Summary
Tight regulation of the balance between self-renewal and differentiation of neural stem cells is crucial to assure proper neural development. In this context, Notch signaling is a well-known promoter of stemness. In contrast, the bifunctional brain-enriched microRNA miR-9/9∗ has been implicated in promoting neuronal differentiation. Therefore, we set out to explore the role of both regulators in human neural stem cells. We found that miR-9/9∗ decreases Notch activity by targeting NOTCH2 and HES1, resulting in an enhanced differentiation. Vice versa, expression levels of miR-9/9∗ depend on the activation status of Notch signaling. While Notch inhibits differentiation of neural stem cells, it also induces miR-9/9∗ via recruitment of the Notch intracellular domain (NICD)/RBPj transcriptional complex to the miR-9/9∗_2 genomic locus. Thus, our data reveal a mutual interaction between bifunctional miR-9/9∗ and the Notch signaling cascade, calibrating the delicate balance between self-renewal and differentiation of human neural stem cells.
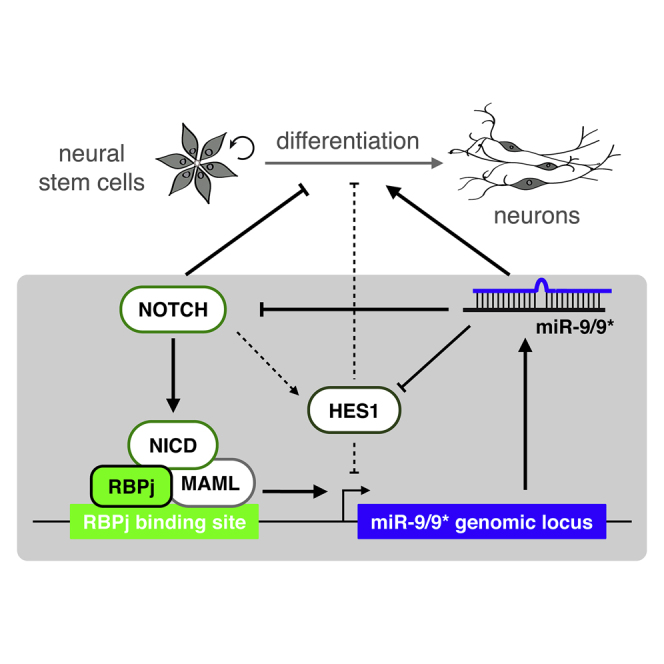
Graphical Abstract
Highlights
-
•
MiR-9/9∗ regulate Notch signaling by targeting NOTCH2 and HES1
-
•
Notch directly regulates transcription of the miR-9_2 genomic locus
-
•
Notch-miR-9 reciprocal regulation calibrates NSC self-renewal and differentiation
In this article, Brüstle and colleagues show that a mutual interaction between microRNA-9 and Notch calibrates the delicate balance between self-renewal and differentiation of human neural stem cells (hNSC). While Notch maintains stemness and proliferation of hNSC, it also directly induces expression of miR-9, which in turn promotes hNSC differentiation by targeting NOTCH2 and HES1.
Introduction
Embryonic and induced pluripotent stem cell-derived neural stem cells represent promising in vitro model systems to explore early stages of human neural development. Our group has recently established homogeneous populations of long-term self-renewing neuroepithelial-like stem (lt-NES) cells from both human embryonic stem cells (ESCs) and induced pluripotent stem (iPS) cells. Lt-NES cells can be continuously propagated in the presence of fibroblast growth factor 2 (FGF2) and epidermal growth factor (EGF). Upon growth factor withdrawal, they show stable and robust neurogenesis and gliogenesis independent of their passage number (Falk et al., 2012, Koch et al., 2009). We previously demonstrated that lt-NES cells depend on Notch signaling for their maintenance, and that blocking Notch activity delays their G1/S-phase transition and commits them to neuronal differentiation (Borghese et al., 2010).
Notch is a transmembrane receptor whose signaling is initiated by binding to ligands (Delta or Jagged) on an adjacent cell. In humans, four isoforms of the Notch receptor exist, namely NOTCH1–4 (Lasky and Wu, 2005). Upon ligand binding, the Notch intracellular domain (NICD) is released by proteolytic cleavage steps carried out sequentially by an ADAM protease and the γ-secretase complex. NICD translocates into the nucleus where it associates with the DNA binding recombination signal binding protein for immunoglobulin kappa J region (RBPj) and recruits co-activator of transcription Mastermind-like (MAML) to assemble a transcriptional complex that activates gene expression (reviewed in Louvi and Artavanis-Tsakonas, 2006). This leads to transcription of downstream targets such as HES and HES-related (HESR or HEY) basic helix-loop-helix transcription factors as well as cell-cycle regulators (reviewed in Andersson et al., 2011). Notch signaling is known to impact on neural progenitor maintenance and cell fate (reviewed in Pierfelice et al., 2011).
More recently, microRNAs (miRNAs) have emerged as critical regulators of neural progenitor development and cell fate (Coolen and Bally-Cuif, 2009, Hebert and De Strooper, 2009, Schratt, 2009). miRNAs are endogenously expressed RNA molecules 22–25 nucleotides in length that are transcribed from intragenic and intergenic regions in the genome. The primary transcript (pri-miRNA) is usually processed into a hairpin-shaped precursor (pre-miRNA) by Drosha/DGCR8 and exported to the cytosol, where it is further converted into the mature miRNA by Dicer and incorporated into the RISC complex (reviewed in Bartel, 2004, Winter et al., 2009). Within the miRISC, miRNAs may regulate the expression of hundreds of mRNA targets at a posttranscriptional level by binding to their 3′ UTRs and triggering either transcript degradation or translational inhibition (reviewed in Bartel, 2009).
miRNAs often take part in fine-tuning major signaling pathways (Inui et al., 2010). The interaction between Notch signaling and an miRNA, namely miR-9a, was first described in Drosophila in the context of sensory organ precursor development (Li et al., 2006). More recently, the existence of an interaction between Notch signaling and miR-9 has been proposed also in the context of vertebrate neural development by studies in zebrafish, frog, and mouse (Bonev et al., 2011, Bonev et al., 2012, Coolen et al., 2012). MiR-9 is evolutionarily conserved from insects to humans (Yuva-Aydemir et al., 2011). Data from mouse revealed that it is specifically expressed in the brain (Lagos-Quintana et al., 2002). The pri-forms of miR-9 are bifunctional, as all three primary transcripts coding for miR-9 also encode a second miRNA referred to as miR-9∗, which shows a lower expression rate (Yuva-Aydemir et al., 2011). MiR-9 has been intensively studied and shown to play important developmental roles in neural stem cell proliferation, migration, and differentiation, depending on the spatial and temporal context (reviewed in Gao, 2010, Stappert et al., 2014). Although less intensively studied, its sister strand miR-9∗ has also been implicated in neural development (Packer et al., 2008) and in modulating dendritic growth (Yoo et al., 2009) as well as stemness of glioma cells (Jeon et al., 2011). Recently, we found that bifunctional miR-9/9∗ contributes to the switch of lt-NES cells from self-renewal to neuronal differentiation (Roese-Koerner et al., 2013).
Here, we show that Notch and miR-9/9∗ have opposing effects on human neural stem cell proliferation and differentiation but also directly regulate each other. While Notch contributes to the transcription of miR-9/9∗, mature miR-9 and miR-9∗ negatively regulate the Notch pathway by targeting NOTCH2 and HES1, thus providing a reciprocally regulated system for calibrating human neural stem cell proliferation and differentiation.
Results
miR-9 and miR-9∗ Target Multiple Components of the Notch Pathway
In previous work, we screened for the expression profile of 330 human miRNAs in human ESCs (hESCs) from the I3 line, lt-NES cells derived from those hESCs, and their differentiated neuronal progeny after 15 and 30 days of growth factor withdrawal (Stappert et al., 2013). We found that miR-9 and miR-9∗ are expressed in lt-NES cells and that their levels increase during neuronal differentiation. We also showed that bifunctional miR-9/9∗ contributes to the transition of these lt-NES cells from self-renewal to neuronal differentiation (Roese-Koerner et al., 2013). Target prediction analyses show that target sites for miR-9 and miR-9∗ can be found in the 3′ UTRs of several members of the Notch signaling pathway (Table S1).
We particularly focused our attention on NOTCH1, NOTCH2, and HES1. Mouse Hes1 and its homologs in frog (Hairy1) and zebrafish (Her6) were all found to be targets of miR-9 (Bonev et al., 2011, Bonev et al., 2012, Coolen et al., 2012). Data generated in mouse mesenchymal stem cells and human breast cancer cells showed that NOTCH1 is regulated by miR-9 (Jing et al., 2011, Mohammadi-Yeganeh et al., 2015). In contrast to Hes1 and Notch1, there were no previous data indicating Notch2 as a target of miR-9/9∗. We chose NOTCH2 as an interesting novel candidate for different reasons. First, NOTCH2 has the longest 3′ UTR of all Notch receptors (NOTCH1, 1,627 nt; NOTCH2, 3,751 nt; NOTCH3, 1,028 nt; NOTCH4, 593 nt), which could suggest an miRNA-based regulation of its expression. Second, targeting of the NOTCH2 3′ UTR by miR-9 and miR-9∗ was predicted by several algorithms (Table S1). Third, previous reports point to opposing roles of miR-9/9∗ and Notch2 in neurogenesis. While overexpression of miR-9/9∗ was shown to promote neuronal fate and reduce the number of glial fibrillary acidic protein-positive cells during neural differentiation of mouse ESCs (Krichevsky et al., 2006), Notch2-ICD (N2ICD) expression was shown to support expansion of the neurogenic niche in vivo (Tchorz et al., 2012) and to induce the differentiation of astrocytes at the expense of neurons and oligodendrocytes in cultured neural stem cells (Tchorz et al., 2012).
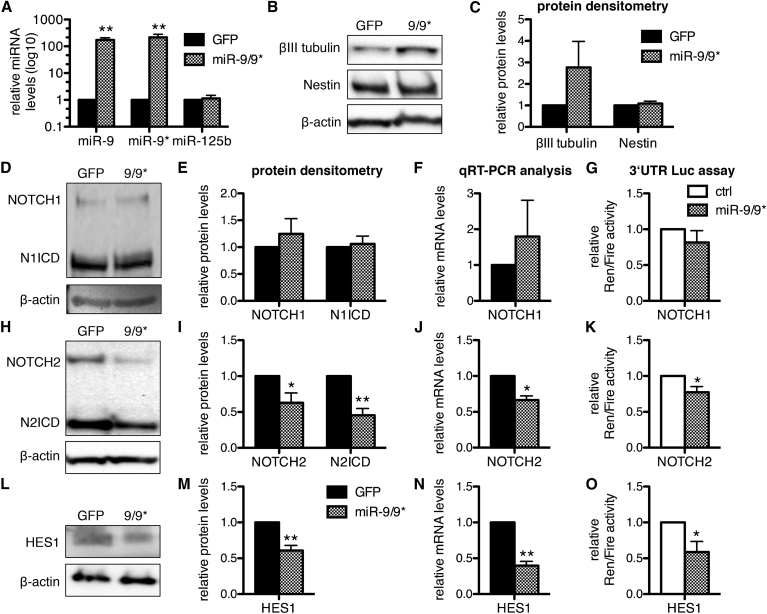
To explore whether human NOTCH1, NOTCH2, and HES1 could be targets of miR-9 and miR-9∗, we overexpressed the genomic sequence of the miR-9_1 locus in lt-NES cells derived from I3 hESCs in a doxycycline-inducible manner and assessed changes in the expression levels of NOTCH1, NOTCH2, and HES1 by western blotting and real-time qRT-PCR analyses. After 4 days of doxycycline treatment, we found a robust increase in the expression of mature miR-9 and miR-9∗ in I3 lt-NES cells cultured in the presence of FGF2 and EGF (Figure 1A). Under these conditions, βIII-tubulin protein levels were slightly increased in miR-9/9∗-overexpressing cultures (Figures 1B and 1C), which is in line with our earlier observation of an enhanced rate of spontaneous neuronal differentiation upon miR-9/9∗ overexpression (Roese-Koerner et al., 2013). However, Nestin protein levels were unchanged (Figures 1B and 1C), indicating that Nestin is less responsive to miR-9/9∗ overexpression. Levels of NOTCH1 did not significantly change (Figures 1D–1F), while we observed a significant decrease in NOTCH2 mRNA levels and NOTCH2 protein variants, i.e., full-length NOTCH2 and N2ICD (Figures 1H–1J). Likewise, both transcript and protein levels of HES1 were reduced upon miR-9/9∗ overexpression (Figures 1L–1N).
Figure 1.
miR-9/9∗ Target NOTCH2 and HES1
(A) qRT-PCR analyses of miR-9, miR-9∗, and miR-125b in lt-NES cells overexpressing the miR-9-1 genomic sequence (9/9∗) or GFP (used as control) after 4 days of doxycycline treatment. Data are normalized to miR-16 reference levels and presented as average changes + SEM relative to expression in GFP-expressing lt-NES cells (GFP, equal to 1; n ≥ 3; ∗∗p ≤ 0.01, Student's t test).
(B, D, H, L) Representative western blot analyses of βIII-tubulin and Nestin (B), full-length NOTCH1 (D), NOTCH2 (H), and their respective intracellular domains (ICD), as well as HES1 (L) protein levels in I3 lt-NES cells overexpressing the miR-9/9∗ locus induced by 4 days of doxycycline treatment compared with a GFP control construct (n ≥ 3). β-Actin is shown as loading control.
(C, E, I, M) Corresponding densitometric analyses of βIII-tubulin and Nestin (C), NOTCH1 and N1ICD (E), NOTCH2 and N2ICD (I), and HES1 (M) protein levels normalized to β-actin. Data are presented as mean ± SEM relative to expression in I3 lt-NES cells overexpressing GFP (equal to 1; n ≥ 3; ∗p ≤ 0.05, ∗∗p ≤ 0.01, Student's t test).
(F, J, N) qRT-PCR analyses of NOTCH1 (F), NOTCH2 (J), and HES1 (N) transcript levels in the conditions described above. Data are normalized to 18S rRNA reference levels and presented as average changes + SEM relative to expression in lt-NES cells overexpressing GFP (equal to 1; n = 6; ∗p ≤ 0.05, ∗∗p ≤ 0.01, Student's t test).
(G, K, O) Analyses of luciferase activity in I3 lt-NES cells expressing the 3′ UTRs of NOTCH1 (G), NOTCH2 (K), and HES1 (O) cloned downstream of Renilla (Ren) luciferase and transfected with synthetic mimics for miR-9 and miR-9∗ (9/9∗) or a short RNA scrambled control (ctrl). Data are normalized to firefly (Fire) luciferase activity and presented as average changes + SEM relative to activity in I3 lt-NES cells transfected with the scrambled control (ctrl, equal to 1; n = 5; ∗p ≤ 0.05, Student's t test). All experiments were performed in I3 lt-NES cells cultured under self-renewing conditions, i.e., in the presence of EGF and FGF2.
To assess whether the regulation of Notch genes by miR-9/9∗ is a peculiarity of lt-NES cells rather than a more general phenomenon, we analyzed the impact of miR-9/9∗ overexpression in another neural stem cell system (Figure S1). We chose the recently described small-molecule neural precursor cells (smNPC; Reinhardt et al., 2013), which are considered to represent an early pre-rosette neural plate border fate compared with the lt-NES cells that grow as small neural rosettes reminiscent of the early neural tube (Koch et al., 2009). smNPCs transduced with miR-9/9∗ lentivirus (Figure S1A) already showed a downregulation of NOTCH2 and HES1 mRNA after 48 hr of doxycycline-induced overexpression, confirming the data gathered in lt-NES cells (Figure S1B). At this time point the amount of βIII-tubulin-positive cells was not affected, indicating that the effect on NOTCH2 and HES1 after short-term overexpression of miR-9/9∗ is due to a direct effect rather than a change in cell fate (Figure S1C). However, overexpression of miR-9/9∗ over 4 days induced a slight increase in neuronal differentiation of smNPCs (Figure S1C), similar to what we had earlier observed in lt-NES cells (Roese-Koerner et al., 2013).
To follow up on these observations, we generated reporter constructs carrying a Renilla luciferase fused to the 3′ UTRs of NOTCH1, NOTCH2, or HES1 and an unregulated firefly luciferase as expression control. These constructs were transfected separately into I3 lt-NES cell lines expressing either the genomic sequence of the miR-9_1 locus or a scrambled control. Upon overexpression of miR-9/9∗, we found a significant decrease in Renilla luciferase activity in lt-NES cells for the constructs harboring NOTCH2 and HES1 3′ UTRs (Figures 1K and 1O). In contrast, no significant change in luciferase activity was observed when testing the NOTCH1 3′ UTR (Figure 1G). Together, these data strongly suggest that NOTCH2 and HES1, but not NOTCH1, are direct targets of miR-9/9∗ activity in human neural stem cells.
Modulation of the Balance between miR-9/9∗ and Notch Activity Results in an Altered Differentiation Potential of lt-NES Cells
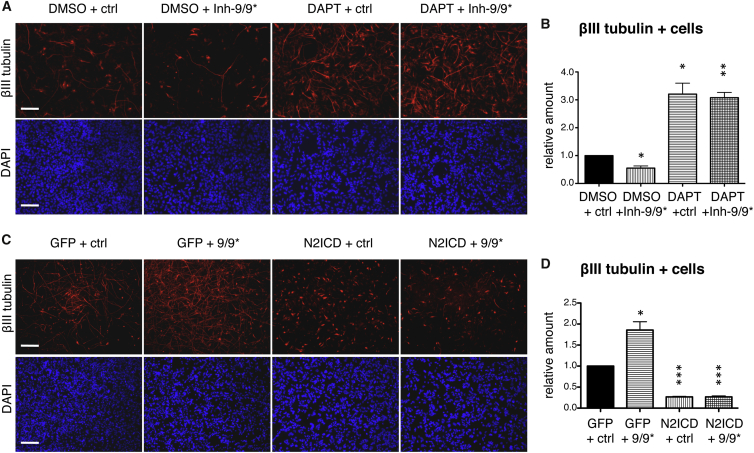
We previously showed that Notch signaling is crucial for maintaining lt-NES cells in a self-renewing undifferentiated state, while its inhibition leads to a delay in G1/S-phase transition and premature onset of neuronal differentiation (Borghese et al., 2010). To understand the biological relevance of a potential targeting of Notch pathway components by miR-9/9∗, we performed loss- and gain-of-function studies in differentiating lt-NES cells. Differentiation was induced by growth factor withdrawal for 7 days. When we inhibited miR-9/9∗ activity by consecutive transfection of synthetic inhibitors, the number of βIII-tubulin-positive cells was reduced by 50% compared with vehicle control cultures treated with DMSO alone and transfected with a short scrambled control RNA (Figures 2A and 2B). In contrast, inhibition of Notch activity via N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine (DAPT; administered every day during the 7 days differentiation paradigm) led to a ∼3-fold increase in the number of βIII-tubulin-positive cells compared with control cultures (Figures 2A and 2B). Interestingly, when combining these two conditions we observed that the impairment of neuronal differentiation induced by inhibition of miR-9/9∗ activity was abolished by DAPT-mediated inhibition of Notch activity (Figures 2A and 2B). Similar results could be obtained in lt-NES cells derived from another parental hESC line (H9.2; Figure S2).
Figure 2.
The Impact of miR-9/9∗ on lt-NES Cell Differentiation Is Mediated by Targeting of the Notch Signaling Pathway
(A) Representative immunostainings for βIII-tubulin in I3 lt-NES cells transfected with synthetic inhibitors for miR-9 and miR-9∗ (Inh-9/9∗) or a short scrambled control RNA (ctrl) and treated with DAPT or DMSO (vehicle control), after 7 days of in vitro differentiation.
(B) Corresponding quantifications of the relative number of βIII-tubulin-positive cells in lt-NES cultures after 7 days of in vitro differentiation in the conditions described above.
(C) Representative immunostainings for βIII-tubulin in I3 lt-NES cells transduced with lentiviral vectors overexpressing N2ICD or GFP control and transfected with synthetic mimics for miR-9 and miR-9∗ (9/9∗) or a short scrambled control RNA (ctrl), after 7 days of in vitro differentiation.
(D) Corresponding quantifications as described in (B). DAPI stains nuclei.
Scale bars indicate 100 μm. All data are presented as mean + SEM (n = 3; ∗p ≤ 0.05, ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001, Student's t test).
When lt-NES cells were co-transfected with synthetic mimics for miR-9 and miR-9∗ the number of βIII-tubulin-positive cells increased by ∼2-fold compared with control cultures (Figures 2C and 2D). In contrast, overexpression of constitutively active N2ICD (Capobianco et al., 1997) led to a ∼5-fold decrease in the number of βIII-tubulin-positive cells compared with control cultures (Figures 2C and 2D). When combining the two conditions, we observed that forced activation of Notch signaling by overexpression of N2ICD, which lacks the NOTCH2 3′ UTR with the binding sites for miR-9 and miR-9∗, abolished the promotion of neuronal differentiation induced by ectopic miR-9/9∗ activity.
Taken together with the miR-9-based repression of NOTCH2 and HES1 shown above, these data strongly indicate that in lt-NES cells the Notch pathway is a functional target of miR-9/9∗ activity and that miR-9/9∗-based promotion of neuronal differentiation is, at least in part, mediated via the Notch pathway.
miR-9 Expression is Decreased upon γ-Secretase Inhibition
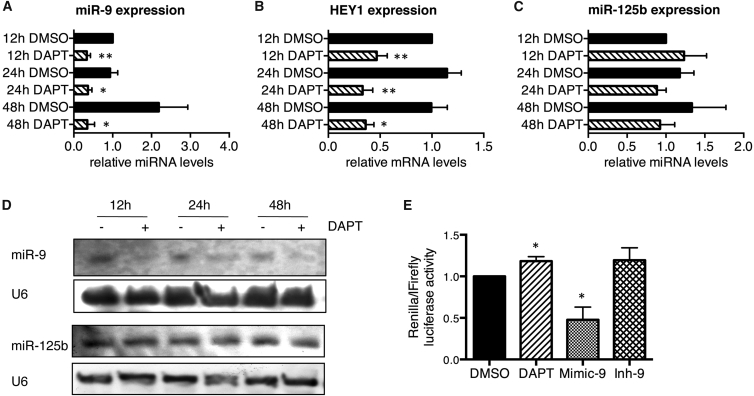
Previous studies have shown that mice lacking Presinilin1 (Psen1), which is the catalytic component of the γ-secretase complex, exhibit decreased miR-9 expression (Krichevsky et al., 2003). In line with this, more recent data in zebrafish also showed decreased miR-9 expression in response to pharmacological inhibition of the γ-secretase complex (Coolen et al., 2012). As miR-9 is expressed in lt-NES cells, we asked whether treating these cells with DAPT, a γ-secretase inhibitor, could lead to similar results. To avoid any differentiation bias in this experiment, we added DAPT to the normal lt-NES cell maintenance medium (containing FGF2 and EGF) for 12 to 48 hr only. This short-term application of DAPT resulted in a robust ∼2-fold downregulation of miR-9 expression in I3 lt-NES cells and also reduced the expression of the known Notch downstream target HEY1 (Figures 3A and 3B). In contrast, expression of mature miR-125b, another brain-enriched miRNA shown to be expressed in lt-NES cells (Roese-Koerner et al., 2013), was not affected by DAPT treatment (Figure 3C). The decrease of mature miR-9 but not miR-125b was validated by northern blot analyses in the same conditions (Figure 3D). This result confirms the specificity of the impact of γ-secretase inhibition on miR-9 expression. Similar data were obtained in proliferating H9.2 lt-NES cells and smNPCs (Figures S3A–S3C), indicating that the impact of DAPT on miR-9 expression is independent of the cell line used.
Figure 3.
Impact of Pharmacological Notch Inhibition on miR-9 Expression in Proliferating lt-NES Cells
(A–C) qRT-PCR analyses monitoring mature miR-9 (A), HEY1 (B), and miR-125b (C) in I3 lt-NES cells treated with DAPT (+) or DMSO (−, vehicle control) for 12, 24, and 48 hr in the presence of EGF and FGF2. Data are normalized to miR-16 (A, C) or 18S rRNA (B) reference levels and presented as average changes + SEM relative to expression in I3 lt-NES cells treated with DMSO for 12 hr (equal to 1; n = 4 [A, C]; n = 6 [B]; ∗p ≤ 0.05, ∗∗p ≤ 0.01, Student's t test).
(D) Northern blot analyses showing the expression of miR-9 or miR-125b in the samples described above. U6 small nuclear RNA was used as loading control.
(E) Analyses of Renilla luciferase activity in I3 lt-NES cells treated with DAPT or transfected with a synthetic mimic or inhibitor of miR-9. Data are normalized to firefly luciferase activity and presented as average changes + SEM relative to activity in DMSO-treated I3 lt-NES cells transfected with a short scrambled control RNA (ctrl, equal to 1; n = 4; ∗p ≤ 0.05, Student's t test).
To further prove that miR-9 expression in lt-NES cells is affected by γ-secretase inhibition, we monitored the impact of DAPT on the activity of a luciferase reporter carrying miR-9 binding sites downstream of the Renilla luciferase cDNA. To validate the sensitivity of the reporter system in lt-NES cells, we modulated endogenous levels of miR-9 by transfection of a synthetic mimic or inhibitor of miR-9. Transfection of I3 lt-NES cells with an miR-9 mimic resulted in a relative decrease of Renilla luciferase activity to 47% ± 7% compared with the baseline luciferase activity (set to 100%) of reporter cells treated with DMSO as vehicle control and transfected with a short scrambled control RNA (Figure 3E). In contrast, treatment with a miR-9 inhibitor increased the Renilla luciferase activity to 119% ± 17% (Figure 3E). Similar to miR-9 inhibition, treatment with DAPT induced a slight, yet significant increase (118% ± 6%; Figure 3E) in luciferase activity. From these data we conclude that, as previously shown for mouse (Krichevsky et al., 2003) and zebrafish (Coolen et al., 2012), expression of miR-9 is also affected by γ-secretase activity in human neural stem cells.
Expression of miR-9 and miR-9∗ is Affected by Modulation of Notch Activity
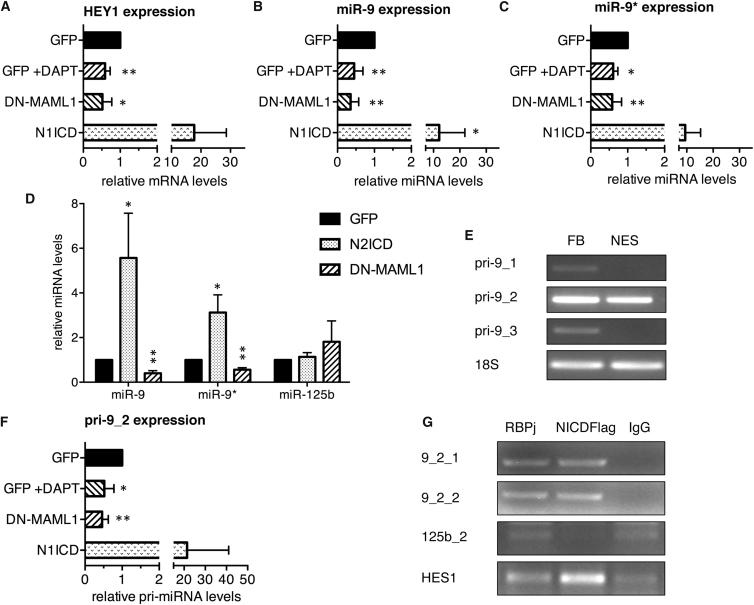
While Notch is a well-known substrate of γ-secretase and DAPT is able to inhibit Notch activity in lt-NES cells ((Borghese et al., 2010); Figure 3B), DAPT-based inhibition of γ-secretase may also impact on other substrates. To confirm that the observed impact of DAPT on miR-9 expression results from modulation of the Notch pathway, we went on to assess expression of mature miR-9 and miR-9∗ in conditions of specific Notch loss and gain of function. For specific interference with Notch activity in lt-NES cells we conditionally overexpressed a truncated form of the NICD co-activator of transcription MAML1 that was previously shown to act in a dominant-negative manner (Weng et al., 2003); hence, we refer to it as DN-MAML1. To achieve Notch gain of function we conditionally overexpressed the human NOTCH1-ICD (N1ICD) construct (Capobianco et al., 1997) in I3 lt-NES cells under proliferating conditions. Efficiency of conditional Notch gain and loss of function was assessed by monitoring mRNA levels of the known Notch transcriptional target HEY1 (Figure 4A). Like HEY1 mRNA, expression of both miR-9 and miR-9∗ was strongly increased upon NICD overexpression and significantly decreased upon DN-MAML1 overexpression (Figures 4B and 4C). In contrast, expression of miR-125b did not change upon modulation of Notch activity (Figures 4D and S4A). Elevated levels of miR-9 and miR-9∗ were also observed after transduction of I3 lt-NES cells with a human NOTCH2-ICD (N2ICD) construct (Figure 4D). Taken together with the finding that miR-9/9∗ targets NOTCH2 (Figures 1H–1K), these data indicate that miR-9 and NOTCH2 mutually regulate each other.
Figure 4.
miR-9/9∗ Expression in Proliferating lt-NES Cells upon Modulation of Notch Pathway Activity
(A–C) qRT-PCR analyses of HEY1 mRNA (A), miR-9 (B), and miR-9∗ (C) in I3 lt-NES cells transduced with lentiviral vectors overexpressing in a doxycycline-dependent manner GFP, NICD, or DN-MAML1 after 4 days of doxycycline treatment in the presence or absence of DAPT. Data are normalized to 18S rRNA (A) and miR-16 (B, C) reference levels and presented as average changes + SEM relative to expression in GFP-expressing I3 lt-NES cells (GFP, equal to 1; n ≥ 4; ∗p ≤ 0.05, ∗∗p ≤ 0.01, Student's t test).
(D) qRT-PCR analyses of miR-9, miR-9∗, and miR-125b in I3 lt-NES cells transduced with lentiviral vectors overexpressing in a doxycycline-dependent manner GFP, N2ICD, or DN-MAML1 after 4 days of doxycycline treatment. Data are normalized to RNU5A reference levels and presented as average changes + SEM relative to expression in GFP-expressing lt-NES cells (GFP, equal to 1; n = 3; ∗p ≤ 0.05, ∗∗p ≤ 0.01, Student's t test).
(E) Representative images of PCR analyses for expression of the three pri-miR-9 forms (pri-9_1, pri-9_2, and pri-9_3) in I3 lt-NES cells (NES) and fetal brain RNA (FB). 18S rRNA levels were used as loading control.
(F) qRT-PCR analyses of pri-miR-9_2 transcript levels (pri-9_2) in the samples described above. Data are normalized to 18S rRNA reference levels and are presented as average changes + SEM relative to expression in GFP-expressing I3 lt-NES cells (GFP, equal to 1; n ≥ 4; ∗p ≤ 0.05, ∗∗p ≤ 0.01, Student's t test).
(G) Representative images of PCR analyses of the predicted RBPj binding sites in the genomic regions 10 kb upstream of pre-miR-9_2 and pre-miR-125b_2 from immunoprecipitates with antibodies against FLAG (FLAG-tagged NICD) and RBPj compared with IgG negative control (n = 3).
Since transcriptional regulation is not the only mechanism impacting on the levels of mature miRNAs, we assessed the expression of the primary transcript produced by the miR-9_2 locus (pri-miR-9_2) in I3 lt-NES cells under conditional gain and loss of Notch activity. Using PCR analysis, we were able to show that in I3 lt-NES cells as well as in smNPCs only the miR-9_2 locus is expressed in a detectable manner (Figures 4E and S3D). As shown in Figure 4F, pri-miR-9_2 levels were significantly decreased upon DN-MAML1 overexpression, and dramatically increased upon NICD overexpression. Similarly, DAPT treatment of smNPCs also resulted in a drastic decrease of pri-miR-9_2 expression (Figure S3E). These data suggest that Notch signaling regulates miR-9 and miR-9∗ expression at the transcriptional level in both lt-NES cells and smNPCs.
The NICD/RBPj Complex Interacts Physically with Regions Upstream of the miR-9/9∗ Loci
In bioinformatics analyses we identified RBPj binding sites in the genomic regions upstream of all three miR-9/9∗ loci and one binding site upstream of the miR-125b_2 locus (Figure S4B). As pri-miR-9_2 was the only precursor strongly detectable in lt-NES cells and smNPCs (Figures 4E and S3D), we focused on the two binding sites predicted in the upstream region of the miR-9_2 genomic locus. To explore whether there is a direct interaction of the NICD/RBPj complex with these sites, we performed chromatin immunoprecipitation (ChIP) assays with antibodies against RBPj and FLAG tag using extracts from lt-NES cells overexpressing a FLAG-tagged NICD construct. The predicted binding sites upstream of the miR-9_2 locus were both enriched according to semi-quantitative PCR analyses following immunoprecipitation with both anti-FLAG and anti-RBPj antibodies compared with an immunoprecipitation with an immunoglobulin G (IgG) control antibody (Figure 4G). Similar results were observed for the HES1 promoter region (Figure 4G). In contrast, the unique RBPj binding site upstream of the miR-125b_2 locus was not enriched (Figure 4G). Overall these data show a recruitment of the NICD/RBPj complex to the miR-9_2 genomic locus, thereby strongly suggesting that Notch is a direct transcriptional modulator of miR-9/9∗.
Discussion
Notch Signaling is Important for Neural Stem Cell Maintenance
Notch signaling is an important player in stem cell self-renewal in general. However, the impact of Notch on neural stem cells strictly depends on the level of its activation (Guentchev and McKay, 2006). Reduced levels of Notch signaling commit neural stem cells to neuronal differentiation (Borghese et al., 2010, Lutolf et al., 2002) and induce severe defects in development of the CNS (reviewed in Chiba, 2006). In turn, too much Notch activity can lead to quiescence, e.g., in adult neural stem cells (Chapouton et al., 2010), or in some other context may induce tumorigenesis (Harper et al., 2003, Pierfelice et al., 2008, Radtke and Raj, 2003, Tchorz et al., 2012). Due to these dose-dependent effects of Notch signaling, a tight control of its activity is crucial to ensure homeostatic stem cell self-renewal.
miR-9/9∗ Target Notch Signaling at Different Levels
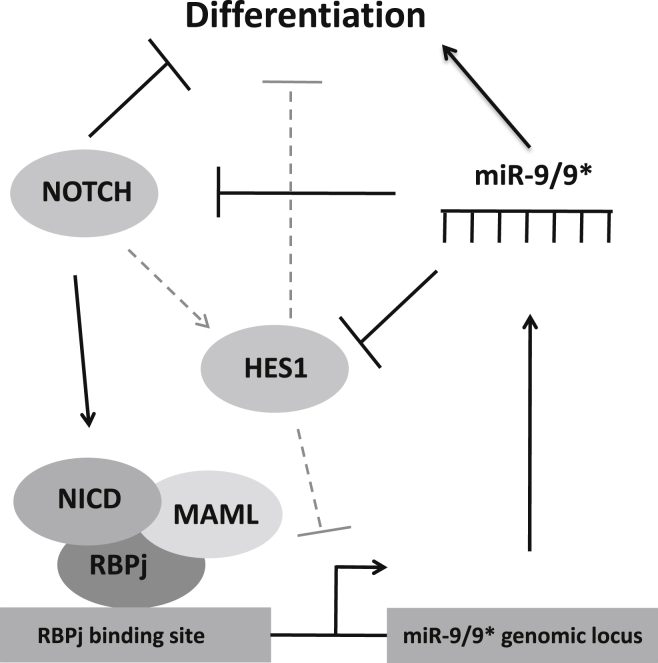
Considering their ability to regulate more than one target gene at once, miRNAs represent optimal candidates to fine-tune key signaling pathways such as Notch in a stringent manner. In addition, previous studies in animal models indicated a potential connection between the Notch signaling cascade and brain-enriched miR-9/9∗. In mouse, expression of Notch1 and Hes1 was shown to change depending on miR-9 levels (Bonev et al., 2012, Jing et al., 2011). We were interested in whether this interaction is also operational in human cells. Interestingly, our data show that, upon miR-9/9∗ overexpression, NOTCH1 mRNA levels are not downregulated in the human neural stem cells studied here. On the other hand, we confirmed that also in human neural cells HES1 is a target of miR-9, in line with what has been described for its homologs in frog (Bonev et al., 2011), mouse (Bonev et al., 2012), and zebrafish (Coolen et al., 2012). Particularly in mouse, Bonev et al. (2012) have proposed the existence of a double-negative feedback loop between Hes1 and miR-9, which regulates their oscillatory expression and activity during neural development. We further identified NOTCH2 as an additional target of miR-9/9∗, which adds a second loop to the miR-9/9∗-based regulation of Notch activity in neural stem cells (Figure 5).
Figure 5.
Reciprocal Interaction between miR-9/9∗ and the Notch Pathway
Proposed model for the identified interactions between miR-9/9∗ and the Notch pathway according to recent findings. Dashed lines indicate information integrated from studies in other model organisms (zebrafish: Coolen et al., 2012; mouse: Bonev et al., 2012).
The phenotype induced by modulating the activity of a single miRNA is rarely due to a single target. The effect of a miRNA is rather the result of concomitantly fine-tuning the expression of several targets. In this regard our study is emblematic, showing how bifunctional miR-9/9∗ mildly (2-fold downregulation) impact on multiple members of the Notch pathway (i.e., NOTCH2 and HES1; Figure 5). Yet the cumulative effect on these different targets turns out to be functionally significant for neural stem cell homeostasis. In addition, according to target prediction analysis, miR-9/9∗ might even regulate more components of the Notch signaling cascade (Table S1) or cooperative factors, such as FoxO1, which was recently identified as a target of miR-9 in mouse adult neural stem cells (Kim et al., 2015). These potential additional targets could further contribute to the effects elicited by miR-9/9∗ overexpression. Together with studies in other model organisms, our work points to bifunctional miR-9/9∗ as an evolutionarily conserved modulator of the Notch signaling pathway in the context of neural stem cell development and maintenance. However, in addition to regulating Notch signaling, miR-9 and miR-9∗ may modulate other major pathways as was shown, e.g., for JAK-STAT (Kim et al., 2011, Zhao et al., 2015). Thus, miR-9/9∗ might function as regulatory node mediating crosstalk among different signaling cascades within the same cell.
Reciprocal Regulation of miR-9/9∗ Expression by Notch Activity
Because miR-9/9∗ is a known pro-neurogenic factor (Laneve et al., 2010, Packer et al., 2008, Yoo et al., 2009, Zhao et al., 2009), its positive regulation by anti-neurogenic Notch in human neural stem cells was initially unexpected. However, this is not the first case whereby a developmentally relevant gene was found to promote expression of its own negative regulator as shown for SOX2 and the miR-200 family (Peng et al., 2012). We were interested in whether Notch directly impacts on miR-9/9∗ expression in a similar fashion. As modulation of Notch activity resulted in changes in levels of miR-9, miR-9∗, and their primary transcript pri-miR-9_2 in both lt-NES cells and smNPCs, we searched for transcription factor binding sites in the promoter regions driving miR-9/9∗ expression. In previous studies, it was reported that the distance of Pol-II enrichment in the genomic regions near pre-miRNAs varies between 0 and 40 kb (Corcoran et al., 2009). For our investigation of predicted RBPj binding sites we particularly searched within the 10-kb region upstream of pre-miR-9/9∗, as this was proposed to be the average distance of the Pol-II peaks from pre-miR locations (Corcoran et al., 2009). We identified two RBPj binding sites upstream of the miR-9_2 genomic locus (Figure S4B) and demonstrated by ChIP assay that the NICD/RBPj complex is recruited to these sites. Taken together, these data strongly suggest that Notch contributes to the transcriptional control of miR-9/9∗ in human neural stem cells, similar to the findings recently made in zebrafish (Coolen et al., 2012).
Relevance of Fine-Tuning Effects of Notch Activity in Development and Homeostasis
In a recent report we have shown that in lt-NES-cell overexpression of miR-9/9∗ leads to impaired self-renewal and premature neuronal differentiation (Roese-Koerner et al., 2013). Interestingly, similar effects were observed upon inhibition of the Notch signaling cascade (Borghese et al., 2010). Antagonistic roles for miR-9/9∗ and Notch were also suggested by a number of studies in model organisms. Knockout of the miR-9_2 and miR-9_3 genomic loci leads to a reduction in early-born neurons and increased proliferation of neural progenitors in the murine subpallium (Shibata et al., 2011). In turn, ablation of Notch activity in murine neuroepithelial cells leads to premature neuronal differentiation (Lutolf et al., 2002).
So far, little is known about these regulatory mechanisms in humans due to limited access to the developing human nervous system. In this context, our study employs the potential of lt-NES cells to address such questions—at a reductionist level—in human neuroepithelial stem cells. This robust cell-culture model opened a window enabling us to catch a glimpse of mechanisms underlying human neurogenesis integrating knowledge gained in model organisms. The data gathered in lt-NES cells demonstrate the existence of a negative feedback mechanism between bifunctional miR-9/9∗ and the whole Notch pathway in human neural stem cells. To explore whether this feedback mechanism also applies to other neural stem cell populations, we extended our study to a recently described small-molecule-induced neural precursor cell population (smNPCs; Reinhardt et al., 2013). Although both cell populations represent multipotent neural stem cells, they differ with respect to their growth pattern, regional identity, and differentiation propensity. Compared with lt-NES cells, smNPCs do not form rosettes but grow as dense, non-polarized cell clusters. In addition, smNPCs readily give rise to both CNS and neural crest derivatives (Reinhardt et al., 2013). The fact that both cell populations show the same reciprocal interaction between miR-9/9∗ and Notch suggests that this regulatory loop is not a specific feature of lt-NES cells but is a more general mechanism calibrating the balance between self-renewal and differentiation of neural stem cells.
While Notch contributes to pri-miR-9/9∗ transcription, mature miR-9 and miR-9∗ repress, for example, NOTCH2, HES1, and perhaps further members of the Notch pathway (Table S1). The functional relevance of this interaction is reflected by the fact that enhancement of neuronal differentiation induced by overexpression of miR-9/9∗ in lt-NES cells is abolished by overexpression of Notch2-ICD. In line with this, the impairment of neuronal differentiation resulting from inhibition of miR-9/9∗ activity is abolished by inhibition of Notch signaling via DAPT treatment. This reciprocal regulation may help to tightly control the delicate balance between neural stem cell self-renewal and differentiation, thereby likely contributing to regulate the time point when cells commit to neurogenesis. It is tempting to speculate that it might even prevent stem cells from crossing the border between self-renewing proliferation and tumorigenic growth.
In summary, our data generated in different human neural stem cell populations show that miR-9/9∗ target the Notch downstream effector HES1 and that NOTCH2 is a functional target of miR-9/9∗. Conversely, our experimental observations strongly suggest that Notch activity regulates miR-9/9∗ expression via direct interaction of its transcriptional complex with the promoter region of the miR-9_2 genomic locus. Together, these data indicate that the Notch signaling pathway and miR-9/9∗ engage in reciprocal regulation, which directs human neural stem cell self-renewal and differentiation (Figure 5).
Experimental Procedures
Cell Culture and Generation of Stably Transduced Cell Lines
Human lt-NES cells (derived from I3 or H9.2 ES cell lines) were derived as previously described (Koch et al., 2009) with some modifications as described in Supplemental Experimental Procedures. Lt-NES cells were propagated on polyornithine/laminin-coated dishes in proliferation medium (N2-medium containing 1:1,000 B27 supplement [Invitrogen], 10 ng/ml FGF2, and 10 ng/ml EGF [both R&D systems]) and split every 2–3 days with trypsin in a ratio of 1:2 to 1:3. Neuronal differentiation was performed on Matrigel-coated dishes (BD Biosciences) in medium devoid of growth factors, as previously described (Koch et al., 2009). Human iPS cell-derived smNPCs (small-molecule neural precursor cells) were established and propagated as described by Reinhardt et al. (2013) (see also Supplemental Experimental Procedures).
For pharmacological inhibition of the Notch signaling pathway, DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester, Sigma-Aldrich) was dissolved in DMSO (Sigma-Aldrich) and used at a final concentration of 5 μM. DMSO (0.1%) was used as vehicle control.
We genetically modified our cells to overexpress the miR-9_1 locus, human NOTCH1-ICD, NOTCH2-ICD (Capobianco et al., 1997), DN-MAML1-GFP (Weng et al., 2003), or GFP alone as control using doxycycline-inducible lentiviral vectors. For further details on cloning and lentiviral transduction, please refer to Supplemental Experimental Procedures.
Luciferase Assay
The dual luciferase constructs were generated by cloning an miR-9 binding site or the 3′ UTR NOTCH1, NOTCH2, and HES1, respectively into the multiple cloning site downstream of the Renilla luciferase cassette in the psiCHECK2 vector (Promega). The primers used for cloning are listed in Table S2. Cells were transfected with 25 ng/ml of the suitably modified psiCHECK2 vectors described above, using Lipofectamine 2000 (Invitrogen). After 24 hr the cells were lysed using passive lysis buffer. The lysates were then analyzed on a microplate luminometer (Berthold) using substrates for firefly luciferase (0.5 mM D-luciferin, PJK) and Renilla luciferase (0.04 mM colenterazine, PJK). Changes in Renilla luciferase activity were normalized to firefly activity.
Transfection of Synthetic miRNA Mimics or Inhibitors
Lt-NES cells were seeded to reach 70% confluence at the time of transfection. Cells were transfected with a final concentration of 5 nM miScript miRNA mimic (Qiagen) or AllStars negative control siRNA (Qiagen), or 50 nM miScript miRNA inhibitor (Qiagen) or negative control inhibitor (Qiagen), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Medium was changed 4 hr after transfection. Transfections were performed every other day.
Real-Time qRT-PCR Analyses
Total RNA samples were extracted using peqGOLD TriFast (Peqlab), DNaseI treated (Invitrogen), and quantified by Nanodrop (Thermo Scientific) before generating cDNA by reverse transcription. Real-time qRT-PCRs were performed on an Eppendorf Mastercycler using the SYBR Green detection method. Specifications for miRNA and mRNA qRT-PCR analyses are described in Supplemental Experimental Procedures.
Semi-quantitative PCR Analyses of Pri-miRNAs
Semi-quantitative PCR analyses were carried out on cDNA produced with an iScript Reverse Transcription Kit (Bio-Rad) using Taq-DNA Polymerase (Invitrogen) according to the manufacturer's protocol. Fetal brain mRNA was purchased from Stratagene (Agilent). Primers used are listed in Table S3.
Chromatin Immunoprecipitation
ChIP was carried out in triplicates with the Magna ChIP G Kit (Millipore) according to the manufacturer's protocol. The antibodies used were anti-RPBj (Abcam, ab25949; 1:100) and anti-Flag-Tag (Millipore, MAB3118; 1:100).
Subsequent semi-quantitative PCR analyses were carried out with Taq-DNA polymerase (Invitrogen) according to the manufacturer's protocol. Primers used are listed in Table S4.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde (Sigma-Aldrich), permeabilized with 0.1% Triton X-100 (Sigma-Aldrich), blocked with 10% fetal calf serum (FCS) (Invitrogen), and stained with primary antibody (βIII-tubulin, Covance, MMS-435P; 1:2,000) and secondary antibody (Cy3-goat-anti-mouse, Jackson ImmunoResearch, 115-165-044; 1:1,000) in 5% FCS in PBS (Invitrogen) overnight. Fixed samples were counterstained with DAPI (Sigma-Aldrich). Specifications for image analysis are given in Supplemental Experimental Procedures.
Western Blot
Protein samples were obtained from I3 lt-NES cells after 4 days of doxycycline treatment using RIPA buffer containing Proteinase Inhibitor cocktail (Calbiochem). 40 μg of protein extracts were loaded onto an SDS-PAGE gel, run at 100 V for 2 hr, and blotted onto a nitrocellulose membrane (Carl-Roth) at 70 V for 2 hr. The membrane was blocked with 5% milk powder and incubated with the primary antibody. Detection was carried out with a peroxidase-conjugated secondary antibody and Millipore ECL solutions according to the manufacturer's protocol. Further details for the specific antibodies are given in Supplemental Experimental Procedures.
Northern Blot
Non-radioactive northern blot was carried out with 20–40 μg of total RNA sample as described in Stappert et al. (2013). SnRNA U6 was used as loading control.
Statistical Analysis
All experiments were carried out in at least three independent experiments (n). Statistical analysis was carried out using the GraphPad Prism 6 (GraphPad Software). Results from Student's t test are stated as ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001. The number of independent replicates (n) as well as statistical analyses for each experiment is stated in the figure legends.
Target Prediction
miRNA target prediction was performed with the MiRWalk database, which is available online (Dweep et al., 2011). Five prediction algorithms (miRanda, miRDB, miRWALK, RNA22, and Targetscan) were chosen for comparison. Table S1 lists the number of algorithms predicting binding of the miRNA analyzed to the target mRNA.
Author Contributions
B.R.-K., conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; L.S., conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; T.B., collection and assembly of data, data analysis and interpretation; N.C.B., collection and assembly of data, data analysis and interpretation; M.V., collection and assembly of data, data analysis; J.J., provision of study material and cell-culture protocols, manuscript reviewing; B.O.E., provision of study material, data analysis and interpretation, manuscript review; M.P., provision of study material, manuscript review; L.B., conception and design, data analysis and interpretation, manuscript writing, financial support; O.B., conception and design, data interpretation, manuscript writing, financial support, final approval of the manuscript.
Acknowledgments
We thank Prof. J. Itskovitz-Eldor (Technion, Israel Institute of Technology, Haifa, Israel) for providing the human ES cell lines I3 and H9.2 from which the lt-NES cells used in this work were derived, Prof. A.J. Capobianco (The Wistar Institute, Philadelphia, PA, USA) for donating the N1ICD and N2ICD constructs, Prof. J.C. Aster (Brigham and Women's Hospital, Boston, MA, USA) for donating the DN-MAML1 construct, Dr. J. Mertens for providing the modified pLVX-tetOn Advanced vector, L. Roese-Koerner for help with the bioinformatics analysis, and Beatrice Weykopf and Fabian Gather for their help with the Western blot analysis. We also thank Kaveri Banerjee and Katja Hamann for outstanding technical support. This work was supported by the EU (FP7-HEALTH-2007-B-22943-NeuroStemcell; HEALTH-F4-2013-602278-NeuroStemCellRepair), BMBF 0315799 (BIODISC), the Ministry of Innovation, Science and Research of the State of North Rhine Westphalia (project StemCellFactory II; #005-1403-0102), the DFG (EV143/1-1), BONFOR, and the Hertie Foundation. O.B. is co-founder of and has stock in LIFE & BRAIN GmbH.
Published: July 14, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.06.008.
Supplemental Information
References
- Andersson E.R., Sandberg R., Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Pisco A., Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Stanley P., Papalopulu N. MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A., Brüstle O. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells. 2010;28:955–964. doi: 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- Capobianco A.J., Zagouras P., Blaumueller C.M., Artavanis-Tsakonas S., Bishop J.M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P., Skupien P., Hesl B., Coolen M., Moore J.C., Madelaine R., Kremmer E., Faus-Kessler T., Blader P., Lawson N.D. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- Coolen M., Bally-Cuif L. MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Coolen M., Thieffry D., Drivenes O., Becker T.S., Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell. 2012;22:1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Corcoran D.L., Pandit K.V., Gordon B., Bhattacharjee A., Kaminski N., Benos P.V. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H., Sticht C., Pandey P., Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Falk A., Koch P., Kesavan J., Takashima Y., Ladewig J., Alexander M., Wiskow O., Tailor J., Trotter M., Pollard S. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One. 2012;7:e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F.B. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guentchev M., McKay R.D. Notch controls proliferation and differentiation of stem cells in a dose-dependent manner. Eur. J. Neurosci. 2006;23:2289–2296. doi: 10.1111/j.1460-9568.2006.04766.x. [DOI] [PubMed] [Google Scholar]
- Harper J.A., Yuan J.S., Tan J.B., Visan I., Guidos C.J. Notch signaling in development and disease. Clin. Genet. 2003;64:461–472. doi: 10.1046/j.1399-0004.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- Hebert S.S., De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Inui M., Martello G., Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Jeon H.M., Sohn Y.W., Oh S.Y., Kim S.H., Beck S., Kim S., Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9∗-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- Jing L., Jia Y., Lu J., Han R., Li J., Wang S., Peng T. MicroRNA-9 promotes differentiation of mouse bone mesenchymal stem cells into neurons by Notch signaling. Neuroreport. 2011;22:206–211. doi: 10.1097/WNR.0b013e328344a666. [DOI] [PubMed] [Google Scholar]
- Kim T.M., Huang W., Park R., Park P.J., Johnson M.D. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Hwang I., Muller F.L., Paik J.-H. Functional regulation of FoxO1 in neural stem cell differentiation. Cell Death Differ. 2015;22:2034–2045. doi: 10.1038/cdd.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P., Opitz T., Steinbeck J.A., Ladewig J., Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A.M., King K.S., Donahue C.P., Khrapko K., Kosik K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A.M., Sonntag K.C., Isacson O., Kosik K.S. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Laneve P., Gioia U., Andriotto A., Moretti F., Bozzoni I., Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010;38:6895–6905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky J.L., Wu H. Notch signaling, brain development, and human disease. Pediatr. Res. 2005;57:104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J.A., Gao F.B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lutolf S., Radtke F., Aguet M., Suter U., Taylor V. Notch1 is required for neuronal and glial differentiation in the cerebellum. Development. 2002;129:373–385. doi: 10.1242/dev.129.2.373. [DOI] [PubMed] [Google Scholar]
- Mohammadi-Yeganeh S., Mansouri A., Paryan M. Targeting of miR9/NOTCH1 interaction reduces metastatic behavior in triple-negative breast cancer. Chem. Biol. Drug Des. 2015;86:1185–1191. doi: 10.1111/cbdd.12584. [DOI] [PubMed] [Google Scholar]
- Packer A.N., Xing Y., Harper S.Q., Jones L., Davidson B.L. The bifunctional microRNA miR-9/miR-9∗ regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Li N., Ng Y.K., Zhang J., Meier F., Theis F.J., Merkenschlager M., Chen W., Wurst W., Prakash N. A unilateral negative feedback loop between miR-200 microRNAs and Sox2/E2F3 controls neural progenitor cell-cycle exit and differentiation. J. Neurosci. 2012;32:13292–13308. doi: 10.1523/JNEUROSCI.2124-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierfelice T.J., Schreck K.C., Eberhart C.G., Gaiano N. Notch, neural stem cells, and brain tumors. Cold Spring Harb. Symp. Quant. Biol. 2008;73:367–375. doi: 10.1101/sqb.2008.73.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierfelice T., Alberi L., Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Radtke F., Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Hoing S., Moritz S., Parga J.A., Wagner L., Bruder J.M. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roese-Koerner B., Stappert L., Koch P., Brüstle O., Borghese L. Pluripotent stem cell-derived somatic stem cells as tool to study the role of MicroRNAs in early human neural development. Curr. Mol. Med. 2013;13:707–722. doi: 10.2174/1566524011313050003. [DOI] [PubMed] [Google Scholar]
- Schratt G. Fine-tuning neural gene expression with microRNAs. Curr. Opin. Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Shibata M., Nakao H., Kiyonari H., Abe T., Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert L., Borghese L., Roese-Koerner B., Weinhold S., Koch P., Terstegge S., Uhrberg M., Wernet P., Brüstle O. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PLoS One. 2013;8:e59011. doi: 10.1371/journal.pone.0059011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert L., Roese-Koerner B., Brüstle O. The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 2014;359:47–64. doi: 10.1007/s00441-014-1981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchorz J.S., Tome M., Cloetta D., Sivasankaran B., Grzmil M., Huber R.M., Rutz-Schatzmann F., Kirchhoff F., Schaeren-Wiemers N., Gassmann M. Constitutive Notch2 signaling in neural stem cells promotes tumorigenic features and astroglial lineage entry. Cell Death Dis. 2012;3:e325. doi: 10.1038/cddis.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A.P., Nam Y., Wolfe M.S., Pear W.S., Griffin J.D., Blacklow S.C., Aster J.C. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Yoo A.S., Staahl B.T., Chen L., Crabtree G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuva-Aydemir Y., Simkin A., Gascon E., Gao F.B. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8:557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Lin Q., Kim K.J., Dardashti F.D., Kim J., He F., Sun Y. Ngn1 inhibits astrogliogenesis through induction of miR-9 during neuronal fate specification. Elife. 2015;4:e06885. doi: 10.7554/eLife.06885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.