Abstract
Background
Anopheles funestus is one of the major malaria vectors in tropical Africa, notably in Senegal. The highly anthropophilic and endophilic behaviours of this mosquito make it a good target for vector control operations through the use of insecticide treated nets, long-lasting insecticide nets and indoor residual spraying. However, little is known about patterns of resistance to insecticides and the underlying resistance mechanisms in field populations of this vector in Senegal.
Methods
Here, we assessed the susceptibility status of An. funestus populations from Gankette Balla, located in northern Senegal and investigated the potential resistance mechanisms.
Results
WHO bioassays indicated that An. funestus is resistant to lambda-cyhalothrin 0.05 % (74.64 % mortality), DDT 4 % (83.36 % mortality) and deltamethrin 0.05 % (88.53 % mortality). Suspected resistance was observed to permethrin 0.75 % (91.19 % mortality), bendiocarb 0.1 % (94.13 % mortality) and dieldrin 4 % (96.41 % mortality). However, this population is fully susceptible to malathion 5 % (100 % mortality) and fenitrothion 1 % (100 % mortality). The microarray and qRT-PCR analysis indicated that the lambda-cyhalothrin resistance in Gankette Balla is conferred by metabolic resistance mechanisms under the probable control of cytochrome P450 genes among which CYP6M7 is the most overexpressed. The absence of overexpression of the P450 gene, CYP6P9a, indicates that the resistance mechanism in Senegal is different to that observed in southern Africa.
Conclusions
This study represents the first report of pyrethroid and DDT resistance in An. funestus from Senegal and shows that resistance to insecticides is not only confined to An. gambiae as previously thought. Therefore, urgent action should be taken to manage the resistance in this species to ensure the continued effectiveness of malaria control.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-016-1735-7) contains supplementary material, which is available to authorized users.
Keywords: Anopheles funestus, Insecticide resistance, Lambda-cyhalothrin, Resistance mechanisms, Senegal
Background
The burden of malaria remains heaviest in the WHO African region, where an estimated 90 % of all malaria deaths occur, and in children aged under five years, who account for 78 % of all deaths [1]. In Senegal, malaria is a major cause of morbidity and mortality and a high priority for the government, even though the number of reported cases of malaria has dropped significantly since 2007–2008 [2]. As in many African countries, malaria control in Senegal relies heavily on vector control through the use of long-lasting insecticide nets (LLINs) and indoor residual spraying (IRS). However, resistance to the main insecticides in the major malaria vectors such as Anopheles funestus is threatening the success of these control interventions. Resistance to different classes of insecticides used in public health is increasingly reported across Africa in An. funestus with fear that this could disrupt control programs against this vector. Indeed, resistance to pyrethroids, DDT and carbamates has been reported in different regions of Africa including southern Africa [3–6], Central Africa [7], East Africa [8, 9] and West Africa [10, 11]. In Senegal, pyrethroid resistance until recently was mainly reported in An. gambiae [12–14] while little is known about the susceptibility of An. funestus to insecticides. Senegal is currently scaling up its malaria control program through LLINs and IRS [15, 16]. It is crucial that information on susceptibility to main insecticides used in public health and the underlying mechanisms be investigated. This will properly inform control programs of the most suitable insecticides to use and facilitate the design of appropriate resistance management strategies. In this study, we report the assessment of the susceptibility of one An. funestus population from northern Senegal to several insecticides used in public health and also investigate the underlying molecular mechanisms conferring resistance to lambda-cyhalothrin, a key pyrethroid insecticide used for IRS. This information will fill the gap in our knowledge on the resistance distribution in An. funestus and help to improve future control programs on this species in Senegal.
Methods
Study site and mosquito collection
Blood-fed An. funestus adult females resting indoor were collected in houses between 7.00 am and 15.00 pm in November 2011, in Gankette Balla (15°58′N, 15°55′W), which is located in the Lake Guiers area, in northern Senegal, West Africa. Blood-fed and gravid mosquitoes resting inside houses were collected using aspirators and torches and kept in small cups, covered by a non-treated net, until fully gravid and transported to the insectary where they were allowed to lay eggs [17] and hatch in larvae bowls for rearing. The egg batches were pooled and reared together and the F1 adults generated were randomly mixed in cages for subsequent experiments.
Species identification
All females used for individual oviposition were morphologically identified as belonging to the An. funestus group [18]. Genomic DNA was extracted from head and thorax using the Livak protocol as previously described [19]. A cocktail PCR was carried to confirm that all females that laid eggs were An. funestus (s.s.) [20].
Adult mosquito susceptibility assays
Insecticide susceptibility assays were carried out using 2–5 day-old F1 adults following the WHO protocol [21]. Approximately 20–25 mosquitoes per tube with 4–6 replicates were exposed to insecticide-impregnated filter paper for 1 h or control paper and then transferred to a clean holding tube supplied with 10 % sugar and mortality was determined 24 h post-exposure. We tested the following insecticides: the pyrethroids permethrin (0.75 %), lambda-cyhalothrin (0.05 %) and deltamethrin (0.05 %); the carbamate bendiocarb (0.01 %); the organophosphate malathion (5 %) and fenitrothion (5 %) and the organochlorines DDT (4 %) and dieldrin (4 %).
Microarray analysis
A custom microarray chip [3] containing 8 × 60 k probes (60mer) (A-MEXP-2374) was used to identify the set of genes associated with lambda-cyhalothrin resistance [22]. RNA was extracted from three batches of 10 An. funestus females that were 2–5 day-old from the following sample sets: alive after exposure to 0.75 % lambda-cyhalothrin (R); unexposed to insecticides and thus representative of the wild-type population (C); and unexposed mosquitoes from the fully susceptible laboratory strain FANG (S). RNA was isolated using the Picopure RNA isolation kit (Arcturus, Applied Biosystems, Carlsbad, CA, USA). The quantity and quality of extracted RNA were assessed using a NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Bioanalyzer (Agilent, Santa Clara, CA, USA), respectively. cRNA of each sample was amplified using the Agilent Quick Amp labeling kit (two-colour) following the manufacturer’s protocol. cRNA from the resistant samples (R) were labelled with cy5 dye and cRNA from the control (C) were labelled with both cy3 and cy5, whereas the susceptible strain FANG (S) was labelled with the cy3 dye. cRNA quantity and quality were checked after labelling using the NanoDrop spectrophotometer and Bioanalyzer. Labelled cRNAs were hybridized to the arrays for 17 h at 65 °C according to the manufacturer’s protocol. Five hybridizations for each of the comparisons (R-S, R-C and C-S) were carried out by swapping the biological replicates. Microarray data were analysed using Genespring GX 12.0 software. To identify differentially expressed genes, a cut-off of 1.5 (R-C) and 2-fold-change (FC) (R-S and C-S) and a statistical significance of P ≤ 0.05 with Benjamin-Hochberg correction for multiple testing were applied.
Transcription profiling of candidate metabolic resistance genes (qRT-PCR)
The main candidates resistance genes (Additional file 1: Table S1) that were highly overexpressed from the microarray analysis were assessed by quantitative Reverse Transcriptase PCR (qRT-PCR) to validate their expression pattern using three biological replicates (10 females F1 for each replicate) for lambda-cyhalothrin resistant mosquitoes (R) (alive after 24 h exposure to lambda-cyhalothrin), control mosquitoes (C) (not exposed to any insecticide) and susceptible FANG mosquitoes (S). One microgram of total RNA from each of the three biological replicates for resistant (R), control (C), and FANG (S) was used as a template for cDNA synthesis using the Super-Script III (Invitrogen, Carlsbad, CA, USA) with oligo-dT20 and RNase H, according to the manufacturer’s instructions. A serial dilution of cDNA was used to establish standard curves for each gene to assess PCR efficiency and quantitative differences between samples. The q-RT-PCR amplification was carried out in a MX3005 real-time PCR system (Agilent, Santa Clara, CA, USA) using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent). A total of 10 ng of cDNA from each sample was used as template in a three-step program involving a denaturation at 95 °C for 3 min followed by 40 cycles of 10 s at 95 °C and 10 s at 60 °C and a last step of 1 min at 95 °C, 30 s at 55 °C, and 30 s at 95 °C. The relative expression and fold-change of each target gene in R and C relative to S was calculated according to the 2−ΔΔCT method incorporating PCR efficiency [23] after normalization with the housekeeping RSP7 (ribosomal protein S7; AFUN007153-RA) and the Actin (AFUN006819) genes.
Investigation of the role of knockdown resistance mutation in lambda-cyhalothrin resistance
A fragment spanning a portion of the voltage-gated sodium channel gene (VGSC), including the 1014 codon associated with resistance in An. gambiae, was amplified using the KdrFunF2/KdrFunR2 primers [9, 11, 24] and sequenced (KdrFunR2) in ten field-collected An. funestus female mosquitoes from Gankette Balla. PCR, sequencing and analysis were carried out as previously described [9, 11, 24].
Results
Mosquito collection
More than 450 blood-fed An. funestus females were collected inside houses in the village of Gankette Balla over a period of four days in November 2011. Around 150 laid eggs and all were confirmed to be An. funestus (s.s.) by PCR.
Insecticide susceptibility
A total of 2,322 F1An. funestus adults from Gankette Balla were generated and exposed to various insecticides (Additional file 2: Table S2). The bioassay performed indicated that the An. funestus population females were resistant to the type II pyrethroids lambda-cyhalothrin and deltamethrin as well as to the organochlorine DDT with mortality rates of 74.64 %, 88.53 % and 83.36 %, respectively. Suspected resistance was observed to permethrin, bendiocarb and dieldrin with mortality rates of 91.19 %, 94.13 % and 96.41 %, respectively. However, this population was fully susceptible to the organophosphates malathion and fenitrothion with a mortality rate of 100 %. The males of this An. funestus population were generally susceptible to exposed insecticides except for DDT and lambda-cyhalothrin for which a moderate resistance was observed with mortality rates of 90.30 % and 97.30 %, respectively (Fig. 1).
Fig. 1.
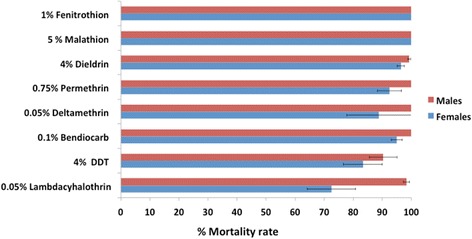
Insecticide resistance profile of Anopheles funestus populations from Gankette Balla
Genome-wide microarray-based transcription analysis of lambda-cyhalothrin resistance
The total number of differentially expressed probes for the comparison between lambda-cyhalothrin resistant samples and the susceptible strain FANG (R-S) was 7,897 (3,789 overexpressed); 7,768 (4,055 overexpressed) for the comparison between the control wild type samples (C) and the susceptible strain FANG (C-S); 115 (59 overexpressed) for the comparison between lambda-cyhalothrin resistant samples and the control wild type samples R-C) (Fig. 2). Fourteen probes were commonly differentially expressed in the three types of comparison (R-S, C-S and R-C). However, 5,388 probes were commonly differentially expressed in the R-S and C-S comparisons, whereas 44 and 18 probes were respectively commonly differentially expressed in the pairs of comparisons R-S vs R-C and C-S vs R-C (Fig. 2).
Fig. 2.
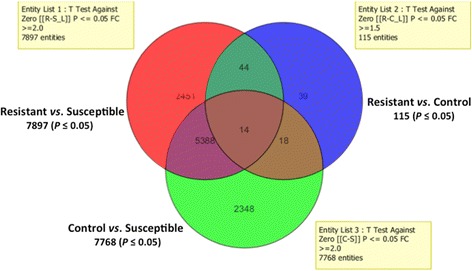
Summary of probes differentially expressed in Lambda-cyhalothrin resistance
Genes commonly overexpressed in R-S, C-S and R-C comparisons
A probe for the CYP6M7 transcript, belonging to the cytochrome P450 gene family, was the most commonly overexpressed detoxification gene in the R-S (FC 101.64) and C-S (FC 120.09) comparisons. This gene was also significantly overexpressed in the R-C comparison but with a much lower FC value (2.59) (Table 1). Other cytochrome P450s genes that were also commonly overexpressed in the three comparisons were three other P450 genes including CYP6AH1 (combined_c1486), CYP304b1 and CYP4C36 (Afun007127). The transcripts Afun008293 and Afun009227 belonging to trypsin-related protease and argininosuccinate lyase genes respectively, were commonly highly overexpressed in R-S and C-S comparisons and significantly overexpressed in R-C. Transcripts belonging to others detoxification genes family such as glutathione transferase (GSTd3), abc transporter (Afun015523), carboxylesterase (Afun011942) were also significantly commonly overexpressed in the three comparisons (Table 1).
Table 1.
Transcripts from detoxification genes with lambda-cyhalothrin resistance upregulated in various comparisons (R-C, R-S and C-S) in Gankette Balla
| Probe name | Transcript | R-C | R-S | C-S | Description |
|---|---|---|---|---|---|
| CUST_7663_PI426302897 | CYP6M7 | 2.59 | 101.64 | 120.09 | cytochrome p450 |
| CUST_2949_PI406199769 | combined_c1486 (CYP6AH1) | 1.52 | 5.59 | 5.27 | cytochrome p450 |
| CUST_12197_PI426302897 | CYP304b1 | 1.61 | 4.44 | 2.74 | cytochrome p450 |
| CUST_7127_PI426302897 | Afun007127 (CYP4C36) | 2.17 | 3.23 | 2.90 | cytochrome p450 |
| CUST_8293_PI426302897 | Afun008293 | 1.78 | 79.77 | 63.27 | trypsin-related protease |
| CUST_13921_PI426302897 | Afun013921 | 2.21 | 34.28 | 25.55 | chymotrypsin 1 |
| CUST_8354_PI426302897 | GSTD3 | 1.52 | 6.33 | 4.69 | glutathione transferase (agap004382-pa) |
| CUST_7773_PI426302897 | Afun007773 | 1.57 | 6.06 | 4.00 | microsomal glutathione s-transferase |
| CUST_9227_PI426302897 | Afun009227 | 1.79 | 29.78 | 42.82 | argininosuccinate lyase |
| CUST_15523_PI426302897 | Afun015523 | 1.55 | 8.39 | 8.34 | abc transporter |
| CUST_14150_PI426302897 | Afun014150 | 1.64 | 7.51 | 4.70 | oxidative stress-induced growth |
| CUST_11942_PI426302897 | Afun011942 | 1.63 | 3.47 | 3.20 | carboxylesterase |
| CUST_376_PI406199788 | gb-CYP4H25 | 10.60 | 4.99 | cytochrome p450 | |
| CUST_12777_PI426302897 | CYP4C27 | 9.98 | 21.93 | cytochrome p450 | |
| CUST_4223_PI426302897 | CYP4H17 | 9.82 | 4.86 | cytochrome p450 | |
| CUST_12343_PI426302897 | CYP4H17 | 6.46 | 3.90 | cytochrome p450 | |
| CUST_12197_PI426302897 | Afun012197 (CYP304B1) | 4.44 | 2.74 | cytochrome p450 | |
| CUST_7369_PI426302897 | Afun007369 (CYP6P9b) | 4.04 | 4.68 | cytochrome p450 | |
| CUST_9312_PI426302897 | Afun009312 | 25.95 | 38.54 | high affinity gaba transporter | |
| CUST_3489_PI406199769 | combined_c1762 | 4.48 | 3.52 | abc transporter | |
| CUST_5545_PI426302897 | Afun005545 | 14.97 | 18.72 | ankyrin repeat domain protein | |
| CUST_3386_PI426302897 | Afun003386 | 5.50 | 4.54 | ankyrin repeat domain-containing protein 50 | |
| CUST_10406_PI426302897 | Afun010406 | 4.15 | 3.86 | ankyrin repeat domain-containing protein 50 | |
| CUST_1930_PI426302897 | Afun001930 | 4.39 | 3.88 | ankyrin repeat-containing | |
| CUST_3672_PI426302897 | Afun003672 | 4.35 | 4.19 | multiple ankyrin repeats single kh d. protein | |
| CUST_1459_PI406199769 | combined_c738 | 9.17 | 10.93 | short-chain dehydrogenase | |
| CUST_2520_PI406199772 | CD578141.1 | 5.54 | 6.72 | short-chain dehydrogenase | |
| CUST_10105_PI426302897 | Afun010105 | 3.38 | 2.44 | short-chain dehydrogenase | |
| CUST_12461_PI426302897 | Afun012461 | 8.67 | 3.72 | alcohol dehydrogenase | |
| CUST_665_PI406199788 | gb-NADH_dehyd | 2.86 | 4.87 | nadh dehydrogenase | |
| CUST_10836_PI426302897 | Esterase b1 | 7.33 | 3.75 | esterase b1 | |
| CUST_11942_PI426302897 | Afun011942 | 3.47 | 3.20 | carboxylesterase | |
| CUST_34_PI406199775 | COEAE6O | 2.41 | 2.05 | carboxylesterase | |
| CUST_13332_PI406199769 | combined_c6826 | 2.06 | 2.33 | esterase b1 | |
| CUST_295_PI406199798 | AGAP000177-RA | 7.06 | 16.73 | cuticle protein 7 | |
| CUST_3736_PI406199772 | CD577515.1 | 5.94 | 3.38 | cuticle protein | |
| CUST_8354_PI426302897 | GSTD3 | 6.33 | 4.69 | glutathione transferase (agap004382-pa) | |
| CUST_7773_PI426302897 | Afun007773 | 6.06 | 4.00 | microsomal glutathione s-transferase | |
| CUST_1870_PI406199769 | combined_c944 | 4.25 | 2.09 | microsomal glutathione s-transferase | |
| CUST_10360_PI426302897 | Afun010360 | 2.63 | 2.84 | glucosyl glucuronosyl transferases | |
| CUST_7499_PI426302897 | GSTd1-5 | 2.55 | 2.48 | glutathione transferase | |
| CUST_8698_PI426302897 | Afun008698 | 5.74 | 25.76 | heat shock protein 70 b2 | |
| CUST_7498_PI426302897 | Afun007498 | 3.69 | 2.81 | heat shock cognate 70 protein | |
| CUST_7302_PI426302897 | Afun007302 | 3.34 | 3.86 | heat shock 70 kda protein cognate 4 | |
| CUST_5336_PI426302897 | Afun005336 | 2.94 | 4.69 | heat shock cognate 70 kda protein | |
| CUST_2701_PI406199769 | combined_c1362 | 2.77 | 2.35 | heat shock protein 70 -interacting protein | |
| CUST_2184_PI406199772 | CD578312.1 | 2.30 | 3.18 | 82 kda heat shock protein | |
| CUST_3246_PI426302897 | Afun003246 | 5.12 | 5.83 | aldehyde oxidase | |
| CUST_11963_PI426302897 | Afun011963 | 4.56 | 4.55 | aldehyde oxidase | |
| CUST_718_PI406199788 | gb-PX4B | 4.32 | 4.43 | oxidase peroxidase | |
| CUST_199_PI426302897 | Afun000199 | 3.04 | 3.46 | chorion peroxidase | |
| CUST_7400_PI426302897 | Afun007400 | 2.57 | 3.12 | thioredoxin-dependent peroxidase | |
| CUST_8347_PI426302897 | Afun008347 | 5.07 | 3.21 | chymotrypsin 1 | |
| CUST_11697_PI426302897 | Afun011697 | 3.04 | 4.34 | chymotrypsin bii | |
| CUST_7894_PI426302897 | Afun007894 | 5.50 | 4.34 | trypsin delta gamma | |
| CUST_1313_PI426302897 | Afun001313 (CYP9J5) | 2.93 | 2.67 | cytochrome p450 | |
| CUST_11293_PI426302897 | Afun011293 | 4.22 | 4.97 | AGAP012443-PA [Anopheles gambiae str. PEST] | |
| CUST_4992_PI426302897 | Afun004992 | 3.46 | 2.61 | AGAP010545-PA [Anopheles gambiae str. PEST] | |
| CUST_9542_PI426302897 | Afun009542 | 2.32 | 2.58 | AGAP000321-PA [Anopheles gambiae str. PEST] | |
| CUST_9600_PI406199769 | combined_c4862 | 1.51 | 2.53 | ankyrin unc44 |
Abbreviations: R-C Resistant-Control, R-S Resistant-Susceptible, C-S Control-Susceptible
Genes commonly overexpressed in R-S and C-S comparisons
Several detoxification genes or resistance-related genes were commonly and significantly overexpressed in the R-S and C-S comparison. A set of five transcripts belonging to cytochrome P450s genes was commonly upregulated in R-S and C-S, with CYP4H25 (FC 10.60; 4.99), CYP4C27 (FC9.98; 21.93) and CYP4H17 (FC9.82; 4.86) being the most overexpressed (Table 1). A transcript (Afun007369) with closest hit to CYP6P9b (92 %) was also overexpressed in both R-S (FC4.04) and C-S (FC4.68). However, none of the three probes designed for CYP6P9a was overexpressed in this population, suggesting a significant difference with southern African populations where this P450 is highly overexpressed. Among the most commonly overexpressed resistance-associated genes in R-S and C-S were the high affinity GABA transporter (Afun009312) and an ankyrin repeat domain protein (Afun005545). Noticeably, the transcripts Afun008698 and AGAP000177-RA belonging to heat shock protein 70 b2 and cuticle protein 7 genes successively were commonly overexpressed with a high FC observed in C-S. Furthermore, several probes from the GSTs gene family were also significantly and commonly overexpressed in R-S and C-S in possible association with the observed DDT resistance in this population. These GST genes include the GSTd1-5 previously also reported in southern African populations of An. funestus [4]. However, GSTe2, the major DDT resistance gene observed in West and Central African countries [4] is overexpressed in this Senegalese population. The other detoxification or resistance related genes are reported in Table 1 and Additional file 3: Table S3 and include short-chain dehydrogenase, esterase b1, aldehyde oxidase and chymotrypsin 1.
Genes commonly overexpressed in R-C and R-S comparisons
Only a limited number of genes were commonly overexpressed in the two comparisons. Indeed, these genes include: a single P450 (CYP9J5), three probes corresponding to An. gambiae orthologous genes, one probe for membrane-associated LPS-inducible TNF-alpha factor protein and zinc metalloproteinase nas-12, successively (Table 1). The top 50 of the most detoxification genes overexpressed only in the comparisons R-S_L (Mosquitoes resistant to lambda-cyhalothrin vs susceptible mosquitoes) and R-C_L (Mosquitoes resistant to lambda-cyhalothrin vs control mosquitoes) are presented in Additional file 4: Tables S4 and Additional file 5: Table S5, respectively.
Genes commonly overexpressed in R-C and C-S comparisons
Only one transcript (combined_c4862) belonging to ankyrin unc44 gene was commonly overexpressed in R-C (FC 1.51) and C-S (FC 2.53) comparisons (Table 1).
Genes commonly underexpressed in lambda-cyhalothrin resistant mosquitoes
The detoxifying gene “CD578215.1” (cuticle protein) was the most commonly underexpressed gene in the R-S_L (FC 24.47), C-S (FC 56.22) and R-C_L (FC 1.52) comparisons. Other genes that were commonly underexpressed in these three comparisons include: combined_c3712 (stress-sensitive b), CD577548.1 (cytochrome c oxidase subunit 1), CD577574.1 (glutathione s-transferase) (Additional file 6: Table S6). The detoxifying gene “Afun013280” (carboxypeptidase n subunit 2) was the most commonly under expressed gene in the R-S_L (FC 29.52) and C-S (FC 16.58) comparisons. Among the others detoxifying gene families commonly underexpressed in R-S_L and CS, we noted the presence of many probes from cytochrome c oxidase subunit III and also from F0 ATP synthase subunit 6 (Additional file 7: Table S7). The top 50 of the most detoxification genes underexpressed only in the comparisons R-S_L and R-C_L are presented in Additional file 8: Table S8 and Additional file 9: Table S9, respectively.
Validation of the microarray upregulation results by qRT-PCR
Quantitative reverse transcription PCR (q-RT-PCR) was used to validate the microarray results for 11 of the most overexpressed detoxification genes. These genes include six cytochrome P450s (CYP6M7, CYP4H17, CYP4C27, CYP6Z3, CYP9J11, CYP304b1), one dehydrogenase (Combined_c738), two glutathione transferases (GSTd3, GSTd1-5), one esterase (EstB1) and one aldehyde oxidase (Ald oxi). The qRT-PCR results confirmed significantly the overexpression of candidate genes at different levels. Same to the microarray results, the cytochrome P450 CYP6M7 was the most overexpressed detoxification gene (FC 11.69) in the R-S comparison (Fig. 3a). A significant correlation between the qRT-PCR and microarray results was observed (R2 = 0.358, P = 0.046) (Fig. 3b).
Fig. 3.
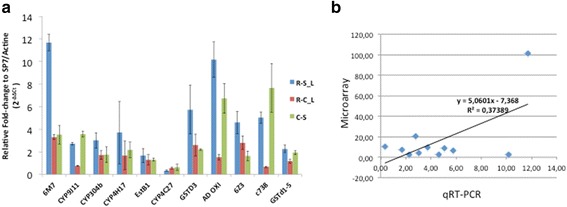
Gene expression analysis. a q-RT PCR expression Profile of selected candidate genes in An. funestus resistant to lambda-cyhalothrin. b Correlation between the microarray and qRT-PCR fold change data for the selected candidates genes
Role of knockdown resistance in lambda-cyhalothrin resistance
PCR products were successfully amplified (994 bp) and sequenced for a fragment (a portion of intron 19 and the entire exon 20, domain II, segment 6) of the VGSC gene in ten field-collected An. funestus female mosquitoes from Gankette Balla. An 868 bp common sequence was aligned for seven individuals detecting 15 polymorphic sites (Fig. 4). Neither the L1014F kdr mutation nor the L1014S mutation was detected in An. funestus from Gankette Balla, as previously reported in other populations of this species [9, 11, 25]. Indeed, the VGSC gene sequencing analysis detected only the TTA 1014 codon indicating that they do not have the L1014F (TTA-to-TTT) or L1014S (TTA-to-TCA) kdr mutation (Additional file 10: Figure S1) commonly found in An. gambiae.
Fig. 4.

Polymorphic sites at the exon 20 fragment of the VGSC gene in wild type An. funestus from Gankette Balla. Only polymorphic sites are shown and these are numbered from the beginning of each aligned sequence. Dots mean identity with the first sequence. A number has been given to each haplotype. The column (N) indicates the number of individuals sharing the haplotype
Discussion
This study provides the first assessment of the susceptibility and mechanisms to the main insecticides used in public health of An. funestus population from Senegal.
Susceptibility / resistance to lambda-cyhalothrin
The results from this study show that An. funestus population from Gankette Balla is resistant to pyrethroids and DDT. In addition, suspected resistance was observed to bendiocarb and dieldrin. Resistance of An. funestus to different classes of insecticides has already been reported in West Africa [10, 11, 26], East Africa [9, 17] and in southern Africa [3, 4, 24, 27]. In Senegal, resistance to pyrethroids and DDT has already been reported in other malaria vectors such as An. gambiae (s.l.) [12–14, 28] and An. pharoensis [28].
On the other hand, our study revealed a resistance pattern where type II pyrethroid (lambda-cyhalothrin / deltamethrin) seems to be more involved compared to type I pyrethroids (permethrin). This resistance profile is different to that observed in An. funestus in Benin [11] where resistance is higher against type I pyrethroids. This difference could suggest the existence of a different resistance mechanism for pyrethroid resistance in Senegal compared to Benin. Indeed, some cytochrome P450s exhibit specificity for either or the two types of pyrethroids [29, 30]. However, the observed resistance profile is similar to that reported in East Africa [17] and South Africa [31]. The full susceptibility observed for malathion is similar for all tests performed so far on An. funestus populations across Africa indicating that this insecticide could be used as an alternative to pyrethroids, carbamates and DDT in Indoor Residual Spraying (IRS) control program directed against this species [32].
The source of the lambda-cyhalothrin resistance observed in Gankette Balla remains unknown although the intense use of this insecticide in the agricultural sector could be a contributing factor. Indeed, due to the permanent availability of water around the Senegal River basin, yearly cultivation is well developed in Gankette Balla and pyrethroids are well represented in the pesticides used. This assumption is reinforced by the fact that the resistance of other malaria vectors to lambda-cyhalothrin was observed in areas of high agricultural pressures in Senegal [28]. Moreover, the link between the resistance of An. gambiae (s.l.) to insecticides and intensive use of pesticides has been already reported in areas where agriculture is highly developed [33–35].
In addition, another source of selection pressure could be likely linked to the use of type two pyrethroids insecticides (Deltamethrin) in treated bed nets in malaria vector control programs. In fact, the village of Gankette Balla was included during the scaling-up coverage with insecticide-treated nets against malaria in Senegal through the “Pal Fleuve” program in 2006.
Furthermore, the IRS campaign using the lambda-cyhalothrin initiated a year later (2007) in the district of Richard Toll (lower valley of the Senegal River) could have been an additional contributing factor. Indeed, two years after this campaign, a lambda-cyhalothrin resistance in An. gambiae (s.l.) was observed in this area [28] and a link between this resistance and that observed in our study is not to exclude as previous studies have shown the existence of strong gene flow between populations of An. funestus in the lower valley and that of the area of Lake Guiers [36]. The resistance of An. funestus to lambda-cyhalothrin and deltamethrin may represent a potential threat to the success of the future vector control program directed against this major vector especially as pyrethroids are the only insecticide used for the impregnation of mosquito nets. This is a serious concern as other vectors of the study area are resistant to pyrethroids [28]. If such resistance is not managed properly, it can still be selected by the current vector control program (Insecticide-treated nets and IRS) to a level that will seriously affect the success of future programs against this major vector. A spread of the observed resistance in Gankette Balla is a concern for the continued effectiveness of pyrethroid-based interventions against An. funestus in Senegal.
Metabolic resistance mechanism is driving lambda-cyhalothrin resistance
Analysis of the transcription profile of the Gankette Balla sample supported the importance of metabolic resistance mechanism in the observed pyrethroid resistance in this An. funestus population as previously reported [32]. The importance of metabolic resistance mechanisms is shown through the consistent overexpression of genes involved in insecticide detoxification such as cytochrome P450 genes, GSTs, aldehyde oxidases and other gene families previously associated in resistance of An. funestus to insecticides.
The P450 CYP6M7 was consistently the most overexpressed detoxification gene in lambda-cyhalothrin exposed mosquitoes compared to the FANG susceptible strain as well as to the unexposed mosquitoes. This gene was shown to be able to metabolise several pyrethroids including lambda-cyhalothrin [37], further supporting the likelihood that it could be the main resistance gene in this An. funestus population. A key role of such P450 gene will be in line with the common implication of the cytochrome P450 family genes in insecticide resistance as previously reported in several populations of An. funestus [3, 37–39], An. gambiae (s.l.) [40], An. albimanus [41] and An. minimus [42]. This is also reported in several other insects [43–45]. More specifically, the role of cytochrome P450 in pyrethroid resistance has been reported in An. funestus from Benin [11], Ghana [10], Uganda [17], Mozambique [24, 27, 46, 47], Kenya [8] and Malawi [3], as well as in the laboratory resistant strain to permethrin [38, 48].
The overexpression of the CYP6M7 gene in lambda-cyhalothrin resistant and non-exposed mosquitoes, compared to the susceptible strain suggests that it plays a key role in pyrethroid metabolic resistance in An. funestus in Gankette Balla similar to recent reports for the important role of this gene in Zambia [6, 37]. Surprisingly, one of the duplicated P450 genes, CYP6P9a, which has been shown to play a main role in pyrethroid resistance in southern populations of An. funestus [3, 38, 39, 49] was not overexpressed at all. The complete absence of overexpression of CYP6P9a indicates that the resistance mechanism in Senegal is different to that observed in southern Africa. A similar difference is also observed for the GSTe2 gene which is not overexpressed at all in the Gankette Balla population whereas it was among the highest overexpressed in a DDT and pyrethroid resistant population from Benin, another West Africa country. It will be interesting to investigate the role of other GST genes overexpressed in the Gankette Balla population such as GSTd3 and GSTd1-5 to see what role they play in the observed DDT resistance.
Furthermore, the overexpression of several other genes from the microarray analysis suggests that in addition to CYP6M7, other genes may also play a role or involved in subsequent phases of pyrethroid detoxification. Further functional characterisation studies will help to establish these roles.
Conclusions
This study has provided the first assessment of the susceptibility to the main insecticides used in public health of an An. funestus population from Senegal and also explored the possible mechanisms responsible for the pyrethroid resistance observed. The resistance profile observed in the Gankette Balla population highlights the need for further studies to assess the extent and the geographical distribution of these resistances in An. funestus populations in Senegal as well as an assessment of its impact on malaria control programs. This will improve the implementation and management of future control programs against this important malaria vector in Senegal and in Africa in general.
Abbreviations
cDNA, complementary deoxyribonucleic acid; cRNA, complementary ribonucleic acid; DDT, dichlorodiphenyltrichloroethane; DNA, deoxyribonucleic acid; FC, fold change; GABA, gamma-aminobutyric acid; GST, glutathione s-transferase; GSTd1-5, glutathione-s-transferase d1-5; GSTd3, glutathione-s-transferase d3; GSTe2: glutathione-s-transferase epsilon 2; IRS, indoor residual spraying; Kdr, Knockdown resistance; LLINs, long-lasting insecticide nets; PCR, polymerase Chain Reaction; QPCR, real-time quantitative PCR; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; RNA, ribonucleic acid; RSP7, ribosomal protein S7; VGSC, voltage-gated sodium channel gene; WHO, World Health Organisation
Acknowledgments
This work was supported by the Malaria Capacity Development Consortium (MCDC) which is funded by the Wellcome Trust (Grant number WT084289MA) and the Bill & Melinda Gates Foundation (Grant number: 51941). We are grateful to Khalifa Thiam, Moussa Fall and Youssou Coulibaly for technical assistance and help during field investigations and the villagers for their cooperation. The authors would like to thank Gareth Weedal for helpful comments and suggestions during sequences analysis.
Funding
This work was supported by the Malaria Capacity Development Consortium (MCDC). The funder had no role in study design, data collection or analysis, interpretation of data, decision to publish or preparation of the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
CSW, BS and OF designed the study. OF, ID, LK and CSW supervised the study. BS carried out the field collections and performed the experiments with assistance of H.I. JMR contributed toward data analysis. BS and CSW analysed the data and wrote the manuscript. All authors read, critically revised, read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
List of primers used in this study. (DOCX 72 kb)
Insecticide resistance profile of Anopheles funestus population from Senegal. (DOCX 54 kb)
Top 50 the most detoxification genes commonly overexpressed in the comparisons R-S_L and C-S (FC ≥2, P ≤ 0.05). (DOCX 108 kb)
Top 50 the most detoxification genes overexpressed in the R-S_L comparisons (FC ≥2, P ≤ 0.05). (DOCX 106 kb)
Top 50 the most detoxification genes overexpressed in the R-C_L comparisons (FC ≥1.5, P ≤ 0.05). (DOCX 96 kb)
Detoxification genes commonly under expressed in the comparisons R-S_L, C-S (FC ≥ 2) and R-C_L (FC ≥ 1.5) P ≤ 0.05. (DOCX 54 kb)
Top 50 the most detoxification genes commonly under expressed in the comparisons R-S_L and C-S (FC ≥2, P ≤ 0.05). (DOCX 110 kb)
Top 50 the most detoxification genes under expressed in the R-S_L comparisons (FC ≥2, P ≤ 0.05). (DOCX 100 kb)
Top 50 the most detoxification genes under expressed in the R-C_L comparisons (FC ≥1.5, P ≤ 0.05). (DOCX 95 kb)
Region flanking the kdr codon 1014 (TTA, indicated in red), showing no variation in wild type An. funestus from Gankette Balla. (TIF 1521 kb)
Contributor Information
Badara Samb, Email: badarasb@gmail.com.
Lassana Konate, Email: konatela@yahoo.fr.
Helen Irving, Email: Helen.Irving@lstmed.ac.uk.
Jacob M. Riveron, Email: Jacob.Riveron@lstmed.ac.uk
Ibrahima Dia, Email: dia@pasteur.sn.
Ousmane Faye, Email: fayeo@orange.sn.
Charles S. Wondji, Email: Charles.Wondji@lstmed.ac.uk
References
- 1.WHO . Global Malaria Program. Geneva: World malaria report; 2014. [Google Scholar]
- 2.PNLP: Rapport statistique 2010–2013. Programme National de Lutte contre le Paludisme (PNLP) Ministère de la Santé et de la Prévention Médicale. 2014. p. 33.
- 3.Riveron JM, Irving H, Ndula M, Barnes KG, Ibrahim SS, Paine MJ, Wondji CS. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc Natl Acad Sci U S A. 2013;110(1):252–257. doi: 10.1073/pnas.1216705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, Ismail HM, Hemingway J, Ranson H, Albert A, et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15(2):R27. doi: 10.1186/gb-2014-15-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi KS, Christian R, Nardini L, Wood OR, Agubuzo E, Muleba M, Munyati S, Makuwaza A, Koekemoer LL, Brooke BD, et al. Insecticide resistance and role in malaria transmission of Anopheles funestus populations from Zambia and Zimbabwe. Parasit Vectors. 2014;7:464. doi: 10.1186/s13071-014-0464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomsen EK, Strode C, Hemmings K, Hughes AJ, Chanda E, Musapa M, Kamuliwo M, Phiri FN, Muzia L, Chanda J, et al. Underpinning sustainable vector control through informed insecticide resistance management. PLoS One. 2014;9(6):e99822. doi: 10.1371/journal.pone.0099822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wondji CS, Dabire RK, Tukur Z, Irving H, Djouaka R, Morgan JC. Identification and distribution of a GABA receptor mutation conferring dieldrin resistance in the malaria vector Anopheles funestus in Africa. Insect Biochem Mol Biol. 2011;41(7):484–491. doi: 10.1016/j.ibmb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawada H, Dida GO, Ohashi K, Komagata O, Kasai S, Tomita T, Sonye G, Maekawa Y, Mwatele C, Njenga SM, et al. Multimodal pyrethroid resistance in malaria vectors, Anopheles gambiae s.s., Anopheles arabiensis, and Anopheles funestus s.s. in western Kenya. PLoS One. 2011;6(8):e22574. doi: 10.1371/journal.pone.0022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulamba C, Riveron JM, Ibrahim SS, Irving H, Barnes KG, Mukwaya LG, Birungi J, Wondji CS. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PLoS One. 2014;9(10):e110058. doi: 10.1371/journal.pone.0110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okoye PN, Brooke BD, Koekemoer LL, Hunt RH, Coetzee M. Characterisation of DDT, pyrethroid and carbamate resistance in Anopheles funestus from Obuasi, Ghana. Trans R Soc Trop Med Hyg. 2008;102(6):591–598. doi: 10.1016/j.trstmh.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Djouaka R, Irving H, Tukur Z, Wondji CS. Exploring mechanisms of multiple insecticide resistance in a population of the malaria vector Anopheles funestus in Benin. PLoS One. 2011;6(11):e27760. doi: 10.1371/journal.pone.0027760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ndiath MO, Sougoufara S, Gaye A, Mazenot C, Konate L, Faye O, Sokhna C, Trape JF. Resistance to DDT and pyrethroids and increased kdr mutation frequency in An. gambiae after the implementation of permethrin-treated nets in Senegal. PLoS One. 2012;7(2):e31943. doi: 10.1371/journal.pone.0031943. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ndiath MO, Cailleau A, Orlandi-Pradines E, Bessell P, Pages F, Trape JF, Rogier C. Emerging knock-down resistance in Anopheles arabiensis populations of Dakar, Senegal: first evidence of a high prevalence of kdr-e mutation in West African urban area. Malar J. 2015;14(1):364. doi: 10.1186/s12936-015-0898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niang el HA, Konate L, Diallo M, Faye O, Dia I. Patterns of insecticide resistance and knock down resistance (kdr) in malaria vectors An. arabiensis, An. coluzzii and An. gambiae from sympatric areas in Senegal. Parasit Vectors. 2016;9(1):71. [DOI] [PMC free article] [PubMed]
- 15.Thwing JI, Perry RT, Townes DA, Diouf MB, Ndiaye S, Thior M. Success of Senegal’s first nationwide distribution of long-lasting insecticide-treated nets to children under five - contribution toward universal coverage. Malar J. 2011;10:86. doi: 10.1186/1475-2875-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PNLP . Rapport d’activités 2012. Programme National de Lutte contre le Paludisme (PNLP) Ministère de la Santé et de la Prévention Médicale. 2013. [Google Scholar]
- 17.Morgan JC, Irving H, Okedi LM, Steven A, Wondji CS. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One. 2010;5(7):e11872. doi: 10.1371/journal.pone.0011872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillies MT, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara (Afrotropical region) Johannesburg: The South African Institute for Medical Research; 1987. [Google Scholar]
- 19.Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107(4):611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66(6):804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 21.WHO . Test procedures for insecticide resistance monitoring in malaria vector mosquitoes. Geneva: World Health Organization; 2013. [Google Scholar]
- 22.Crawford JE, Guelbeogo WM, Sanou A, Traore A, Vernick KD, Sagnon N, Lazzaro BP. De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS One. 2010;5(12):e14202. doi: 10.1371/journal.pone.0014202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 24.Cuamba N, Morgan JC, Irving H, Steven A, Wondji CS. High level of pyrethroid resistance in an Anopheles funestus population of the Chokwe District in Mozambique. PLoS One. 2010;5(6):e11010. doi: 10.1371/journal.pone.0011010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riveron JM, Chiumia M, Menze BD, Barnes KG, Irving H, Ibrahim SS, Weedall GD, Mzilahowa T, Wondji CS. Rise of multiple insecticide resistance in Anopheles funestus in Malawi: a major concern for malaria vector control. Malar J. 2015;14:344. doi: 10.1186/s12936-015-0877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coetzee M, van Wyk P, Booman M, Koekemoer LL, Hunt RH. Insecticide resistance in malaria vector mosquitoes in a gold mining town in Ghana and implications for malaria control. Bull Soc Pathol Exot. 2006;99(5):400–403. [PubMed] [Google Scholar]
- 27.Kloke RG, Nhamahanga E, Hunt RH, Coetzee M. Vectorial status and insecticide resistance of Anopheles funestus from a sugar estate in southern Mozambique. Parasit Vectors. 2011;4:16. doi: 10.1186/1756-3305-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faye O, Konate L, Diop A. Profil entomologique du paludisme au Sénégal. Ministère de la Santé et de la Prévention Médicale. 2011. p. 39.
- 29.Scollon EJ, Starr JM, Godin SJ, DeVito MJ, Hughes MF. In vitro metabolism of pyrethroid pesticides by rat and human hepatic microsomes and cytochrome p450 isoforms. Drug Metab Dispos. 2009;37(1):221–228. doi: 10.1124/dmd.108.022343. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim SS, Riveron JM, Stott R, Irving H, Wondji CS. The cytochrome P450 CYP6P4 is responsible for the high pyrethroid resistance in knockdown resistance-free Anopheles arabiensis. Insect Biochem Mol Biol. 2016;68:23–32. doi: 10.1016/j.ibmb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hargreaves K. KLL, Brooke B.D., Hunt R.H., Mthembu J. and Coetzee M. Anopheles funestus resistant to pyrethroid insecticides in South Africa. Med Vet Entomol. 2000;14(2):181–189. doi: 10.1046/j.1365-2915.2000.00234.x. [DOI] [PubMed] [Google Scholar]
- 32.Coetzee M, Koekemoer LL. Molecular systematics and insecticide resistance in the major African malaria vector Anopheles funestus. Annu Rev Entomol. 2013;58:393–412. doi: 10.1146/annurev-ento-120811-153628. [DOI] [PubMed] [Google Scholar]
- 33.Diabate A, Baldet T, Chandre F, Akoobeto M, Guiguemde TR, Darriet F, Brengues C, Guillet P, Hemingway J, Small GJ, et al. The role of agricultural use of insecticides in resistance to pyrethroids in Anopheles gambiae s.l. in Burkina Faso. Am J Trop Med Hyg. 2002;67(6):617–622. doi: 10.4269/ajtmh.2002.67.617. [DOI] [PubMed] [Google Scholar]
- 34.Djegbe I, Boussari O, Sidick A, Martin T, Ranson H, Chandre F, Akogbeto M, Corbel V. Dynamics of insecticide resistance in malaria vectors in Benin: first evidence of the presence of L1014S kdr mutation in Anopheles gambiae from West Africa. Malar J. 2011;10:261. doi: 10.1186/1475-2875-10-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dabire RK, Namountougou M, Sawadogo SP, Yaro LB, Toe HK, Ouari A, Gouagna LC, Simard F, Chandre F, Baldet T, et al. Population dynamics of Anopheles gambiae s.l. in Bobo-Dioulasso city: bionomics, infection rate and susceptibility to insecticides. Parasit Vectors. 2012;5:127. doi: 10.1186/1756-3305-5-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samb B, Dia I, Konate L, Ayala D, Fontenille D, Cohuet A. Population genetic structure of the malaria vector Anopheles funestus, in a recently re-colonized area of the Senegal River basin and human-induced environmental changes. Parasit Vectors. 2012;5:188. doi: 10.1186/1756-3305-5-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riveron JM, Ibrahim SS, Chanda E, Mzilahowa T, Cuamba N, Irving H, Barnes KG, Ndula M, Wondji CS. The highly polymorphic CYP6M7 cytochrome P450 gene partners with the directionally selected CYP6P9a and CYP6P9b genes to expand the pyrethroid resistance front in the malaria vector Anopheles funestus in Africa. BMC Genomics. 2014;15:817. doi: 10.1186/1471-2164-15-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amenya DA, Naguran R, Lo TC, Ranson H, Spillings BL, Wood OR, Brooke BD, Coetzee M, Koekemoer LL. Over expression of a cytochrome P450 (CYP6P9) in a major African malaria vector, Anopheles funestus, resistant to pyrethroids. Insect Mol Biol. 2008;17(1):19–25. doi: 10.1111/j.1365-2583.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- 39.Wondji CS, Irving H, Morgan J, Lobo NF, Collins FH, Hunt RH, Coetzee M, Hemingway J, Ranson H. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 2009;19(3):452–459. doi: 10.1101/gr.087916.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell SN, Stevenson BJ, Müller P, Wilding CS, Egyir-Yawson A, Field SG, Hemingway J, Paine MJI, Ranson H, Donnelly MJ. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc Natl Acad Sci. 2012;109(16):6147–6152. doi: 10.1073/pnas.1203452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rongnoparut P, Boonsuepsakul S, Chareonviriyaphap T, Thanomsing N. Cloning of cytochrome P450, CYP6P5, and CYP6AA2 from Anopheles minimus resistant to deltamethrin. J Vector Ecol. 2003;28(2):150–158. [PubMed] [Google Scholar]
- 42.Rodpradit P, Boonsuepsakul S, Chareonviriyaphap T, Bangs MJ, Rongnoparut P. Cytochrome P450 genes: molecular cloning and overexpression in a pyrethroid-resistant strain of Anopheles minimus mosquito. J Am Mosq Control Assoc. 2005;21(1):71–79. doi: 10.2987/8756-971X(2005)21[71:CPGMCA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 43.Kasai S, Scott JG. Overexpression of Cytochrome P450 CYP6D1 Is Associated with Monooxygenase-Mediated Pyrethroid Resistance in House Flies from Georgia. Pestic. Biochem. Physiol. 2000;68(1):34–41. doi: 10.1006/pest.2000.2492. [DOI] [Google Scholar]
- 44.Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, Nauen R, Harrison DA, Palli SR. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc Natl Acad Sci U S A. 2010;107(19):8557–8562. doi: 10.1073/pnas.1000059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feyereisen R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim Biophys Acta. 2011;1814(1):19–28. doi: 10.1016/j.bbapap.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Brooke BD, Kloke G, Hunt RH, Koekemoer LL, Temu EA, Taylor ME, Small G, Hemingway J, Coetzee M. Bioassay and biochemical analyses of insecticide resistance in southern African Anopheles funestus (Diptera: Culicidae) Bull Entomol Res. 2001;91(4):265–272. doi: 10.1079/BER2001108. [DOI] [PubMed] [Google Scholar]
- 47.Casimiro SL, Hemingway J, Sharp BL, Coleman M. Monitoring the operational impact of insecticide usage for malaria control on Anopheles funestus from Mozambique. Malar J. 2007;6:142. doi: 10.1186/1475-2875-6-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunt RH, Brooke BD, Pillay C, Koekemoer LL, Coetzee M. Laboratory selection for and characteristics of pyrethroid resistance in the malaria vector Anopheles funestus. Med Vet Entomol. 2005;19(3):271–275. doi: 10.1111/j.1365-2915.2005.00574.x. [DOI] [PubMed] [Google Scholar]
- 49.Christian RN, Matambo TS, Spillings BL, Brooke BD, Coetzee M, Koekemoer LL. Age-related pyrethroid resistance is not a function of P450 gene expression in the major African malaria vector, Anopheles funestus (Diptera: Culicidae) Genet Mol Res. 2011;10(4):3220–3229. doi: 10.4238/2011.December.21.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.


