Abstract
Neutrophils infiltrate tissues during inflammation, and when activated, they release β-glucuronidase. Since inflammation is associated with carcinogenesis, we investigated how extracellular β-glucuronidase changed the in vitro cellular response to the chemical carcinogen benzo(a)pyrene (B[a]P). For this we exposed human liver (HepG2) and lung (A549) cells to B[a]P in the presence or absence of β-glucuronidase. β-Glucuronidase reduced B[a]P-induced expression of CYP1A1 and CYP1B1 at 6 h after exposure, which did not depend on β-glucuronidase activity, because the inhibitor d-saccharic acid 1,4-lactone monohydrate did not antagonize the effect of β-glucuronidase. On the other hand, the inhibitory effect of β-glucuronidase on CYP expression was dependent on signalling via the insulin-like growth factor receptor (IGF2R, a known receptor for β-glucuronidase), because co-incubation with the IGF2R inhibitor mannose-6-phosphate completely abolished the effect of β-glucuronidase. Extracellular β-glucuronidase also reduced the formation of several B[a]P metabolites and B[a]P–DNA adducts. Interestingly, at 24 h of exposure, β-glucuronidase significantly enhanced CYP expression, probably because β-glucuronidase de-glucuronidated B[a]P metabolites, which continued to trigger the aryl hydrocarbon receptor (Ah receptor) and induced expression of CYP1A1 (in both cell lines) and CYP1B1 (in A549 only). Consequently, significantly higher concentrations of B[a]P metabolites and DNA adducts were found in β-glucuronidase-treated cells at 24 h. DNA adduct levels peaked at 48 h in cells that were exposed to B[a]P and treated with β-glucuronidase. Overall, these data show that β-glucuronidase alters the cellular response to B[a]P and ultimately enhances B[a]P-induced DNA adduct levels.
Keywords: Benzo[a]pyrene, Inflammation, β-Glucuronidase, Cytochrome P450 1A1, Carcinogen metabolism, IGF2R, DNA adducts
Introduction
Chronic inflammation is causally associated with cancer development, and therefore, inflammation was considered as the seventh hallmark feature of cancer (Colotta et al. 2009; Shacter and Weitzman 2002). This is illustrated for instance by the relatively high incidence of lung cancer in chronic obstructive pulmonary disease (COPD) patients (Young et al. 2009). Recent studies have pointed out that polymorphonuclear neutrophils (PMN), which are recruited at the site of inflammation play an important role in the initiation and progression of cancer (Fridlender and Albelda 2012; Knaapen et al. 2006). Inhalation of complex air pollutants like tobacco smoke and/or fine particles may result in pulmonary inflammation, which generates reactive oxygen/nitrogen species (ROS/RNS) that can damage lung tissue (van Berlo et al. 2010). At the same time, it has been shown that under inflammatory conditions, PMN enhance the mutagenic potential of chemical carcinogens (Borm et al. 1997; Van Schooten et al. 2004). This is partly explained by the action of the PMN-derived enzyme myeloperoxidase (MPO) that can metabolically activate carcinogens and inhibit DNA repair, leading to higher levels of carcinogen–DNA adducts (Gungor et al. 2010a, b, c). However, there is still little data available on other factors that are released by the relatively high number of PMN during chronic inflammation, including β-glucuronidase, and how these influence the cellular response to chemical carcinogens (Basinska and Florianczyk 2003).
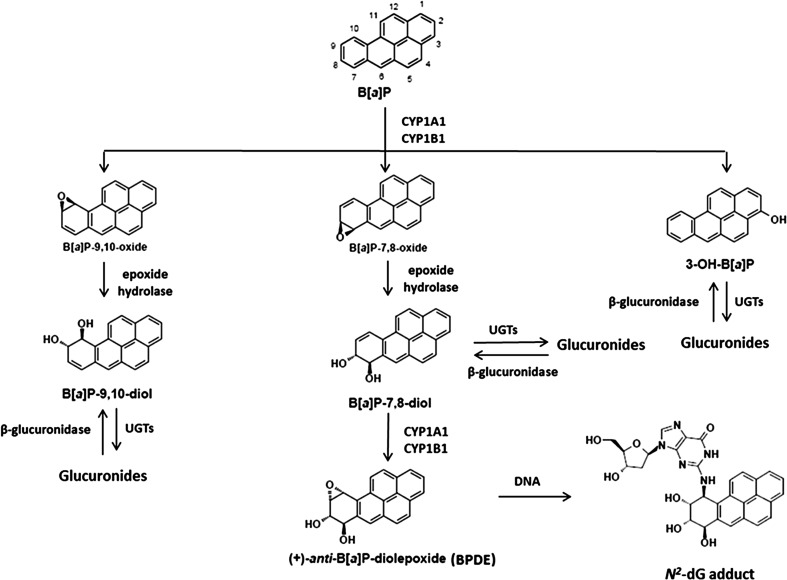
Inhalatory exposure to air pollutants like particles often results in a combination of an inflammatory response in the presence of genotoxic agents, like polycyclic aromatic hydrocarbons (PAHs) (Hoffmann and Hoffmann 1997). PAHs, including benzo[a]pyrene (B[a]P), have gained much attention, because they are abundantly present in the environment (Uppstad et al. 2010), and they have serious adverse genotoxic effects. B[a]P becomes mutagenic and carcinogenic after bioactivation by enzymes, including cytochrome P450 (CYP) and epoxide hydrolase (EH) (Stiborova et al. 2014). It is converted into various metabolites, including oxides, phenols, diols, diol-epoxides, quinones, and radical cations (Shimada and Guengerich 2006). The best studied metabolite is B[a]P-diol epoxide (BPDE) (Fig. 1). The first steps is the conversion of B[a]P by the microsomal NADPH-dependent cytochrome P450 isoforms 1A1 (CYP1A1) and 1B1 (CYP1B1) to yield the B[a]P-7,8-oxide, B[a]P-9,10-oxide or 3-hydroxy-B[a]P (3-OH-B[a]P) (Krais et al. 2015; Wohak et al. 2014). Subsequently, both B[a]P-7,8-oxide and B[a]P-9,10-oxide can be hydrated by microsomal EH to yield the corresponding B[a]P-7,8-trans-dihydrodiols (B[a]P-7,8-diol) and B[a]P-9,10-trans-dihydrodiols (B[a]P-9,10-diol). B[a]P-7,8-diol is further metabolized to the ultimate carcinogen benzo[a]pyrene-7,8-diol-9,10-epoxide (BPDE), which is known as a reactive derivative of B[a]P that can covalently bind to DNA to form adducts preferentially at guanine residues (e.g. 10-(deoxyguanosin-N 2-yI)-7,8,9-trihydroxy-7,8,9,10-tetrahydro benzo[a]pyrene; dG-N 2-BPDE) (Arlt et al. 2008; Kim et al. 1998; Wohak et al. 2014). It is generally accepted that the formation of such DNA adducts leads to mutations relevant for carcinogenesis (Kucab et al. 2015). In addition, the reactive metabolites can be conjugated by an important phase II detoxification enzyme UDP-glucuronosyltransferases (UGTs) that leads to glucuronides which are not mutagenic or carcinogenic (Shimoi and Nakayama 2005; Shimoi et al. 2001). Indeed, UGT1A6 was found to glucuronate a range of B[a]P metabolites including 3-OH-B[a]P, B[a]P-9,10-diol and the pro-carcinogen B[a]P-7,8-diol (Jin et al. 1993; Trushin et al. 2012; Zheng et al. 2002).
Fig. 1.
Critical steps of B[a]P activation and UDP-glucuronosyltransferases (UGTs) detoxification. B[a]P is metabolized to hydroxylated B[a]P including B[a]P-7,8-diol and these B[a]P metabolites are further detoxified by UGTs. β-Glucuronidases are able to hydrolyse glucuronidated B[a]P metabolites and therefore increase the amount of B[a]P-7,8-diol, consequently leading to more BPDE and DNA adducts formation
In order to elucidate the role of β-glucuronidase in inflammatory disease and its effect on chemically induced cancer, it is crucial to understand how β-glucuronidase changes the cellular response towards B[a]P. In a recent study, it was found that lipopolysaccharide (LPS) treatment increased B[a]P-induced DNA adduct levels in lung and liver tissues of B[a]P inhalatory-exposed mice (Arlt et al. 2015). To explain this result, we hypothesise that the LPS induced an inflammatory response resulting in the release of β-glucuronidase that hydrolysed glucuronidated B[a]P metabolites and thus reversing the protective effect of glucuronidation, and subsequently enhanced the binding of B[a]P metabolites to DNA. To test our hypothesis, we studied the impact of extracellular β-glucuronidase on the formation of B[a]P metabolites (3-OH-B[a]P, B[a]P-9,10-diol and B[a]P-7,8-diol), gene expression of some enzymes pivotal in B[a]P metabolism (including CYP1A1, CYP1B1 and UGT1A6), and DNA adduct formation in human liver cell line (HepG2) and human lung cell line (A549).
Materials and methods
Mouse lung and liver tissues
All procedures performed in the study involving animal experiments were conducted at King’s College London under license in accordance with the Institutional Ethics Committee on the protocols approved by the Home Office under “The Animals (Scientific Procedures) Act (1986).” Mice were divided into four groups (n = 3 in each group): (a) control group; mice were nasally instilled with saline. After 24 h, mice were intratracheally instilled with tricaprylin. (b) The lipopolysaccharide (LPS) treatment group; each mouse was nasally instilled with 20 μg LPS (dissolved in saline). After 24 h, mice were intratracheally instilled with tricaprylin. (c) The B[a]P-treated group; mice were nasally instilled with saline and intratracheally instilled with 0.5 mg B[a]P (dissolved in 25 μl tricaprylin) after 24 h. (d) The B[a]P- and LPS-treated group; each mouse was nasally instilled with 20 μg of LPS. After 24 h, mice were intratracheally instilled with 0.5 mg B[a]P. All mice were sacrificed at 48 h after the intratracheal exposure. The collection of bronchoalveolar lavage fluid (BAL fluid) and the isolation of cytosolic fractions from lung and liver were performed as described in Arlt et al. (2015).
Cell lines and cell treatment
Human liver hepatocellular carcinoma HepG2 cells and human epithelial lung adenoma carcinoma A549 cells were obtained from the American Tissue Culture Collection. HepG2 cells were cultured in minimum essential medium (MEM) plus glutamax containing 10 % (v/v) foetal calf serum (FCS, Gibco invitrogen, Breda, the Netherlands), 1 % (v/v) sodium pyruvate, 1 % (v/v) penicillin/streptomycin (Sigma, Zwijndrecht, the Netherlands), and 1 % (v/v) non-essential amino acids (Sigma). A549 cells were cultured in RPMI (Sigma) supplemented with 5 % (v/v) FCS and 1 % (v/v) penicillin/streptomycin. All cells were cultured under humidified atmosphere containing 5 % CO2 at 37 °C. Cell passages between 20 and 29 were used for experiments. All chemicals were purchased from Sigma-Aldrich unless stated otherwise.
Cells with 80 % confluency were exposed to 1 μM B[a]P in the presence or absence of β-glucuronidase (4 U/ml) for 6, 24 or 48 h. B[a]P was dissolved in dimethylsulphoxide (DMSO) and added to the medium with a final DMSO concentration of 0.5 % (v/v). β-Glucuronidase was dissolved in 0.1 M sodium acetate buffer (pH 5.5). Mannose-6-phosphate (M6P) was dissolved in 0.1 M sodium acetate buffer (pH 5.5) and the final concentration in cell culture is 100 μM. d-Saccharic acid 1,4-lactone monohydrate was dissolved in 0.1 M sodium acetate buffer (pH 5.5), and the final concentration in cell culture is 100 μM. Before incubation, all cell media were adjusted to pH 5.5 by using 1 M HCl in order to mimic the microenvironment of inflammation. After incubation, the medium and cells were stored at −20 °C until further analysis. Experiments were performed at least with three replicates in three independent cultures.
Measurement of β-glucuronidase activity
β-Glucuronidase (Helix pomatia-type H5, ≥400,000 Units/g solid) activity was determined by fluometrically monitoring the hydrolysis of 4-methylumbelliferyl-β-d-glucuronide (4MUgIA) according to the method described by (Bartholome et al. 2010) with some modifications. Briefly, the reaction mixture in a total volume of 140 μl contained 0.1 M sodium acetate buffer (pH 5.5), 2 mM 4MUgIA, and sample containing β-glucuronidase. The reaction was initiated by mixing 4MUgIA and the sample. The hydrolysis of 4MUgIA was measured in a thermostated plate reader (Spectra max m2, MDS, CA) at 37 °C and 320-/460-nm excitation/emission wavelengths. A standard curve of β-glucuronidase [0.4–400 Units/ml (U/ml)] was generated to quantitate formation of fluorescence in the presence of 4MUgIA. d-Saccharic acid 1,4-lactone monohydrate, a β-glucuronidase inhibitor, was used to inhibit β-glucuronidase activity.
For testing the interaction between B[a]P and β-glucuronidase, we used similar condition as mentioned above. A total volume of 140 μl contained 0.1 M sodium acetate buffer (pH 5.5), different concentrations of 4MUgIA (e.g. 500, 250, 100, 50, 10 and 1 μM), and 4 U/ml β-glucuronidase with additional 1 μl of 200 μM B[a]P or 1 μl DMSO. The measurement of fluorescence was performed for 10 h at 37 °C.
HPLC fluorescence analysis of B[a]P and B[a]P metabolites
B[a]P and its metabolites were extracted from 5 ml cell medium by mixing with 1 ml ethylacetate for 20 min and followed by centrifugation (10 min, 980g). The top layer was transferred to a new tube. This procedure was repeated twice. The top layers were evaporated under nitrogen, and the residue was redissolved in 0.5 ml methanol (Biosolve Chemicals, Valkenswaard, the Netherlands). Samples were subsequently analysed by HPLC-FD using a Gynkotek P580A HPLC system (Separations Analytical Instruments, Hendrik-Ido-Ambacht, the Netherlands) consisting of a Spark SP830 autosampler (Spark Holland, Emmen, The Netherlands) and a Perkin Elmer LS-30 programmable fluorescence detector (Perkin Elmer, Foster City, CA, USA) operated at excitation/emission wavelengths 257/>350 nm. The samples were injected onto a Hypersil 5-μm ODS HPLC column (250 mm × 3 mm) (Supelco 54933, Bellefonte, PA, USA) with a flow rate of 0.5 ml/min. Separation was performed using a mixture of two mobile phases: A (100 % methanol) and B (40 % methanol in water) in the following multi-step gradient conditions: 0–5 min, 30/70 (A/B, v/v); 5–30 min, gradient from 30/70 (A/B, v/v) to 90/10 (A/B, v/v); 30–35 min, 90/10 (A/B, v/v); 35–37 min, gradient from 90/10 (A/B, v/v) to 30/70 (A/B, v/v); and 37–40 min, 30/70 (A/B, v/v). For quantitation of the specific metabolites, a standard mix which contained 50 ng/ml B[a]P-9,10-diol 50 ng/ml B[a]P-7,8-diol and 50 ng/ml 3-OH-B[a]P (Midwest Research Institute, Kansas City, MO, USA) were injected, and the area of each metabolite peak in the chromatogram was determined.
Quantitative real-time PCR
Gene expression levels were measured by quantitative real-time reverse transcriptase PCR (RT-qPCR) using a MyiQ Single Colour real-time PCR detection system (BioRad, Veenendaal, The Netherlands). Total RNA was isolated and purified by using the RNeasy® Mini Kit (Qiagen Westburg, Leusden, The Netherlands) in combination with DNase treatment (Qiagen). cDNA was generated from 500 ng total RNA by using the iScript™ cDNA synthesis kit protocol (BioRad). Primers were purchased from Operon (Leiden, The Netherlands) for the following genes: β-actin, CYP1A1, CYP1B1 and UGT1A6 [see (Schults et al. 2014)]. The reaction contained SYBR© Green Supermix (Bio-Ras), 5 μl (40 times diluted) cDNA and 0.3 μM primers in a total volume of 25 μl. PCR was conducted under the following condition: denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 55 °C for 45 s. All PCR reactions included a cDNA dilution curve to assess PCR efficiency, and all reactions were followed by a melt curve (55–95 °C). Data were analysed by using MyiQ Software system (BioRad), and the amount of target cDNA in each sample was determined by a fractional PCR threshold cycle number (Ct value) and compared to the corresponding Ct value for the housekeeping gene β-actin. The relative gene expression level for each gene was calculated by using the 2−ΔΔCt method (Livak and Schmittgen 2001).
32P-Postlabelling of B[a]P–DNA adducts
DNA harvested from cells was isolated using a phenol–chloroform–isoamylalcohol extraction procedure as described by Schults et al. (2013). Briefly, after incubation, cells were resuspended in 450 μl lysis buffer [10 mM Tris, 10 mM TEMPO, 1 mM EDTA and 1 % (w/v) sodium dodecyl sulphate (SDS); pH 8] and incubated with proteinase K (10 μg/ml) at 37 °C overnight. The mixture was extracted with 1 volume Tris-saturated phenol, 1 volume Tris-saturated phenol–chloroform–isoamyl alcohol (25:24:1 by volume), and 1 volume chloroform–isoamyl alcohol (24:1, v/v). The DNA was precipitated with 1/30 volume 3 M NaAc pH 5.2 and 2 volumes of cold 100 % ethanol. Precipitated DNA was washed with 70 % ethanol and dried under nitrogen. The DNA was dissolved in 2 mM Tris (pH 8.0) with final concentration 0.5 μg/μl.
DNA digestion and 32P-postlabelling were performed as described by Van Schooten et al. (1997). In short, DNA samples (10 μg) were digested with micrococcal nuclease (Sigma) (0.25 U/μl) and spleen phosphodiesterase (Sigma) (2 μg/μl) for 4 h at 37 °C in a total volume of 9.5 μl. For DNA adduct enrichment, samples were treated with nuclease P1 (Sigma) (2.5 μg/μl) at 37 °C for 30 min. The nuclease P1 reaction was terminated by addition of 1 µl 1 M Tris (pH 9.6). DNA adducts were subsequently labelled with [ɣ-32P]ATP (50 μCi/sample; PerkinElmer, Indianapolis) using T4-polynucleotide kinase (10 U/μl) for 30 min at 37 °C. The 32P-labelled adducts were separated on PEI–cellulose sheets (Machery Nagel, Düren, Germany) by multi-directional thin-layer chromatography (TLC).
The TLC sheets were scanned using Phosphor-Imaging technology (Fujifilm FLA-3000) and DNA adducts levels were calculated from two B[a]PDE–DNA standards with known adducts levels (1 adduct/106 and 1 adduct/107 nucleotides). The major B[a]P–DNA adduct that was used for quantitation purposes in both HepG2 and A549 cells migrated to the same position as the major adduct of the BPDE-DNA adduct standard. In addition, the B[a]PDE-DNA adduct levels were corrected for the amount of DNA in the sample which was assessed by HPLC–UV analysis.
Statistical analysis
Data were expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed using Graphpad Prism 6. To examine differences between the different treatments at each time point, a two-way analysis of variance test (ANOVA) with Bonferroni post hoc multiple comparison was used. Differences were considered to be statistically significant if the p value was less than 0.05 (p < 0.05).
Results
Activity of β-glucuronidase in lung and liver tissues of mice
Intranasal exposure of the mice to LPS resulted in an approximately twofold induction of β-glucuronidase activity in lung tissue compared to control (Table 1). Similarly, when LPS treatment was combined with B[a]P treatment, a significant 1.5-fold higher activity of β-glucuronidase was observed compared with the B[a]P-treated group without LPS. In addition, bronchoalveolar lavage fluid (BAL fluid) was collected, and the β-glucuronidase activity in BAL fluid was lower than in the tissues, but LPS treatment did result in a significant increase in β-glucuronidase when compared to animals that were not treated with LPS (i.e. irrespective of B[a]P exposure). On the other hand, in liver tissue a 1.2-fold lower activity of β-glucuronidase was observed in LPS-treated animals when compared to control and B[a]P-treated mice, respectively. These changes in β-glucuronidase activity after LPS treatment were in the range of 2–30 U/ml β-glucuronidase, and therefore, 4 U/ml was used as biologically relevant dose in the subsequent cell culture experiments (see Table 1).
Table 1.
Activity of β-glucuronidase in mouse liver, lung tissues and BAL fluids
| Control (U/ml) | LPS (U/ml) | B[a]P (U/ml) | B[a]P and LPS (U/ml) | |
|---|---|---|---|---|
| Liver cytosol | 27.1 ± 1.5 | 23.3 ± 0.4* | 27.6 ± 0.9 | 22.5 ± 1.0** |
| Lung cytosol | 7.6 ± 0.8 | 15.4 ± 1.3* | 7.4 ± 0.8 | 11.1 ± 1.3** |
| BAL fluids | 2 ± 0.2 | 2.2 ± 0.1# | 2.0 ± 0.1 | 2.2 ± 0.1# |
* Significantly different from control animals (p < 0.05)
** Significantly different from B[a]P-treated animals (p < 0.05)
# p < 0.05 if both LPS-treated groups were combined when compared to non-LPS-treated animals
Phenotypes after B[a]P exposure with or without β-glucuronidase
Expression of CYP1A1 and CYP1B1
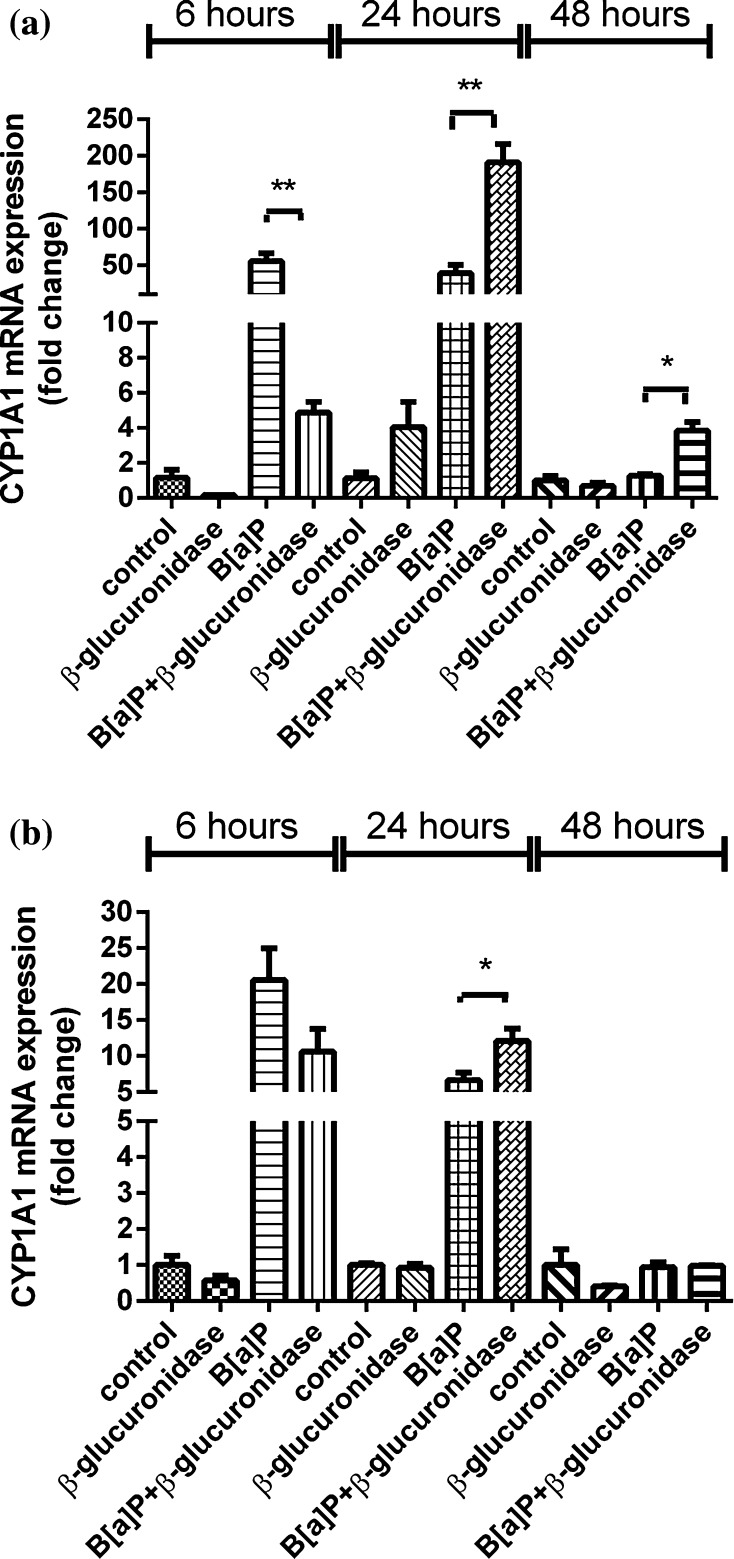
Exposure to B[a]P significantly induced the expression of CYP1A1 and CYP1B1 (Figs. 2a, 3a). In A549 cells, the expression of these genes at 6 h after exposure was increased 56-fold and fivefold, respectively. Surprisingly, co-incubation with β-glucuronidase inhibited the induction of expression with approximately 50–90 %. Expression of CYP1A1 and CYP1B1 in cells that were treated with β-glucuronidase without additional exposure to B[a]P was also reduced when compared to the expression observed in control cells, but this difference did not reach statistical significance. At 24 h, the expression of CYP1A1 and CYP1B1 was induced by B[a]P to 39-fold and threefold, respectively. Surprisingly, the induction of both genes was now strongly increased by the presence of β-glucuronidase (191-fold and sixfold, respectively). At t = 48 h, the induction of CYP1A1 and CYP1B1 in cells that were co-exposed to β-glucuronidase remained higher than in cells that were only exposed to B[a]P. The same pattern of changes in gene expression of CYP1A1 and CYP1B1 by β-glucuronidase was observed in the absence of B[a]P, although less pronounced. Moreover, changes in gene expression of CYP1A1 and CYP1B1 were essentially similar in HepG2 cells (Figs. 2b, 3b), but the fold changes that were reached were lower than in A549 cells.
Fig. 2.
RT-qPCR analysis of gene expression CYP1A1 in both A549 (a) and HepG2 (b) cells after exposure to β-glucuronidase and/or B[a]P. Cells were exposed to 1 μM B[a]P with or without 4 U/ml β-glucuronidase and harvested after the times indicated. Cells exposed to DMSO and sodium acetate buffer were used as a vehicle control. All values are given as the mean ± SEM (n = 4 per data point). (*p < 0.05; **p < 0.01; ***p < 0.001)
Fig. 3.
RT-qPCR analysis of gene expression CYP1B in both A549 (a) and HepG2 cells (b) after exposure to β-glucuronidase and/or B[a]P. Cells were exposed to 1 μM B[a]P with or without 4 U/ml β-glucuronidase and harvested after the times indicated. Cells exposed to DMSO and sodium acetate buffer were used as a vehicle control. All values are given as the mean ± SEM (n = 4 per data point). (*p < 0.05; **p < 0.01; ***p < 0.001)
Expression of UGT1A6
As shown in Fig. 4b, in HepG2 cells the expression of UGT1A6 was threefold and 11-fold induced by B[a]P when compared to unexposed cells at 6 and 24 h after exposure, respectively. Addition of β-glucuronidase inhibited UGT1A6 expression after 6 h when compared to cells treated with B[a]P alone. However, a significant increase in UGT1A6 expression was observed 24 h after exposure (61-fold and sixfold compared to unexposed cells and only B[a]P-treated cells, respectively). At 48 h, the expression of UGT1A6 remained enhanced by B[a]P and B[a]P with additional β-glucuronidase, but the induction levels were three times lower (ninefold and 28-fold, respectively). In addition, a similar pattern of changes in gene expression of UGT1A6 were shown by β-glucuronidase in the absence of B[a]P.
Fig. 4.
RT-qPCR analysis of gene expression UGT1A6 in A549 (a) and HepG2 (b) cells after exposure to β-glucuronidase and/or B[a]P. Cells were exposed to 1 μM B[a]P with or without 4 U/ml β-glucuronidase and harvested after the times indicated. Cells exposed to DMSO and sodium acetate buffer were used as a vehicle control. All values are given as the mean ± SEM (n = 4 per data point). (*p < 0.05; **p < 0.01; ***p < 0.001)
On the other hand, in A549 cells, there was no significant induction or inhibition of UGT1A6 expression by B[a]P nor by β-glucuronidase, but at t = 48 h, all treatments induced the expression of UGT1A6 compared to unexposed cells (see Fig. 4a).
B[a]P and its metabolites
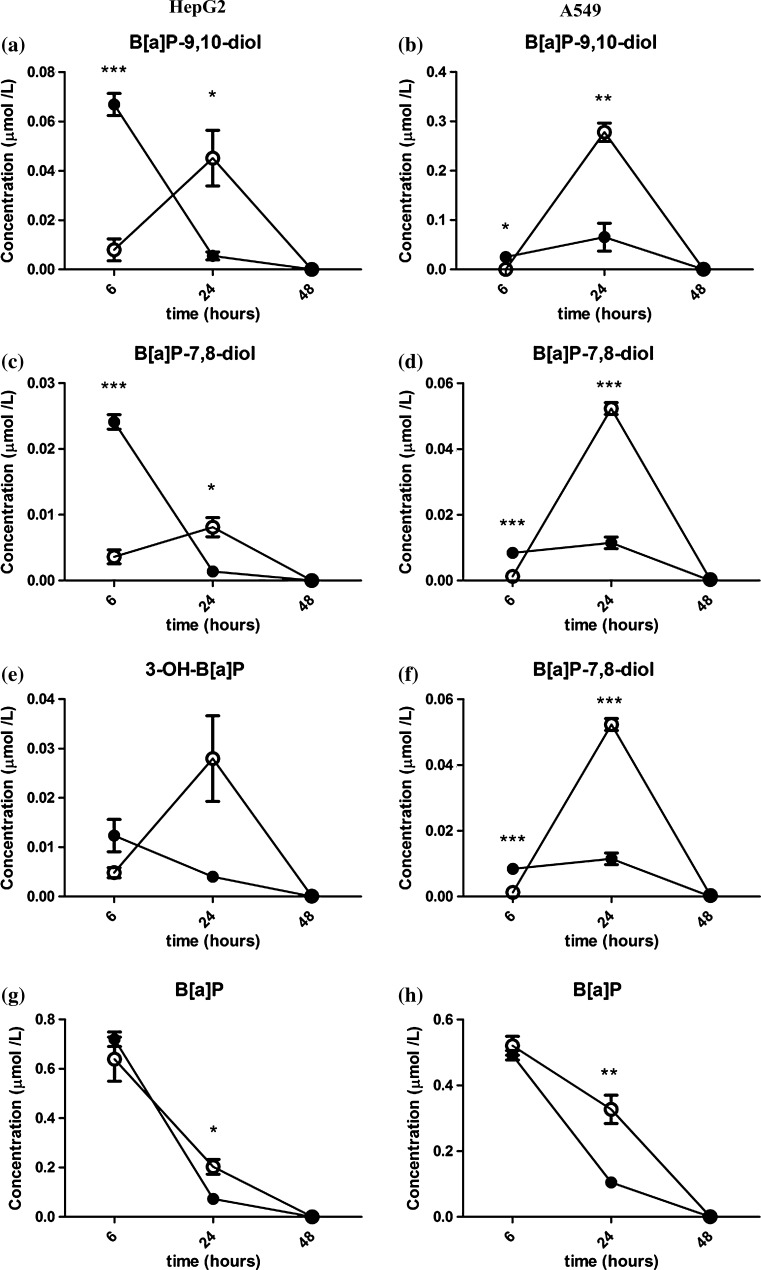
Extracellular B[a]P metabolite (B[a]P-7,8-diol and B[a]P-9,10-diol) concentrations of A549 cells that were exposed to 1 μM B[a]P with β-glucuronidase (10 µg/ml) for 6 h significantly decreased, when compared to A549 cells that were exposed to B[a]P only (Fig. 5, right column). However, when cells were exposed for 24 h, the concentrations of extracellular B[a]P metabolites in the medium of β-glucuronidase-treated cells significantly increased and were higher than in A549 cells that were exposed to B[a]P alone. At 48 h, the extracellular B[a]P metabolite levels were below the detection limit in both treatments (presence or absence of β-glucuronidase).
Fig. 5.
HPLC fluorescence analysis of B[a]P-7,8-diol, B[a]P-9,10-diol, 3-OH-B[a]P and B[a]P in HepG2 cells (left column) and A549 cells (right column) after exposure to β-glucuronidase and/or B[a]P. Cells were exposed to 1 μM B[a]P with or without 4 U/ml β-glucuronidase and cell medium was harvested after the time indicated. Cells exposed to DMSO and sodium acetate buffer were used as a vehicle control. All values are given as the mean ± SEM (n = 3 per data point). Filled circle without β-glucuronidase, open circle with β-glucuronidase
The results were essentially similar for HepG2 cells (Fig. 5, left column). However, the initial difference at t = 6 h was more pronounced, and statistically significant for B[a]P-7,8-diol and B[a]P-9,10-diol.
Moreover, the concentration of unmetabolized B[a]P in the medium showed similar time-dependent patterns in both cell lines (Fig. 5g, h). The concentration of unmetabolized B[a]P gradually declined with time. However, after 24 h of exposure, the concentration of the parent compound in A549 cells was about threefold higher (p < 0.05) in the samples that included β-glucuronidase than in cells without β-glucuronidase. For HepG2 cells, the concentration of B[a]P in the presence of β-glucuronidase was approximately twofold higher than in samples that were treated with B[a]P alone. For both cell lines, B[a]P was almost fully metabolized 48 h after exposure.
B[a]P–DNA adducts level
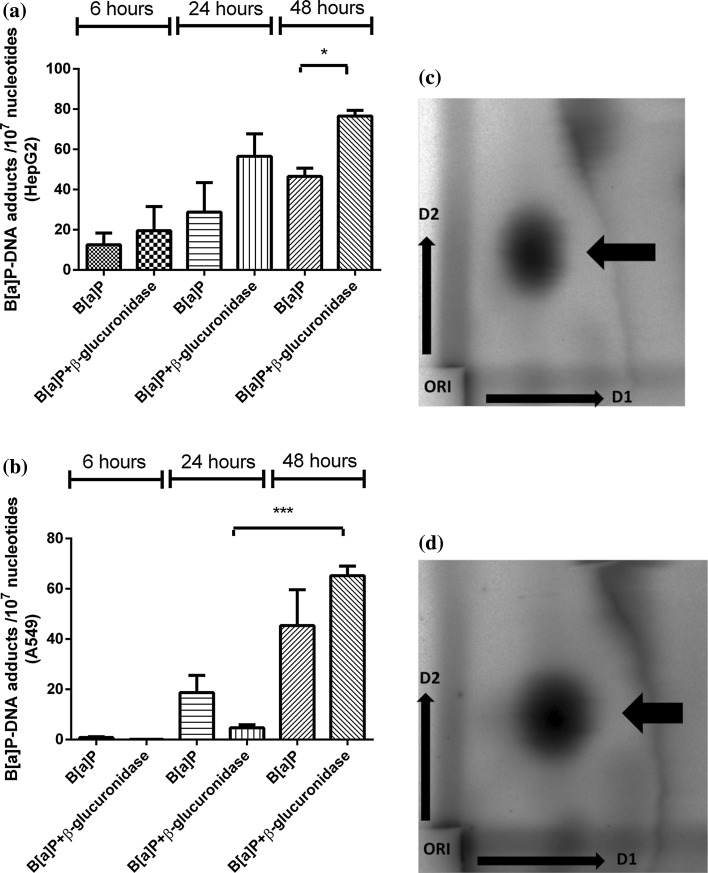
B[a]P exposure resulted in a time-dependent increase in B[a]P–DNA adduct levels in both cell lines (Fig. 6). However, the presence of β-glucuronidase altered the kinetics in which DNA adducts were formed: in A549 cells, at t = 6 and t = 24 h, DNA adduct levels were initially lower in cells that were treated with B[a]P and β-glucuronidase. However, a strong increase in DNA adduct levels from 5 adducts per 107 nucleotides at t = 24 h to 65 adducts per 107 nucleotides at 48 h (p < 0.0001), was found in β-glucuronidase-treated A549. This was not the case in the samples with B[a]P only. Consequently, at t = 48 h, B[a]P–DNA adduct levels were 1.4-fold higher in cells that were treated with β-glucuronidase compared to treatment with B[a]P only. In HepG2 cells, the presence of β-glucuronidase resulted in 1.5-fold, twofold, and 1.6-fold higher levels of B[a]P–DNA adducts at 6, 24, and 48 h, respectively, compared to the samples that were treated with B[a]P only (p < 0.05 at 48 h).
Fig. 6.
32P-Postlabelling analysis of DNA adducts level in HepG2 cells and A549 cells after exposure to B[a]P with or without β-glucuronidase. a HepG2 cells were exposed to 1 μM B[a]P with or without 10 μg/ml β-glucuronidase and harvest after the times indicated. b A549 cells were exposed to 1 μM B[a]P with or without 4 U/ml β-glucuronidase and harvest after the times indicated. Data are expressed as number of B[a]P–DNA adducts per 107 nucleotides (n = 5 for HepG2 cells and n = 4 for A549 cells, mean ± SEM) (*p < 0.05; ***p < 0.001). Representative chromatograms obtained by 32P-postlabelling in HepG2 cells (c) and A549 cells (d). The adduct spot (arrow) that migrated during 2D-TLC to the same position as the major DNA adduct in a BPDE-DNA adduct standard was quantitated in all samples. Before phosphorimaging of the TLC plates the origin located at the bottom left-hand corner was excised
Potential mechanisms
Can β-glucuronidase interact with B[a]P to prevent B[a]P from entering the cells?
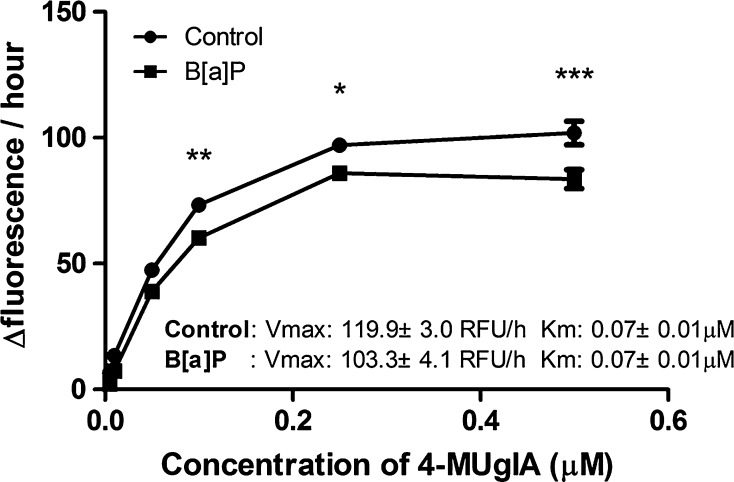
Since B[a]P metabolism seems to be delayed, we studied whether B[a]P could temporarily bind to β-glucuronidase, which could prevent B[a]P from entering the cell. We assumed that binding of B[a]P to β-glucuronidase would interfere with β-glucuronidase activity. Therefore, we assessed the β-glucuronidase hydrolysis of 4MUgIA in the presence B[a]P (Fig. 7). At 37 °C for 10 h, 10 μg/ml of β-glucuronidase and 1 μM B[a]P were mixed with different concentration of substrate (4MUgIA). The addition of B[a]P lowered the changes in fluorescence units per hours compared with control. With increasing concentrations of 4MUgIA, the difference of the Δfluorescence/hour between these two groups became larger. This difference was significant at 100 μM (p < 0.01), 250 μM (p < 0.05) and 500 μM (p < 0.001) of 4MUgIA. The largest difference was observed at the highest concentration of 4MUgIA and was approximately 18 % lower than control. In addition, we performed Michaelis–Menten equation to determine the V max and K m for both reactions. Although the K m in both reactions is same (0.07 ± 0.01 µM and 0.07 ± 0.01 µM for control and B[a]P-treated, respectively), the V max in control is significantly larger than in the B[a]P-treated sample (p < 0.05). Therefore, we concluded that there is a non-competitive inhibition reaction.
Fig. 7.
4MUgIA assay was applied to assess the possibility of B[a]P to bind to β-glucuronidase. A total volume of 140 μl contained 0.1 M sodium acetate buffer (pH 5.5), different concentration of 4MUgIA (e.g. 500, 250, 100, 50, 10 and 1 μM), and 4 U/ml β-glucuronidase with 1 μl of 200 μM B[a]P or 1 μl DMSO. The measurement of fluorescence [Relative Fluorescence Unit (RFU)] was performed for 10 h at 37 °C. B[a]P-treated samples were compared with control at each concentration, respectively. (*p < 0.05; **p < 0.01; ***p < 0.001)
Is the inhibitory effect of β-glucuronidase on CYP1A1 expression dependent on its activity?
In order to gain further insight into the role of β-glucuronidase activity in influencing CYP1A1 expression, an β-glucuronidase inhibitor (d-saccharic acid 1,4-lactone monohydrate) was added to the B[a]P and β-glucuronidase incubations in A549 cells (Fig. 8). A concentration of 100 μM fully inhibited β-glucuronidase activity (Fig. 8a). As shown in Fig. 8b, the presence of this inhibitor in incubations with B[a]P and β-glucuronidase did not change the inhibitory effect of β-glucuronidase on CYP1A1 expression (0.13 ± 0.03 and 0.08 ± 0.01 in the absence or presence of inhibitor, respectively) at t = 6 h. On the contrary, at t = 24 h, the presence of this inhibitor significantly lowered CYP1A1 expression when compared to cells that were exposed to B[a]P and β-glucuronidase without inhibitor (p < 0.01).
Fig. 8.
a 4MUgIA assay was applied to assess the inhibition of β-glucuronidase by d-saccharic acid 1,4-lactone monohydrate. A total volume of 140 μl contained 0.1 M sodium acetate buffer (pH 5.5), 2 mM 4MUgIA, 4 U/ml β-glucuronidase and different concentration of d-saccharic acid 1,4-lactone monohydrate (10 μM, 100 and 500 μM). 100 μM of d-saccharic acid 1,4-lactone monohydrate was used in the following incubation. b A549 cells were exposed to 1 μM B[a]P with or without β-glucuronidase, d-saccharic acid 1,4-lactone monohydrate for 6 and 24 h. Cells exposed to 1 μM B[a]P and sodium acetate buffer was used as a control. All values are given as the mean ± SEM (n = 4 per data point) (*p < 0.05; **p < 0.01; ***p < 0.001)
Involvement of the insulin-like growth factor 2/mannose-6-phosphate pathway
Extracellular enzymes like β-glucuronidase are known to bind to the mannose-6-phosphate (M6P) receptor (Gonzalez-Noriega et al. 2001), which is also known as IGF2 receptor. The IGF2 receptor can be inhibited by high concentrations of M6P (El-Shewy and Luttrell 2009). β-Glucuronidase inhibited the induction of CYP1A1 expression by B[a]P at 6 h of incubation to approximately 10 % (Fig. 9), whereas in the presence of M6P, β-glucuronidase was unable to inhibit CYP1A1 expression at 6 h. As shown previously, β-glucuronidase significantly induced CYP1A1 expression at 24 h when co-incubated with B[a]P (p < 0.01). CYP1A1 expression was still enhanced with additional M6P at 24 h, but the changes in expression were less pronounced.
Fig. 9.
RT-qPCR analysis of gene expression CYP1A1 in A549 cells after exposure to β-glucuronidase and B[a]P with the IGF2R inhibitor M6P. Cells were exposed to 1 μM B[a]P with or without 4 U/ml β-glucuronidase and with or without 100 μM M6P. Cells were harvested after the times indicated. Cells exposed to B[a]P and sodium acetate buffer were used as vehicle control. All values are given as the mean ± SEM (n = 4 per data point) (*p < 0.05; **p < 0.01; ***p < 0.001)
Discussion
It remains to be established how the cellular response to B[a]P is affected by the presence of extracellular β-glucuronidase, which is released during inflammation. In this study, we demonstrated that β-glucuronidase initially (6 h after exposure) inhibited gene expression of enzymes that are pivotal in B[a]P metabolism, including CYP1A1, CYP1B1, and UGT1A6. As a result, subsequent formation of B[a]P metabolites and the formation of B[a]P–DNA adducts was delayed in the presence of β-glucuronidase. However, at 24 h of exposure, CYP expression was significantly enhanced in β-glucuronidase-treated cells, probably because more B[a]P remained unmetabolized and β-glucuronidase converted B[a]P-derived glucuronide metabolites into active B[a]P metabolites. The higher concentration of active B[a]P metabolites continued to trigger the Ah receptor for gene expression of CYP1A1 (in both cell lines) and CYP1B1 (in A549 only). Consequently, the formation of active B[a]P metabolites and DNA adducts at t = 24 h was further increased in β-glucuronidase-treated cells. Because of this delayed metabolism of B[a]P, DNA adduct levels could accumulate in cells that were treated with β-glucuronidase and peaked at 48 h after the initial exposure.
A recent study observed that increased B[a]P–DNA adduct levels in mice that were exposed to B[a]P and intranasally instilled with LPS (Arlt et al. 2015). As LPS can induce an inflammatory response and stimulate the release of β-glucuronidase from neutrophils (Basinska and Florianczyk 2003; Ngkelo et al. 2012), we determined the β-glucuronidase activity in lung and liver tissues from these mice (Table 1). Indeed, β-glucuronidase activity was significantly enhanced in lung and BAL fluid of all LPS-treated animals when compared to the control group. On the other hand, we found that the β-glucuronidase activity was significantly decreased in liver tissue 3 days after being intranasally instilled with LPS. LPS treatment results in the recruitment of neutrophils from the liver to the lung, which could explain the lower β-glucuronidase activity in the liver after LPS treatment (Reutershan et al. 2005). The significant increase in β-glucuronidase activity in the lung samples by LPS treatment was approximately 4 U/ml, which we also subsequently used in our in vitro experiments.
B[a]P is known to bind to the AhR which stimulates its own metabolism by inducing the expression of CYP1A1 and CYP1B1 (Spink et al. 2002). CYP1A1 and CYP1B1 play an important role in both B[a]P activation and detoxification (Moserova et al. 2009), and B[a]P metabolites are further detoxified by glucuronidation. β-Glucuronidase will hydrolyse glucuronidated B[a]P metabolites and therefore will increase the concentration of active B[a]P metabolites (Shimoi and Nakayama 2005). Higher concentrations of these B[a]P metabolites (e.g. B[a]P-7,8-diol and B[a]P-9,10-diol) enhance the expression of CYP1A1 and CYP1B1 (Almahmeed et al. 2004; Spink et al. 2008). Therefore, we expected that addition of β-glucuronidase would increase CYP1A1 and CYP1B1 expression. However, we found that β-glucuronidase inhibited CYP expression shortly after B[a]P exposure (i.e. 6 h) in both A549 and HepG2 cells. This initial inhibition of gene expression by β-glucuronidase was independent of β-glucuronidase activity, because the β-glucuronidase inhibitor d-saccharic acid 1,4-lactone monohydrate did not change the results. Several studies report the binding of β-glucuronidase to insulin-like growth factor 2 receptors (IGF2R), which are located in the cell membrane (Gonzalez-Noriega and Michalak 2001; Urayama et al. 2004; Vogler et al. 2005). Moreover, IGF2 induced AhR in MCF-7 cells (Tomblin and Salisbury 2014). Therefore, we studied the involvement of IGF2R signalling by adding its inhibitor mannose-6-phosphate (M6P), and we found that the inhibitory effect of β-glucuronidase on CYP expression was also blocked. Therefore, we suggest a connection between IGF2R as receptor of extracellular β-glucuronidase and intracellular AhR-signalling after B[a]P exposure.
In addition, UGT1A6 can be induced by AhR ligands in order to detoxify reactive B[a]P derivatives (Jin et al. 1993). UGT1A6 is an important enzyme for the detoxification of B[a]P metabolites and is predominantly located in human liver nuclear membranes (Radominska-Pandya et al. 2002; Zheng et al. 2002). This could explain the different kinetics of expression of UGT1A6 between the two cell lines in this study. Hence, our mRNA expression data of UGT1A6 exhibited a similar pattern as CYP1A1 in HepG2 cells, but not in A549 cells. It is known that lung cells have a lower expression of UGT’s than liver cells (Ohno and Nakajin 2009), and therefore, it was expected that the effects of β-glucuronidase on the metabolism of B[a]P would be more pronounced in the liver-derived HepG2 cells than in the lung-derived A549 cells. Indeed, B[a]P–DNA adducts were already higher in β-glucuronidase-treated HepG2 cells at t = 24 h, whereas in A549 cells an additional 24 h was needed for further accumulation of DNA adducts. At the 24-h time point, the effects of β-glucuronidase on CYP expression were at least partly dependent on β-glucuronidase activity. Thus, we suggest that de-glucuronidated metabolites of B[a]P in combination with the higher concentrations of unmetabolized B[a]P, continued to trigger the Ah receptor and the subsequent expression of CYP1A1 and CYP1B1. Therefore, our results indicate that there are two possible underlying mechanisms resulting in altered B[a]P metabolism and subsequent DNA adduct formation; (1) changes in gene expression by IGF2 signalling and (2) de-glucuronidation of glucuronidated metabolites.
Moreover, we studied whether B[a]P could temporarily bind to β-glucuronidase which would postpone B[a]P from entering the cell. Indeed, the presence of B[a]P decreased the capacity of β-glucuronidase to deconjugate 4MUgIA, suggesting an interaction between β-glucuronidase and B[a]P. However, this effect is unlikely to explain the present data, because the changes in β-glucuronidase activity by B[a]P are relatively small (<20 %), and in our experiments B[a]P is added in excess. However, this interaction may become more relevant at sites of inflammation and low B[a]P exposure.
Since CYP1A1 is considered to be the major enzyme for the activation and detoxification of B[a]P (Arlt et al. 2008), our measured B[a]P metabolites, including B[a]P-7,8-diol, paralleled the pattern of CYP1A1 expression in both cell lines (HepG2 and A549). In addition, it is known that inhibition of CYP1A1 decreased B[a]P–DNA adduct formation in vitro (Endo et al. 2008), but not in vivo (Ma and Lu 2007). However, we showed that with additional β-glucuronidase, the metabolism of B[a]P is delayed, which prolonged the effective exposure of cells to unmetabolized B[a]P, ultimately producing more toxic metabolites instead of excretable derivatives of B[a]P. A study using Cyp1a1(‒/‒) mice revealed that slower metabolic clearance of B[a]P may indeed lead to greater formation of B[a]P-mediated DNA adducts (Uno et al. 2001).
In summary, in this study we showed that β-glucuronidase alters the cellular response towards B[a]P by changing gene expression of CYP1A1 in both lung- and liver-derived cells, ultimately causing higher DNA adduct levels. Moreover, we identified that β-glucuronidase may bind to IGF2R, thereby delaying B[a]P metabolism. This study exemplifies the complexity of the effect of inflammation on B[a]P-induced carcinogenesis, which deserves further attention.
Acknowledgments
This work was supported by a personal grant of the Chinese Scholarship Council (Grant Number CSC201307720049). Work at King’s College London is supported by Cancer Research UK (Grant C313/A14329) and in part by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Health Impact of Environmental Hazards at King’s College London in partnership with Public Health England (PHE). The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, the Department of Health or PHE.
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest in this study.
Contributor Information
Q. Shi, Email: q.shi@maastrichtuniversity.nl
R. W. L. Godschalk, Phone: +31 43 3881104, Email: r.godschalk@maastrichtuniversity.nl
References
- Almahmeed T, et al. Benzo[a]pyrene phenols are more potent inducers of CYP1A1, CYP1B1 and COX-2 than benzo[a]pyrene glucuronides in cell lines derived from the human aerodigestive tract. Carcinogenesis. 2004;25:793–799. doi: 10.1093/carcin/bgh078. [DOI] [PubMed] [Google Scholar]
- Arlt VM, et al. Metabolic activation of benzo[a]pyrene in vitro by hepatic cytochrome P450 contrasts with detoxification in vivo: experiments with hepatic cytochrome P450 reductase null mice. Carcinogenesis. 2008;29:656–665. doi: 10.1093/carcin/bgn002. [DOI] [PubMed] [Google Scholar]
- Arlt VM, et al. Pulmonary inflammation impacts on CYP1A1-mediated respiratory tract DNA damage induced by the carcinogenic air pollutant benzo[a]pyrene toxicological sciences. Off J Soc Toxicol. 2015 doi: 10.1093/toxsci/kfv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholome R, et al. Deconjugation kinetics of glucuronidated phase II flavonoid metabolites by beta-glucuronidase from neutrophils. Drug Metab Pharmacokinet. 2010;25:379–387. doi: 10.2133/dmpk.DMPK-10-RG-002. [DOI] [PubMed] [Google Scholar]
- Basinska A, Florianczyk B. Beta-glucuronidase in physiology and disease. Ann Univ Mariae Curie Sklodowska Med. 2003;58:386–389. [PubMed] [Google Scholar]
- Borm PJ, Knaapen AM, Schins RP, Godschalk RW, Schooten FJ. Neutrophils amplify the formation of DNA adducts by benzo[a]pyrene in lung target cells. Environ Health Perspect. 1997;105(5):1089–1093. doi: 10.1289/ehp.97105s51089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- El-Shewy HM, Luttrell LM. Insulin-like growth factor-2/mannose-6 phosphate receptors. Vitam Horm. 2009;80:667–697. doi: 10.1016/S0083-6729(08)00624-9. [DOI] [PubMed] [Google Scholar]
- Endo K, et al. Inhibition of aryl hydrocarbon receptor transactivation and DNA adduct formation by CYP1 isoform-selective metabolic deactivation of benzo[a]pyrene. Toxicol Appl Pharmacol. 2008;230:135–143. doi: 10.1016/j.taap.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Fridlender ZG, Albelda SM. Tumor-associated neutrophils: friend or foe? Carcinogenesis. 2012;33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Noriega A, Michalak C. Mannose 6-phosphate-independent endocytosis of beta-glucuronidase. II. Purification of a cation-dependent receptor from bovine liver. Biochim Biophys Acta. 2001;1538:152–161. doi: 10.1016/S0167-4889(00)00139-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Noriega A, Michalak C, Cruz-Perez JR, Masso F. Mannose 6-phosphate-independent endocytosis of beta-glucuronidase by human fibroblasts. I. Evidence for the existence of a membrane-binding activity. Biochim Biophys Acta. 2001;1538:141–151. doi: 10.1016/S0167-4889(00)00140-3. [DOI] [PubMed] [Google Scholar]
- Gungor N, Haegens A, Knaapen AM, Godschalk RW, Chiu RK, Wouters EF, van Schooten FJ. Lung inflammation is associated with reduced pulmonary nucleotide excision repair in vivo. Mutagenesis. 2010;25:77–82. doi: 10.1093/mutage/gep049. [DOI] [PubMed] [Google Scholar]
- Gungor N, et al. Genotoxic effects of neutrophils and hypochlorous acid. Mutagenesis. 2010;25:149–154. doi: 10.1093/mutage/gep053. [DOI] [PubMed] [Google Scholar]
- Gungor N, Pennings JL, Knaapen AM, Chiu RK, Peluso M, Godschalk RW, Van Schooten FJ. Transcriptional profiling of the acute pulmonary inflammatory response induced by LPS: role of neutrophils. Respir Res. 2010;11:24. doi: 10.1186/1465-9921-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann D, Hoffmann I. The changing cigarette, 1950–1995. J Toxicol Environ Health. 1997;50:307–364. doi: 10.1080/009841097160393. [DOI] [PubMed] [Google Scholar]
- Jin CJ, Miners JO, Burchell B, Mackenzie PI. The glucuronidation of hydroxylated metabolites of benzo[a]pyrene and 2-acetylaminofluorene by cDNA-expressed human UDP-glucuronosyltransferases. Carcinogenesis. 1993;14:2637–2639. doi: 10.1093/carcin/14.12.2637. [DOI] [PubMed] [Google Scholar]
- Kim JH, Stansbury KH, Walker NJ, Trush MA, Strickland PT, Sutter TR. Metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-diol by human cytochrome P450 1B1. Carcinogenesis. 1998;19:1847–1853. doi: 10.1093/carcin/19.10.1847. [DOI] [PubMed] [Google Scholar]
- Knaapen AM, Gungor N, Schins RP, Borm PJ, Van Schooten FJ. Neutrophils and respiratory tract DNA damage and mutagenesis: a review. Mutagenesis. 2006;21:225–236. doi: 10.1093/mutage/gel032. [DOI] [PubMed] [Google Scholar]
- Krais AM, et al. The impact of p53 on DNA damage and metabolic activation of the environmental carcinogen benzo[a]pyrene: effects in Trp53(+/+), Trp53(±) and Trp53(-/-) mice. Arch Toxicol. 2015 doi: 10.1007/s00204-015-1531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucab JE, van Steeg H, Luijten M, Schmeiser HH, White PA, Phillips DH, Arlt VM. TP53 mutations induced by BPDE in Xpa-WT and Xpa-Null human TP53 knock-in (Hupki) mouse embryo fibroblasts. Mutat Res. 2015;773:48–62. doi: 10.1016/j.mrfmmm.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma Q, Lu AY. CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos. 2007;35:1009–1016. doi: 10.1124/dmd.107.015826. [DOI] [PubMed] [Google Scholar]
- Moserova M, Kotrbova V, Aimova D, Sulc M, Frei E, Stiborova M. Analysis of benzo[a]pyrene metabolites formed by rat hepatic microsomes using high pressure liquid chromatography: optimization of the method. Interdiscip Toxicol. 2009;2:239–244. doi: 10.2478/v10102-009-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngkelo A, Meja K, Yeadon M, Adcock I, Kirkham PA. LPS induced inflammatory responses in human peripheral blood mononuclear cells is mediated through NOX4 and Gialpha dependent PI-3kinase signalling. J Inflamm (Lond) 2012;9:1. doi: 10.1186/1476-9255-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Pokrovskaya ID, Xu J, Little JM, Jude AR, Kurten RC, Czernik PJ. Nuclear UDP-glucuronosyltransferases: identification of UGT2B7 and UGT1A6 in human liver nuclear membranes. Arch Biochem Biophys. 2002;399:37–48. doi: 10.1006/abbi.2001.2743. [DOI] [PubMed] [Google Scholar]
- Reutershan J, Basit A, Galkina EV, Ley K. Sequential recruitment of neutrophils into lung and bronchoalveolar lavage fluid in LPS-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289:L807–L815. doi: 10.1152/ajplung.00477.2004. [DOI] [PubMed] [Google Scholar]
- Schults MA, Oligschlaeger Y, Godschalk RW, Van Schooten FJ, Chiu RK. Loss of VHL in RCC reduces repair and alters cellular response to benzo[a]pyrene. Front Oncol. 2013;3:270. doi: 10.3389/fonc.2013.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schults MA, Sanen K, Godschalk RW, Theys J, van Schooten FJ, Chiu RK. Hypoxia diminishes the detoxification of the environmental mutagen benzo[a]pyrene. Mutagenesis. 2014;29:481–487. doi: 10.1093/mutage/geu034. [DOI] [PubMed] [Google Scholar]
- Shacter E, Weitzman SA (2002) Chronic inflammation and cancer. Oncology 16:217–226, 229; discussion 230–212 [PubMed]
- Shimada T, Guengerich FP. Inhibition of human cytochrome P450 1A1-, 1A2-, and 1B1-mediated activation of procarcinogens to genotoxic metabolites by polycyclic aromatic hydrocarbons. Chem Res Toxicol. 2006;19:288–294. doi: 10.1021/tx050291v. [DOI] [PubMed] [Google Scholar]
- Shimoi K, Nakayama T. Glucuronidase deconjugation in inflammation. Methods Enzymol. 2005;400:263–272. doi: 10.1016/S0076-6879(05)00015-7. [DOI] [PubMed] [Google Scholar]
- Shimoi K, Saka N, Nozawa R, Sato M, Amano I, Nakayama T, Kinae N. Deglucuronidation of a flavonoid, luteolin monoglucuronide, during inflammation. Drug Metab Dispos. 2001;29:1521–1524. [PubMed] [Google Scholar]
- Spink DC, et al. Induction of CYP1A1 and CYP1B1 in T-47D human breast cancer cells by benzo[a]pyrene is diminished by arsenite. Drug Metab Dispos. 2002;30:262–269. doi: 10.1124/dmd.30.3.262. [DOI] [PubMed] [Google Scholar]
- Spink DC, Wu SJ, Spink BC, Hussain MM, Vakharia DD, Pentecost BT, Kaminsky LS. Induction of CYP1A1 and CYP1B1 by benzo(k)fluoranthene and benzo(a)pyrene in T-47D human breast cancer cells: roles of PAH interactions and PAH metabolites. Toxicol Appl Pharmacol. 2008;226:213–224. doi: 10.1016/j.taap.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiborova M, et al. Cytochrome b5 and epoxide hydrolase contribute to benzo[a]pyrene-DNA adduct formation catalyzed by cytochrome P450 1A1 under low NADPH:P450 oxidoreductase conditions. Toxicology. 2014;318:1–12. doi: 10.1016/j.tox.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Tomblin JK, Salisbury TB. Insulin like growth factor 2 regulation of aryl hydrocarbon receptor in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2014;443:1092–1096. doi: 10.1016/j.bbrc.2013.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushin N, et al. Comparative metabolism of benzo[a]pyrene by human keratinocytes infected with high-risk human papillomavirus types 16 and 18 as episomal or integrated genomes. J Carcinog. 2012;11:1. doi: 10.4103/1477-3163.92309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Shertzer HG, Genter MB, Warshawsky D, Talaska G, Nebert DW. Benzo[a]pyrene-induced toxicity: paradoxical protection in Cyp1a1(–/–) knockout mice having increased hepatic BaP–DNA adduct levels. Biochem Biophys Res Commun. 2001;289:1049–1056. doi: 10.1006/bbrc.2001.6110. [DOI] [PubMed] [Google Scholar]
- Uppstad H, Ovrebo S, Haugen A, Mollerup S. Importance of CYP1A1 and CYP1B1 in bioactivation of benzo[a]pyrene in human lung cell lines. Toxicol Lett. 2010;192:221–228. doi: 10.1016/j.toxlet.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Urayama A, Grubb JH, Sly WS, Banks WA. Developmentally regulated mannose 6-phosphate receptor-mediated transport of a lysosomal enzyme across the blood–brain barrier. Proc Natl Acad Sci USA. 2004;101:12658–12663. doi: 10.1073/pnas.0405042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo D, et al. Neutrophil-derived ROS contribute to oxidative DNA damage induction by quartz particles. Free Radic Biol Med. 2010;49:1685–1693. doi: 10.1016/j.freeradbiomed.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Van Schooten FJ, et al. 32P-postlabelling of aromatic DNA adducts in white blood cells and alveolar macrophages of smokers: saturation at high exposures. Mutat Res. 1997;378:65–75. doi: 10.1016/S0027-5107(97)00098-5. [DOI] [PubMed] [Google Scholar]
- Van Schooten FJ, et al. Myeloperoxidase (MPO)–463G->A reduces MPO activity and DNA adduct levels in bronchoalveolar lavages of smokers. Cancer Epidemiol Biomark Prev. 2004;13:828–833. [PubMed] [Google Scholar]
- Vogler C, et al. Overcoming the blood–brain barrier with high-dose enzyme replacement therapy in murine mucopolysaccharidosis VII. Proc Natl Acad Sci USA. 2005;102:14777–14782. doi: 10.1073/pnas.0506892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohak LE, et al. Carcinogenic polycyclic aromatic hydrocarbons induce CYP1A1 in human cells via a p53-dependent mechanism. Arch Toxicol. 2014;1:11. doi: 10.1007/s00204-014-1409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RP, Hopkins RJ, Christmas T, Black PN, Metcalf P, Gamble GD. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur Respir J. 2009;34:380–386. doi: 10.1183/09031936.00144208. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Fang JL, Lazarus P. Glucuronidation: an important mechanism for detoxification of benzo[a]pyrene metabolites in aerodigestive tract tissues. Drug Metab Dispos. 2002;30:397–403. doi: 10.1124/dmd.30.4.397. [DOI] [PubMed] [Google Scholar]