These small molecules can easily diffuse through the inner mucus layer and provide the host and its epithelium with energy. In this way, the host recovers some of the energy spent to build and renew the inner mucus layer. That this is a true symbiotic relation is illustrated by the bacterial capacity to stimulate the formation of a denser and less penetrable mucus and by this provide the host with a better protection and bacteria with more food [45]. The details on how the bacteria can signal this to the host is not understood.
The intestinal bacteria are very efficient in degrading the mucus oligosaccharides and eventually also the protein core is degraded as there is very little mucus to be found in normal feces. At the same time, it is important that the degradation of the glycans is not too fast and by this affecting the mucus faster than it is renewed. The importance of sufficiently long O-glycans is illustrated by the Core3−/− mice that are more susceptible to dextran sulfate colitis (DSS) and the Core1−/− mice that can spontaneously develop colitis [59,60]. This is in concordance with the knowledge that mice have largely Core1 glycans, but is also highly dependent on the bacterial composition as the inflammation varies between animal facilities.
Glycans protect mucins from proteases
In order to maintain intact intestinal mucus it is very important that the digestive enzymes are not able to degrade the MUC2 mucin polymer in either the small or large intestine. In fact, the MUC2 mucin is not affected by the digestive proteases from pancreas or epithelium. This is largely dependent on its glycosylation as the mucin domains of MUC2 and other mucins are resistant to proteases. This is achieved by their dense glycosylation that does not allow proteases to reach the protein core. The less glycosylated N- and C-termini of MUC2 are still not degraded as these parts are highly stabilized by numerous disulfide bonds where the specific cleavage sites for the pancreatic endoproteases (trypsin, chymotrypsin and elastase) are hidden. It is also important to realize that most of the three N-terminal vWD assemblies up to the localization of the timerization are not required for maintaining the MUC2 polymeric network (Fig. 1 and 3). If the MUC2 mucin is exposed to trypsin the polymeric network remains intact, but once the disulfide bonds are reduced the whole molecule falls apart to small peptides and two large glycopeptides of about 350 and 700 kDa representing the two mucin PTS domains [42]. Hosts do not secrete any enzymes in the gastrointestinal tract that can degrade the type of O-glycans that covers mucins. On the other hand, commensal bacteria carry numerous of such glycan degrading enzymes [53,57].
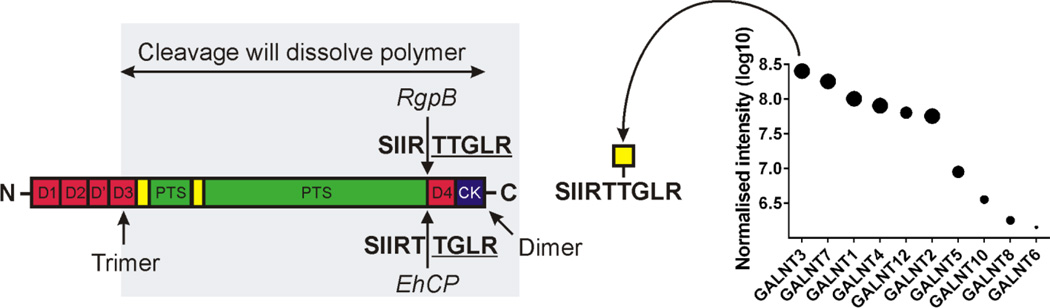
Fig. 3. Protection of human MUC2 susceptible cleavage sites by specific peptidyl-GalNAc transferase 3.
Schematic figure of MUC2 showing the part where proteolytic cleavage might dissolve the MUC2 polymer network. Cleavage sites for the Entamoeba histolytica EhCP and Porphyrinomonas aerogenosa RgpB proteases dissolving the human MUC2 polymer if not glycosylated by GALNT3, the most abundant peptidyl-GalNAc transferase in human colon. Results from Lidell et al. [62] and van der Post et al. [63,64].
Pathogenic organisms have on the other hand developed mechanisms to circumvent the normal intestinal protective systems. For example, the large parasite Entamoeba histolytica that needs to penetrate the inner colon mucus layer to reach the epithelium in order to invade the host [61]. Interestingly, the human MUC2 mucin, in contrast to mouse Muc2, has a more susceptible site for specific proteases located between the trimeric and dimeric positions (Fig. 3). Cleavages anywhere between these two points will dissolve the MUC2 polymeric network. Studies of the cysteine proteases of E. histolytica have shown that EhCP enzyme is capable of cleaving the MUC2 protein core at SIIRT↓TGLR sequence [62]. However, this is not possible if the second Thr is O-glycosylated (Fig. 3). Interestingly, this site is only glycosylated by the peptidyl-GalNAc transferase 3 (GALNT3) (Fig. 3) [63]. This suggests that this specific transferase could be of special importance for humans where it is the most abundant GALNT [64]. It is known that only about 20% of E. histolytica infected individuals get invasive disease, something that could be related to the level of glycosylation by GALNT3. Individuals that totally lack the GALNT3 enzyme have the disease Familial Tumoral Calcinosis [65], a rare disease with hyperphosphatemia and ectopic calcifications where absence of specific O-glycosylation cause an abnormal protease processing [66]. The lack of GALNT3 in these individuals should probably make them more susceptible to E. histolytica infection.
The oral pathogenic bacteria Porfyromonas aerogenosa produce a serine protease (RgpB) capable at cleaving MUC2 one amino acid away from the E. histolytica site, SIIR↓TTGLR (Fig. 3) [63]. Also this cleavage is blocked by glycosylation by GALNT3. This is not a typical colonic bacteria although found there as well as in the mouth, but there are likely also other bacterial proteases capable of cleaving MUC2. Interestingly, none of the two enzymes cleaving MUC2 at this site are able to cleave MUC2 at any other position.
Alterations of mucin glycosylation at disease
Genetic differences in glycosyltransferases cause altered glycans where some are giving severe disease, whereas others do not [67]. There are also numerous publications suggesting glycosylation alterations claimed to be linked to disease [68,69]. This has been the case for the disease Cystic Fibrosis where the absence of the CFTR channel causes severe problems of the respiratory tract. Altered glycosylation of the mucins MUC5B and MUC5AC was earlier pointed out to be caused by absent CFTR channel, but later studies have shown that the glycosylation alterations are due to secondary effects of infection and inflammation [70–72].
Chronic infection by Helicobacter pylori and by this induced gastric tissue inflammation cause an altered glycan repertoire found with increased levels of sialyl-Lewis a/x antigens. The glycan changes are caused by transcriptional up-regulation of the B3GNT5, B3GALT5, and FUT3 genes [73].
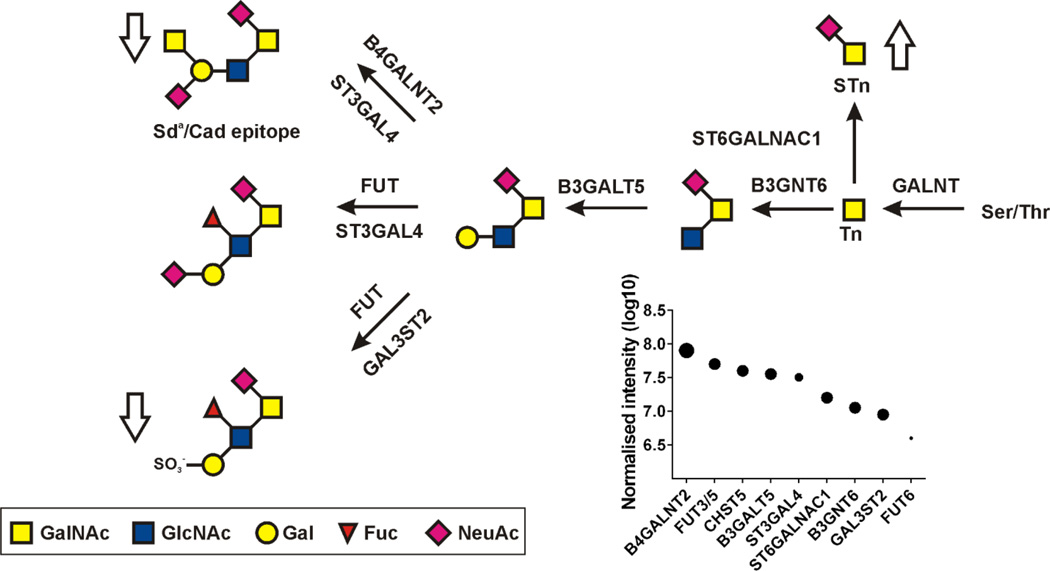
Ulcerative colitis (UC) is also a disease where altered glycosylation has been implicated [69]. A thorough study of the MUC2 mucin O-glycosylation of more than 50 individuals showed that control individuals and most of the UC patients with none or little inflammation had similar glycans [39,74]. The human MUC2 glycans are characterized by NeuAc2-6GalNAc and Core3 glycans terminated by Sda/Cad epitopes and Fuc and sulfate (Fig. 4). In a separate study of colon epithelium, the glycosyltransferases responsible for these normal structures were observed (Fig. 4) [64]. It was only the UC patients with more severe inflammation and active disease that showed altered glycans as indicated by the arrows in Fig. 4. These patients had increased levels of NeuAc2-6GalNAc (STn) and lower levels of more complex glycans as for example the ones carrying Sda/Cad epitope (Fig. 4). Interestingly, when one patient with active disease and altered MUC2 O-glycans was analyzed a second time and with little inflammation in remission, the patient had returned to a normal glycan pattern [74]. This clearly shows that the altered glycosylation also in this disease was due to the inflammation and not caused by the disease itself.
Fig. 4. Reversible alterations of the MUC2 O-glycans upon active inflamed ulcerative colitis (UC).
Major MUC2 O-glycans in human sigmoid colon of control individuals and patients with inactive UC disease. Arrows show alterations in patients with active UC. The relative amounts of the glycosyltransferases used for the biosynthesis of the glycans in human control individuals are shown as dots. Results from Holmen-Larsson et al. [74] and van der Post et al. [64].
Cancer and cancer development is well-known for showing altered glycans where the most characteristic and common features include shortened glycans and increased sialylation. This topic was recently reviewed in depth [75].
Conclusions and Perspectives
The variable O-glycosylation on mucins generated by the high number of glycosyltransferases is important for the protective and bacterial binding capacity of mucus. A major reason for the high number of peptidyl-GalNAc and other transferases involved in O-glycan biosynthesis should be sought in the mucin interplay with microorganisms. Inflammation and infection will trigger alteration in glycosylation and may hint to general mechanisms and roles of epithelial glycans in responding to outside threats.
Studies of the commensal microbiota interaction with the host and the bacterial effects on the host metabolism and other functions will be of highest priority coming years. On a glycobiology point of view, we need to develop better experimental animal models being truly humanized. This means that they carry human mucins with human glycosylation patterns, something that require a number of gene modifications in mice. The mechanisms behind alterations of glycosyltransferase levels and activities and thus glycan repertoire on mucosal surfaces upon inflammation and infection are poorly understood. Too little attention has been placed on transcriptional and other control systems of glycosyltransferase expression.
Acknowledgments
This work was supported by the Swedish Research Council, The Swedish Cancer Foundation, The Knut and Alice Wallenberg Foundation, IngaBritt and Arne Lundberg Foundation, Sahlgren's University Hospital (ALF), Wilhelm and Martina Lundgren’s Foundation, Torsten Söderberg Foundation, The Sahlgrenska Academy, National Institute of Allergy and Infectious Diseases (U01AI095473, the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH), The Swedish Foundation for Strategic Research - The Mucus-Bacteria-Colitis Center (MBC) of the Innate Immunity Program, Cystic Fibrosis Foundation (CFF), and Lederhausen’s Center for CF Research at Univ. Gothenburg.
Abbreviations
- GF
germ-free
- Conv-R
conventionally raised
- vWD
von Willebrand D domain
- PTS
proline-threonine-serine domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTB, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. Embo J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockhausen I. Pathways of O-glycanbiosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 4.Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. Embo Rep. 2006;7:599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corfield AP, Berry M. Glycan variation and evolution in the eukaryotes. Trends Biochem Sci. 2015;40:351–359. doi: 10.1016/j.tibs.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Corfield AP. Mucins: A biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta. 2014;1850:236–252. doi: 10.1016/j.bbagen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, et al. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. Journal of Cell Science. 1998;111:45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- 8.Axelsson MAB, Karlsson NG, Steel D, Ouwendijk J, Nilsson T, Hansson GC. Neutralization of the pH in the Golgi apparatus causes redistribution of glycosyltransferases and changes in the O-glycosylation of mucins. Glycobiology. 2001;11:633–644. doi: 10.1093/glycob/11.8.633. [DOI] [PubMed] [Google Scholar]
- 9.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang T, Hansson GC, Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci USA. 2007;104:16209–16214. doi: 10.1073/pnas.0705984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollingsworth MA, Swanson BJ. Mucin in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 13.Lang T, Alexandersson M, Hansson GC, Samuelsson T. Bioinformatic identification of polymerizing and transmembrane mucins in the puffer fish Fugu rubripes. Glycobiology. 2004;14:521–527. doi: 10.1093/glycob/cwh066. [DOI] [PubMed] [Google Scholar]
- 14.Lang T, Hansson GC, Samuelsson T. An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics. 2006;7:97. doi: 10.1186/1471-2164-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GMH, Schutte A, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macao B, Johansson DGA, Hansson GC, Härd T. Auto-proteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nature Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 17.Pelaseyed T, Zäch M, Petersson AC, Svensson F, Johansson DG, Hansson GC. Unfolding dynamics of the mucin SEA domain probed by force spectroscopy suggest that it acts as a cell protective device. FEBS J. 2013;280:1491–1501. doi: 10.1111/febs.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rong M, Rossi EA, Zhang J, McNeer RR, vandenBrande JMH, Yasin M, et al. Expression and localization of Muc4/sialomucin complex (SMC) in the adult and developing rat intestine: Implications for Muc4/SMC function. J Cell Physiol. 2005;202:275–284. doi: 10.1002/jcp.20121. [DOI] [PubMed] [Google Scholar]
- 19.Sauvanet C, Wayt J, Pelaseyed T, Bretscher A. Structure, Regulation, and Functional Diversity of Microvilli on the Apical Domain of Epithelial Cells. Annu Rev Cell Dev Biol. 2015;31:593–621. doi: 10.1146/annurev-cellbio-100814-125234. [DOI] [PubMed] [Google Scholar]
- 20.Ligtenberg AJ, Karlsson NG, Veerman ECI. Deleted in Malignant Brain Tumors-1 Protein (DMBT1): A Pattern Recognition Receptor with Multiple Binding Sites. Int J Mol Sci. 2010;11:5212–5233. doi: 10.3390/ijms1112521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gum JR, Hicks JW, Toribara NW, Siddiki B, Kim YS. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J Biol Chem. 1994;269:2440–2446. [PubMed] [Google Scholar]
- 22.Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751. [DOI] [PubMed] [Google Scholar]
- 23.Thornton DJ, Rousseau K, McGuckin MA. Structure and Function of the Polymeric Mucins in Airways Mucus. Ann Rev Physiol. 2008;70:459–486. doi: 10.1146/annurev.physiol.70.113006.100702. [DOI] [PubMed] [Google Scholar]
- 24.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godl K, Johansson MEV, Karlsson H, Morgelin M, Lidell ME, Olson FJ, et al. The N-termini of the MUC2 mucin form trimers that are held together within a trypsin-resistant core fragment. J Biol Chem. 2002;277:47248–47256. doi: 10.1074/jbc.M208483200. [DOI] [PubMed] [Google Scholar]
- 26.Ambort D, Johansson MEV, Gustafsson JK, Nilsson H, Ermund A, Johansson BR, et al. Calcium and pH-dependent Packing and Release of the Gel-forming MUC2 Mucin. Proc Natl Acad Sci U S A. 2012;109:5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustafsson JK, Ermund A, Ambort D, Johansson MEV, Nilsson HE, Thorell K, et al. Bicarbonate and functional CFTR channel is required for proper mucin secretion and link Cystic Fibrosis with its mucus phenotype. J Exp Med. 2012;209:1263–1272. doi: 10.1084/jem.20120562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson MEV, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nature Rev Gastroenterol Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ermund A, Schutte A, Johansson MEV, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastroint Liver Physiol. 2013;305:G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson MEV, Phillipson M, Petersson J, Holm L, Velcich A, Hansson GC. The inner of the two Muc2 mucin dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson MEV, Holmen Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansson MEV. Fast Renewal of the Distal Colonic Mucus Layers by the Surface Goblet Cells as Measured by In Vivo Labeling of Mucin Glycoproteins. PLoS ONE. 2012;7:e41009. doi: 10.1371/journal.pone.0041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsson HE, Rodriguez-Pineiro AM, Schütte A, Ermund A, Boysen P, Sommer F, et al. The gut microbiota composition impairs the colon inner mucus layer barrier. EMBO Reports. 2015;16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson MEV, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and in patients with ulcerative colitis. Gut. 2014;213:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravn V, Dabelsteen E. Tissue distribution of histo-blood group antigens. APMIS. 2000;108:1–28. doi: 10.1034/j.1600-0463.2000.d01-1.x. [DOI] [PubMed] [Google Scholar]
- 36.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, et al. Evidence of Regio-specific Glycosylation in Human Intestinal Mucins: Presnece of an acidic gradient along the gastrointestinal tract. J Biol Chem. 2003;278:46337–46348. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- 38.Capon C, Maes E, Michalski JC, Leffler H, Kim YS. Sd(a)-antigen-like structures carried on core 3 are prominent features of glycans from the mucin of normal human descending colon. Biochem J. 2001;358:657–664. doi: 10.1042/bj3580657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmen Larsson JM, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19:756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 40.Campbell F, Appleton MAC, Fuller CE, Greeff MP, Hallgrimsson J, Katoh R, et al. Racial variation in the O-acetylation phenotype of human colonic mucosa. Journal of Pathology. 1994;174:169–174. doi: 10.1002/path.1711740305. [DOI] [PubMed] [Google Scholar]
- 41.Holmen-Larsson JM, Thomsson KA, Rodriguez-Pineiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucinc O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastroint Liver Physiol. 2013;305:G357–G363. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlstedt I, Herrmann A, Karlsson H, Sheehan JK, Fransson L, Hansson GC. Characterization of two different glycosylated domains from the insoluble mucin complex of rat small intestine. J Biol Chem. 1993;268:18771–18781. [PubMed] [Google Scholar]
- 43.Breimer ME, Hansson GC, Karlsson KA, Leffler H. Blood group type glycosphingolipids from the small intestine of different animals analysed by mass spectrometry and thin-layer chromatography. A note on species diversity. J Biochem. 1981;90:589–609. doi: 10.1093/oxfordjournals.jbchem.a133513. [DOI] [PubMed] [Google Scholar]
- 44.Bergstrom KS, Xia L. Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology. 2013;23:1026–1037. doi: 10.1093/glycob/cwt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson MEV, Jacobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodriguez-Pineiro A, et al. Normalization of the host intestinal mucus systems requires long-term colonization. Cell Host Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-Bacterial Mutualism in the Human Intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 48.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Micro. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 49.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 50.Bevins CL. Paneth cell defensins: key effector molecules of innate immunity. Biochem Soc Trans. 2006;34:263–266. doi: 10.1042/BST20060263. [DOI] [PubMed] [Google Scholar]
- 51.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of Mammals and Their Gut Microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Micro. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Micro. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host & Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sommer F, Adam N, Johansson ME, Xia L, Hansson GC, Backhed F. Altered mucus glycosylation in core 1 o-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One. 2014;9:e85254. doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin FP, Wang Y, Sprenger N, Yap IK, Rezzi S, Ramadan Z, et al. Top-down systems biology integration of conditional prebiotic modulated transgenomic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:205. doi: 10.1038/msb.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex Glycan Catabolism by the Human Gut Microbiota: The Bacteroidetes Sus-like Paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, et al. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu J, Wei B, Wen T, Johansson MEV, Xiaowei L, Bradford E, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dharmani P, Srivastava V, Kissoon-Singh V, Chadee K. Role of Intestinal Mucins in Innate Host Defense Mechanisms against Pathogens. J Innate Immun. 2009;1:123–135. doi: 10.1159/000163037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal part and dissolves the protective colonic mucus gel. Proc Nat Acad Sci Usa. 2006;103:9298–9393. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Post S, Subramani DB, Backstrom M, Johansson MEV, Vester-Christensen MB, Mandel U, et al. Site-specific O-glycosylation on the MUC2 mucin inhibits cleavage by the Porphyromonas gingivalis secreted cysteine protease (RgpB) J Biol Chem. 2013;288:14636–14646. doi: 10.1074/jbc.M113.459479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Post S, Hansson GC. Membrane protein profiling of human colon reveals distinct regional differences. Mol Cell Proteomics. 2014;13:2277–2287. doi: 10.1074/mcp.M114.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, et al. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet. 2004;36:579–581. doi: 10.1038/ng1358. [DOI] [PubMed] [Google Scholar]
- 66.Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, et al. Polypeptide GalNAc-transferase T3 and Familial Tumoral Calcinosis. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- 67.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 68.Rhim AD, Stoykova L, Glick MC, Scanlin TF. Terminal glycosylation in cystic fibrosis (CF): A review emphasizing the airway epithelial cell. Glycoconjugate J. 2001;18:649–659. doi: 10.1023/a:1020815205022. [DOI] [PubMed] [Google Scholar]
- 69.Rhodes JM. Unifying hypothesis for inflammatory bowel disease and associated colon cancer: Sticking the pieces together with sugar. Lancet. 1996;347:40–44. doi: 10.1016/s0140-6736(96)91563-9. [DOI] [PubMed] [Google Scholar]
- 70.Holmen JM, Karlsson NG, Abdullah LH, Randell SH, Sheehan JK, Hansson GC, et al. Mucins and their O-Glycans from Human Bronchial Epithelial Cell Cultures. Am J Physiol, Lung, Cell and Mol Physiol. 2004;287:L824–L834. doi: 10.1152/ajplung.00108.2004. [DOI] [PubMed] [Google Scholar]
- 71.Schulz BL, Sloane AJ, Robinson LJ, Sebastian LT, Glanville AR, Song YL, et al. Mucin glycosylation changes in cystic fibrosis lung disease are not manifest in submucosal gland secretions. Biochem J. 2005;387:911–919. doi: 10.1042/BJ20041641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roussel P, Lamblin G. The glycosylation of airway mucins in cystic fibrosis and its relationship with lung infection by Pseudomonas aeruginosa. Adv Exp Med Biol. 2003;535:17–32. doi: 10.1007/978-1-4615-0065-0_2. 17–32. [DOI] [PubMed] [Google Scholar]
- 73.Magalhaes A, Marcos-Pinto R, Nairn AV, dela Rosa M, Ferreira RM, Junqueira-Neto S, et al. Helicobacter pylori chronic infection and mucosal inflammation switches the human gastric glycosylation pathways. Biochim Biophys Acta. 2015;1852:1928–1939. doi: 10.1016/j.bbadis.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holmén Larsson JM, Karlsson H, Gråberg Crespo J, Johansson MEV, Eklund L, Sjövall H, et al. An Altered O-glycosylation Profile of the MUC2 Mucin Occurs in Active Ulcerative Colitis and is Associated with Increased Inflammation. Infl Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 75.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 76.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci U S A. 2002;99:16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]