Summary
Embryonic stem cells (ESCs) are cell lines derived from the mammalian pre-implantation embryo. Here we assess the impact of derivation and culture conditions on both functional potency and ESC transcriptional identity. Individual ESCs cultured in either two small-molecule inhibitors (2i) or with knockout serum replacement (KOSR), but not serum, can generate high-level chimeras regardless of how these cells were derived. ESCs cultured in these conditions showed a transcriptional correlation with early pre-implantation embryos (E1.5–E3.5) and contributed to development from the 2-cell stage. Conversely, the transcriptome of serum-cultured ESCs correlated with later stages of development (E4.5), at which point embryonic cells are more restricted in their developmental potential. Thus, ESC culture systems are not equivalent, but support cell types that resemble distinct developmental stages. Cells derived in one condition can be reprogrammed to another developmental state merely by adaptation to another culture condition.
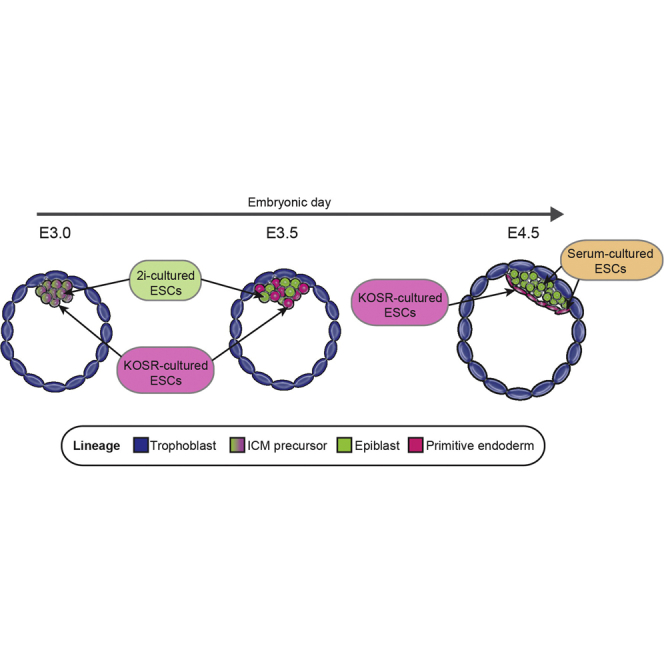
Graphical Abstract
Highlights
-
•
ESC derivation condition does not irreversibly affect functional potency
-
•
ESCs cultured in 2i and KOSR resemble early stages of embryonic development
-
•
ESCs cultured in 2i and KOSR have enhanced functional potency
-
•
ESCs cultured in KOSR resemble primitive endoderm
In this article, Brickman and colleagues show that different embryonic stem cell (ESC) culture conditions select for unique combinations of embryo-like subpopulations resembling distinct developmental stages. ESCs similar to early stages have the capacity to clonally contribute to development and differentiate into extraembryonic lineages in vivo and in vitro.
Introduction
Embryonic stem cells (ESCs) are karyotypically normal, self-renewing cell lines, derived from the inner cell mass (ICM) of the pre-implantation embryo (Evans and Kaufman, 1981, Martin, 1981). ESCs can be derived and expanded using a variety of conditions, including culture with the cytokine leukemia inhibitory factor (LIF) in the presence of serum (Smith et al., 1988, Williams et al., 1988), in serum-free medium with two small-molecule inhibitors (2i) (Ying et al., 2008), or with knockout serum replacement (KOSR) (Ward et al., 2002). ESCs can be maintained indefinitely in vitro, while retaining the capacity to participate in development and generate all cell types of the embryo including the germ cells (Beddington and Robertson, 1989, Gossler et al., 1986, Lallemand and Brulet, 1990, Robertson et al., 1986, Suemori et al., 1990). They are therefore said to be pluripotent.
Although the first ESCs were derived more than 30 years ago, a number of fundamental questions remain unanswered. At the embryonic stages from which ESCs are derived, the blastocyst is composed of several cell types, the epiblast (Epi), primitive endoderm (PrE), and trophoblast, and, during ESC derivation, subpopulations of embryo-derived cells are selected to expand. While these populations are not the same as the parental embryonic cells from which they are derived (Tang et al., 2010), to what degree do they represent embryonic development? ESC cultures are also heterogeneous (Canham et al., 2010, Chambers et al., 2007, Hayashi et al., 2008, Kobayashi et al., 2009, Singh et al., 2007, Toyooka et al., 2008) and this heterogeneity is dynamic, perhaps more dynamic than the blastocyst from which they are derived. However, does this heterogeneity reflect the endogenous cell populations that arise in normal blastocyst development?
The functional potential of ESCs can be assessed using a number of different approaches including in vitro differentiation, teratoma formation, and chimera generation (Beddington and Robertson, 1989, Poueymirou et al., 2007, Robertson et al., 1986, Saburi et al., 1997). Nevertheless, as ESCs are heterogeneous and chimeras are routinely generated by injecting 10–15 ESCs into morula or blastocyst-stage embryos (Bradley et al., 1984, Lallemand and Brulet, 1990) it is difficult to discern the functional properties of individual ESCs or specific ESC subpopulations. Based on the prospective isolation of ESC subpopulations, it has been shown that ESCs cultured in serum and LIF contain dynamic populations of PrE- and Epi-biased cells (Canham et al., 2010). However, these cells are clearly different from the blastocyst from which they are derived, as the PrE-primed cells express elevated levels of PrE RNA, but not protein. ESCs cultured under these conditions also contain a subpopulation that expresses 2-cell embryo (2C)-specific genes (Falco et al., 2007, Macfarlan et al., 2012). Similarly, culture of ESCs in 2i supports a totipotent population of cells that coexpress Epi determinants such as Nanog and the RNA for extraembryonic genes such as Gata6 or Hhex (Morgani et al., 2013).
So, how do the conditions used to maintain ESCs influence the gene-expression state and populations contained within the culture? In this paper we explore this question by testing the impact of culture and derivation conditions on ESC populations, comparing ESC gene expression and heterogeneity, and the capacity of individual ESCs to contribute to full-term embryonic development. We found that ESCs maintained in standard serum culture conditions were comparable to populations of the late blastocyst (embryonic day 4.5 [E4.5]) ICM, at which point cells are specified and restricted in their functional potential. Conversely, ESCs cultured in 2i or KOSR showed a correlation with embryos from as early as the 2C stage, when cells are unrestricted and highly plastic. Consistent with expression data, we observed that single 2i and KOSR, but not serum, cultured ESCs could generate high-level chimeras when injected into either morulae or 2C embryos. This suggests that different ESC culture conditions support the expansion of populations reminiscent of different embryonic stages with distinct functional potentials. We found that populations induced during derivation could be “reprogrammed” by transferring ESCs to a different culture condition.
Results
KOSR and 2i Culture Enhances ESC Single-Cell Potency
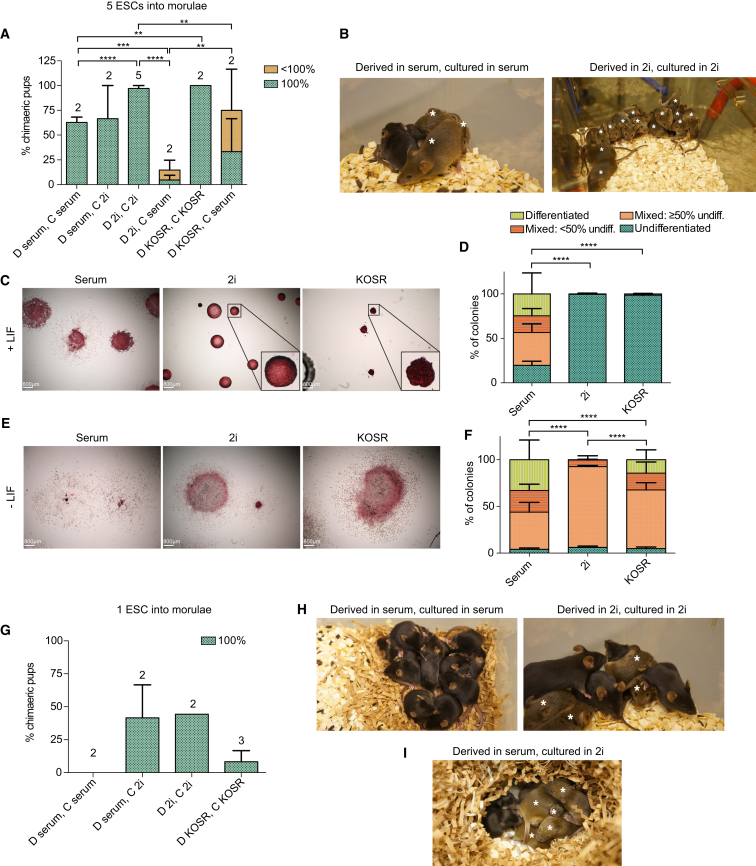
ESC lines have been derived and maintained in a number of different medium compositions (Smith et al., 1988, Ward et al., 2002, Williams et al., 1988, Ying et al., 2008). We sought to compare the functional potential of individual ESCs derived and/or maintained by different protocols. As ESCs of different genetic backgrounds generate chimeras with variable efficiencies (Auerbach et al., 2000, Brook and Gardner, 1997), we wished to start with high-quality ESC lines capable of generating chimeras from single cells. We therefore utilized mice from an F1 hybrid genetic background, reported to generate chimeras with high efficiency based on hybrid vigor (Eggan et al., 2001). We derived F1 129S2;C57BL/6N hybrid ESC lines in conventional serum/LIF (serum) culture conditions, as well as 2i/LIF (2i) and KOSR/LIF (KOSR). We confirmed that resulting ESC lines were of high quality by standard injection of multiple ESCs into morulae. ESC lines generated 100% ESC-derived mice as judged by coat color and internal organs (Figures 1A, 1B, S1A, and S1B; Table 1) and contributed to the germline (n = 15/15, Table 2). However, ESCs derived in 2i, but switched to serum medium, showed a significant reduction in their capacity to generate chimeras in this assay (Figure 1A).
Figure 1.
KOSR and 2i Culture Enhances ESC Single-Cell Potency
ESCs were derived (D) in and cultured (C) under various conditions; D in serum and C in serum, D in serum and C in 2i, D in 2i and C in 2i, and D in KOSR and C in KOSR.
(A and B) Multiple (5) ESCs were injected into morulae and their contribution to full-term chimeras assessed according to coat color. (A) Graph showing the percentage of chimeras born out of the total number of pups and quantification of coat color chimerism. n Values are provided above bars and refer to the number of independent ESC clones tested for each condition. Error bars indicate SEM. Results for 2i conditions are a combination of ESCs cultured with and without mouse embryonic fibroblasts (MEFs) to control for the effect that MEFs may have in serum and KOSR cultures. No difference was observed with or without MEFs. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, Student's unpaired t test. (B) Representative images of litters of mice. Asterisks indicate mice with ESC contribution.
(C and D) ESCs were cultured in serum, 2i, or KOSR and stained for alkaline phosphatase activity. (C) Representative images of ESC colonies. (D) Quantification of ESC colony categories as defined in Figure S1C. Error bars represent SEM of three biological replicates using independent ESC lines. ∗∗∗∗p ≤ 0.0001, two-tailed chi-square test.
(E and F) ESCs were cultured in serum, 2i, or KOSR and then differentiated by removal of LIF for 5 days. Cells were stained for alkaline phosphatase activity. (E) Representative images of differentiated colonies. (F) Quantification of ESC colony categories as defined in Figure S1C. Error bars represent SEM of three biological replicates of independent ESC lines. ∗∗∗∗p ≤ 0.0001, two-tailed chi-square test.
(G–I) Single ESCs were injected into morulae and their contribution to full-term chimeras assessed according to coat color. (G) Graph showing the percentage of chimeras born out of the total number of pups and quantification of coat color chimerism. n Values are provided above bars and refer to the number of independent ESC clones tested for each condition. Error bars indicate SEM. Results for 2i conditions are a combination of ESCs cultured with and without MEFs to control for the effect that MEFs may have in serum and KOSR cultures. No difference was observed with or without MEFs. (H and I) Representative images of litters of mice. Asterisks indicate mice with ESC contribution.
See also Figure S1.
Table 1.
| Condition | Line | Injected/Transferred | Chimeras/Pups |
|---|---|---|---|
| 1 ESC into Morula | |||
| D serum, C serum | 3.4 + MEFS | 25 | 0/11 |
| 3.2 + MEFs | 25 | 0/2 | |
| D serum, C 2i | 3.4 + MEFs | 31 | 6/9 |
| 3.2 + MEFs | 19 | 1/6 | |
| D 2i, C 2i | 2.2H + MEFs | 38 | 4/9 |
| D KOSR, C KOSR | 5.3 + MEFs | 30 | 0/2 |
| 5.45 + MEFs | i. 30 ii. 45 |
i. 0/5 ii. 2/8 |
|
| 5 ESCs into Morula | |||
| D serum, C serum | 3.4 + MEFs | i. 17 ii. 40 |
i. 3/5 ii. 1/2 |
| 3.2 + MEFs | i. 24 ii. 30 |
i. 3/4 ii. 2/3 |
|
| D serum, C 2i | 3.4 + MEFs | 12 | 4/4 |
| 3.2 + MEFs | 20 | 2/6 | |
| D 2i, C 2i | 2.2H + MEFs | i. 25 ii. 56 |
i. 4/5 ii. 21/21 |
| 2.2Ack1#5 + MEFs | 13 | 1/1 | |
| 2.2Ack1#11 + MEFs | 15 | 6/6 | |
| 2.2Ack1#28 + MEFs | 15 | 6/6 | |
| 2.2Ack1#14 + MEFs | 15 | 4/4 | |
| 2.2PTIP#8 + MEFs | 42 | 4/5 (1 < 100%) | |
| 2.2PTIP#9 + MEFs | 30 | 4/4 | |
| D 2i, C serum | 2.5 | 30 | 0/1 |
| 2.2 | 30 | 1/7 | |
| D KOSR, C KOSR | 5.3 + MEFs | 30 | 14/14 |
| 5.45 + MEFs | 25 | 3/3 | |
| D KOSR, C serum | 5.3 + MEFs | 28 | 4/6 |
| 5.45 + MEFs | 25 | 5/6 (5 < 100%) | |
| 1 ESC into 2C Embryo | |||
| D serum, C serum | 3.4 + MEFs | 24 | 0/6 |
| 3.2 + MEFs | 13 | 0/8 | |
| D serum, C 2i | 3.4 + MEFs | 24 | 5/6 |
| 3.2 + MEFs | 14 | 2/6 | |
| D 2i, C 2i | 2.5 | 30 | 2/18 |
| 2.5 + MEFs | 30 | 0/7 | |
| 2.2 | 59 | 6/11 | |
| 2.2 + MEFs | 30 | 3/8 | |
| 2.6 | 26 | 3/18 (1 < 100%) | |
| 2.2H + MEFs | 24 | 7/14 (2 < 100%) | |
| 2.2V + MEFs | 58 | 5/11 (1 < 100%) | |
| D 2i, C serum | 2.2 | 28 | 0/6 |
| 2.5 | 17 | 0/5 | |
| D KOSR, C KOSR | 5.3 + MEFs | 13 | 2/5 |
| 5.45 + MEFs | 13 | 1/3 | |
| 5 ESCs into 2C Embryo | |||
| D serum, C serum + MEFs | 3.4 | 48 | 0/3 |
| D serum, C 2i + MEFs | 3.4 | 40 | 6/7 |
ESCs were derived (D) in and cultured (C) under various conditions. Table shows the number of embryos injected and transferred and the resulting number of chimeras out of total pups born from each cell line under each derivation and culture condition. Individual injection experiments of the same clone are marked as (i) and (ii). Results are shown for single- and multiple-cell injection into morulae and single- and multiple-cell injection into 2-cell (2C) embryos.
Table 2.
| Condition | Line | Experimental Setup Mice Were Generated from: |
|
|---|---|---|---|
| 1 ESC into 2C Embryo | 5 ESCs into Morula | ||
| D serum, C serum | 3.2 | – | 1/1 |
| 3.4 | – | 1/1 | |
| D 2i, C 2i | 2.2 | 3/3 | 3/3 |
| 2.5 | 1/1 | 2/2 | |
| 2.6 | 1/1 | 1/1 | |
| D KOSR, C KOSR | 5.3 | – | 2/2 |
| 5.45 | – | 2/2 | |
F1 129S6;C57BL/6N ESCs were derived (D) in and cultured (C) under various conditions. Chimeric mice were generated by different means (described in table headings) and then tested for ESC germline contribution. Results show the number of mice demonstrating germline transmission out of the number of mice tested per condition.
We also assessed the capacity of these ESC lines for clonal self-renewal in vitro. Single ESCs were expanded and stained for alkaline phosphatase activity, a marker of undifferentiated cell types (Figures 1C–1F and S1C). While the majority of 2i- and KOSR-cultured ESC colonies were entirely undifferentiated, a significant fraction of serum-cultured colonies exhibited some degree of differentiation (Figures 1C and 1D). When challenged to differentiate in the absence of the cytokine LIF, all ESCs efficiently differentiated, although fewer colonies were entirely differentiated when ESCs were previously cultured in 2i or KOSR (Figures 1E and 1F). This suggested that individual cells in these conditions retain some intrinsic capacity to self-renew despite exposure to differentiation-promoting signals. There was no difference in the proliferation rate of ESCs in each condition (Figure S1D).
After this initial validation of ESC lines, we wished characterize the impact of serum, 2i, and KOSR culture on the potency of individual ESCs. To this end we injected single ESCs into morulae. Single ESCs, derived and cultured in 2i, generated 100% ESC-derived mice, as judged by coat color (Figures 1G and 1H; Table 1). ESCs cultured in KOSR also demonstrated this capacity, albeit to a lower extent (Figure 1G and Table 1). Conversely, ESCs derived and cultured in serum did not generate full-term chimeras when injected as single cells into morulae (Figures 1G and 1H; Table 1). We also found that ESCs derived in serum conditions were not inherently limited in their capacity to generate chimeras as, after transient transfer of these cells to 2i (three passages), single cells now contributed to embryos (Figures 1G and 1I; Table 1). This suggests that in chimera assays, as in vitro (Figures 1C and 1D), culture in 2i or KOSR increases the clonogenicity of ESCs compared with standard serum culture.
2i- and KOSR-Cultured ESCs Represent Early Stages of Embryonic Development
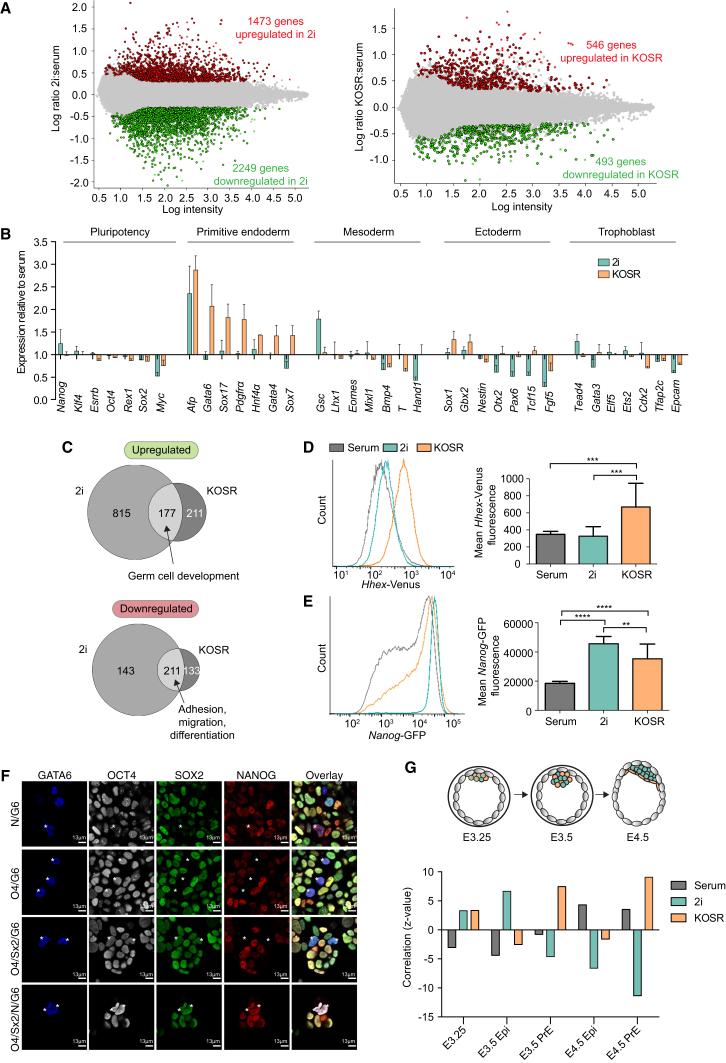
To determine the transcriptional landscape related to these functional differences, we performed microarray expression analysis on ESCs cultured in these conditions (GEO: GSE72445). When compared with serum cultures, 1,473 genes were upregulated and 2,249 genes were downregulated in 2i, and 546 genes were upregulated and 493 genes were downregulated in KOSR (Figure 2A, Tables S1–S4, FDR ≤0.05, fold change ≥2). Genes upregulated in 2i and KOSR showed a strong signature for lipid and glutathione metabolism (Tables S5 and S6), while genes upregulated in KOSR were also associated with meiosis and anterior-posterior patterning (Table S6). Downregulated genes in 2i were associated with extracellular matrix, differentiation, and the mitogen-activated protein kinase pathway (Table S7). Similarly, genes downregulated in KOSR were related to cell junctions, matrix adhesion, and fibroblast growth factor (FGF) signaling (Table S8). Consistent with previous reports (Marks et al., 2012, Morgani et al., 2013), we also observed that 2i cultures downregulated mesoderm and ectoderm markers while having little effect on endodermal gene expression (Figure 2B). However, KOSR culture increased the expression of endoderm markers but had little effect on pluripotent gene expression (Figure 2B). The coexpression of pluripotency and extraembryonic genes is an expression state previously associated with totipotency (Morgani et al., 2013).
Figure 2.
KOSR, Serum, and 2i Support Unique Transcriptional States
(A and B) Microarrays were carried out on 129/Ola ESCs cultured in serum, 2i, and KOSR conditions. At least three biological replicates of independent ESC lines were analyzed per condition. (A) Pairwise comparisons (FDR <0.05, ≥2-fold expression levels) were performed between serum-cultured and 2i- or KOSR-cultured ESCs to reveal non-redundant, significant changes in gene expression. Red dots represent significantly upregulated genes, green dots represent significantly downregulated genes, and gray dots represent genes that are not significantly changing in expression. (B) Expression changes in lineage markers in 2i- and KOSR-cultured ESCs relative to the expression in standard serum cultures.
(C) Venn diagrams illustrating the overlap between genes up- and downregulated in 2i and KOSR compared with standard serum cultures (identified in A).
(D and E) Flow cytometry profiles and corresponding mean fluorescence values for PECAM-1+ HV (D) or Nanog-GFP TNGB (E) ESCs cultured in serum, 2i, and KOSR. n = 3 independent experiments. Error bars indicate SD of the mean. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, Student's unpaired t test.
(F) Confocal optical section of ESCs cultured in serum, 2i, and KOSR immunostained for GATA6, OCT4, SOX2, and NANOG. Asterisks indicate coexpressing ESCs. The same observation was made in two independent cell lines.
(G) Graph showing the correlation of serum-, 2i-, and KOSR-cultured ESCs with published microarray data of different embryonic stages (Ohnishi et al., 2014).
See also Figure S2.
As 2i- and KOSR-cultured ESCs both generated high-level chimeras from single cells, we asked whether they shared a common gene-expression signature. As serum-cultured ESCs did not demonstrate this property, we overlapped genes that were differentially expressed in 2i or KOSR compared with standard serum cultures (Tables S1–S4) and found a highly significant, non-random overlap (Figure 2C). In KOSR-cultured ESCs of annotated genes that were significantly changing in expression, 46% of upregulated (representation factor 5.5, p < 8.1 × 10−84) and 62% of downregulated genes (representation factor 5.0, p < 6.7 × 10−101) overlapped with those up- or downregulated in 2i (Figure 2C and Table S9). Common upregulated genes were associated with germ cell development, e.g., Dazl, Hormad1, and Sohlh2, while downregulated genes were enriched for adhesion, migration, and differentiation markers, e.g., Lamb3, Egfr, and Neurod1 (Table S10).
As the correlation between KOSR-cultured ESCs and PrE had not previously been reported, we investigated this in more detail. We confirmed by qPCR that the PrE markers Gata4, Gata6, Dab2, and FoxA2 were upregulated in KOSR compared with either serum or 2i conditions (Figure S2A). KOSR culture also resulted in elevated expression of a Hhex-Venus (HV) transgene, which marks a PrE-primed ESC population (Canham et al., 2010) (Figure 2D). We observed that expression of the Nanog-GFP reporter (Chambers et al., 2007) was increased in KOSR cultures compared with serum cultures, although not to levels as high as in 2i (Figure 2E). In serum cultures, the PrE marker Hhex and the Epi marker Nanog are expressed mutually exclusively, whereas in 2i they are coexpressed (Canham et al., 2010, Morgani et al., 2013). Similar observations were made in KOSR, where the Hhex-expressing subpopulation contained a high number of Nanog-positive cells (Figure S2B). We also observed that KOSR cultures contained cells that coexpressed OCT4, SOX2, and GATA6 protein not observed in either serum or 2i cultures (Figures 2F and S2C). Similar observations were made by flow cytometry, as we observed a fraction of ESCs expressing the PrE marker platelet-derived growth factor receptor α (PDGFRα) in KOSR, but not serum or 2i (Figures S2D and S2E).
As there were marked transcriptional and functional differences between ESCs maintained in serum, 2i, or KOSR, we asked how these expression states compared with gene expression during embryonic development. We compared our microarray data with expression data from pre-implantation embryo populations (Ohnishi et al., 2014) using ExAtlas (FDR ≤0.05, fold change ≥2) (Sharov et al., 2015). We observed a correlation between ESCs cultured in 2i and KOSR and early embryos (E3.25) (Figure 2G and Table 3, z = 3.5 and 3.6, respectively), when ICM cells are unspecified and have a wide functional potency (Grabarek et al., 2012, Rossant and Lis, 1979). This is consistent with our previous observation that 2i cultures contain totipotent cells (Morgani et al., 2013). In addition, 2i-cultured ESCs showed a correlation with E3.5 early Epi (Figure 2G and Table 3, z = 7.4), which may reflect a second population of Epi-like Nanog-positive, Hhex-negative cells. As KOSR-cultured ESCs express endoderm markers, it is not surprising that gene expression in these conditions correlates with all stages of PrE development (Figure 2G and Table 3, E3.5 PrE z = 7.5, E4.5 PrE z = 9.4). Conversely, gene expression in serum-cultured ESCs is correlated with later E4.5 embryos (Figure 2G and Table 3, E4.5 Epi z = 4.5, E4.5 PrE z = 3.3) when Epi and PrE are fully specified and functionally restricted (Grabarek et al., 2012). As ESCs cultured in serum undergo a degree of spontaneous differentiation into cell types reminiscent of the later embryo, these differentiated cells could also contribute to the increased correlation of the serum transcriptome to later developmental stages. This spontaneous differentiation is inhibited by culture in either 2i or KOSR.
Table 3.
Lists of Genes Correlated between 2i- and KOSR-Cultured ESCs and Different Stages of Embryonic Development, Related to Figure 2
| Symbols | EPFP | Gene Title | RefSeq |
|---|---|---|---|
| E3.25 versus 2i (z Value, 3.508; Correlation, 0.1001) | |||
| Bhmt2 | 0.0237 | betaine-homocysteine methyltransferase 2 | NM_022884 |
| Spic | 0.0283 | Spi-C transcription factor (Spi-1/PU.1 related) | NM_011461 |
| Jakmip1 | 0.1569 | janus kinase and microtubule interacting protein 1 | NM_178394 |
| Ly6a | 0.2471 | lymphocyte antigen 6 complex, locus A | NM_010738 |
| Gsc | 0.2702 | goosecoid homeobox | NM_010351 |
| Rab38 | 0.2745 | RAB38, member of RAS oncogene family | NM_028238 |
| Khdc1a | 0.2745 | KH domain-containing 1A | NM_183322 |
| Fxyd4 | 0.2859 | FXYD domain-containing ion transport regulator 4 | NM_033648 |
| Bdh1 | 0.319 | 3-hydroxybutyrate dehydrogenase, type 1 | NM_175177 |
| Bcl2l14 | 0.3263 | BCL2-like 14 (apoptosis facilitator) | NM_025778 |
| Cdkn1a | 0.4118 | cyclin-dependent kinase inhibitor 1A (P21) | NM_007669 |
| Gss | 0.4612 | glutathione synthetase | NM_008180 |
| Pdp2 | 0.4887 | pyruvate dehydrogenase phosphatase catalytic subunit 2 | NM_001024606 |
| E3.25 versus KOSR (z Value, 3.59; Correlation, 0.1024) | |||
| Pdgfra | 0.1396 | platelet-derived growth factor receptor, α polypeptide | NM_011058 |
| Cphx1 | 0.3658 | cytoplasmic polyadenylated homeobox 1 | NM_175342 |
| Gata6 | 0.3658 | GATA binding protein 6 | NM_010258 |
| Pnliprp2 | 0.3658 | pancreatic lipase-related protein 2 | NM_011128 |
| E3.5 Epi versus 2i (z Value, 7.351; Correlation, 0.2063) | |||
| Spic | 0.0087 | Spi-C transcription factor (Spi-1/PU.1 related) | NM_011461 |
| Gsc | 0.1293 | goosecoid homeobox | NM_010351 |
| Aspa | 0.1293 | aspartoacylase | NM_023113 |
| Gsta2 | 0.1293 | glutathione S-transferase, α 2 (Yc2) | NM_008182 |
| Mal | 0.3835 | myelin and lymphocyte protein, T cell differentiation protein | NM_010762 |
| Khdc1a | 0.4185 | KH domain-containing 1A | NM_183322 |
| E3.5 PrE versus KOSR (z Value, 7.538; Correlation, 0.2113) | |||
| Pdgfra | 0.1232 | platelet-derived growth factor receptor, α polypeptide | NM_011058 |
| Cubn | 0.1416 | cubilin (intrinsic factor-cobalamin receptor) | NM_001081084 |
| Tcf23 | 0.1416 | transcription factor 23 | NM_053085 |
| Gata4 | 0.236 | GATA binding protein 4 | NM_008092 |
| Cpn1 | 0.236 | carboxypeptidase N, polypeptide 1 | NM_030703 |
| Gata6 | 0.2364 | GATA binding protein 6 | NM_010258 |
| Gdpd5 | 0.2364 | glycerophosphodiester phosphodiesterase domain-containing 5 | NM_201352 |
| Srgn | 0.2364 | serglycin | NM_011157 |
| Meis1 | 0.2364 | Meis homeobox 1 | NM_010789 |
| Sox17 | 0.2364 | SRY (sex determining region Y)-box 17 | NM_011441 |
| Pth1r | 0.2389 | parathyroid hormone 1 receptor | NM_011199 |
| Bmp6 | 0.247 | bone morphogenetic protein 6 | NM_007556 |
| Aqp8 | 0.26 | aquaporin 8 | NM_007474 |
| E4.5 PrE versus KOSR (z Value, 9.405; Correlation, 0.2604) | |||
| Cubn | 0.0421 | cubilin (intrinsic factor-cobalamin receptor) | NM_001081084 |
| Sox7 | 0.3368 | SRY (sex determining region Y)-box 7 | NM_011446 |
| Srgn | 0.3378 | serglycin | NM_011157 |
| Nostrin | 0.3378 | nitric oxide synthase trafficker | NM_181547 |
| Pdgfra | 0.3378 | platelet-derived growth factor receptor, α polypeptide | NM_011058 |
| Sox17 | 0.3439 | SRY (sex determining region Y)-box 17 | NM_011441 |
| Gata4 | 0.3536 | GATA binding protein 4 | NM_008092 |
| Pth1r | 0.3953 | parathyroid hormone 1 receptor | NM_011199 |
| Ctsh | 0.4678 | cathepsin H | NM_007801 |
Pearson correlation of our microarray data of ESCs cultured in serum, 2i, and KOSR and microarrays on different stages of embryonic development (Ohnishi et al., 2014) was determined using ExAtlas online tool for gene expression meta-analysis. The list of correlated genes (FDR ≤0.1, fold change ≥1.5), z value, and correlation value is shown. EPFP, expected proportion of false positives.
2i- and KOSR-Cultured ESCs Can Contribute to 2C Embryos
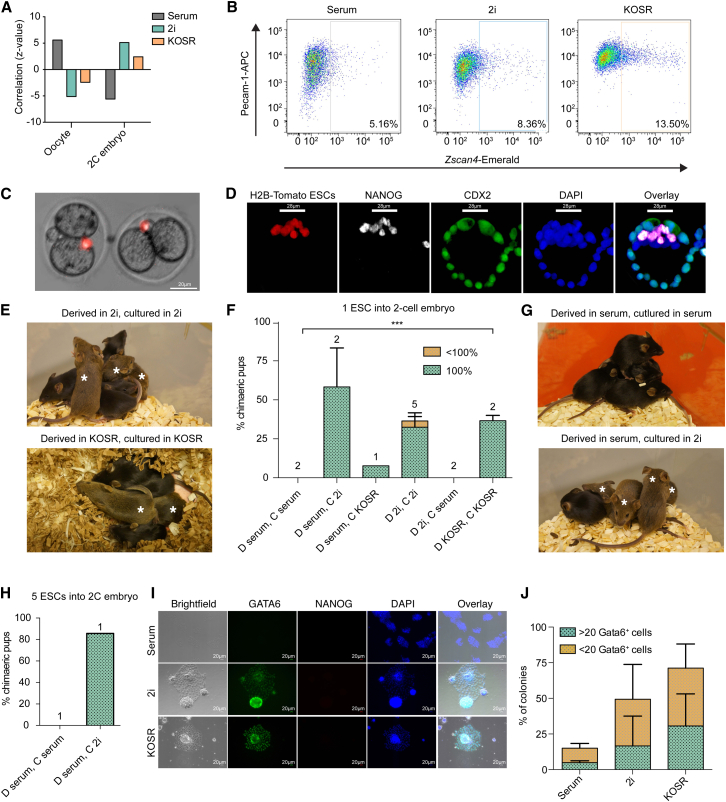
As gene-expression profiles of 2i and KOSR-cultured ESCs correlated with the early blastocyst (Figure 2G), in addition to containing subpopulations that coexpressed embryonic and extraembryonic determinants (Figure 2E) (Morgani et al., 2013), reminiscent of early developmental stages, we asked whether they showed a correlation to even earlier stages of development. We compared our microarray data with RNA-sequencing data on oocytes and 2C embryos (Macfarlan et al., 2012) (ExAtlas, FDR ≤0.05, fold change ≥2). Gene expression of ESCs cultured in 2i and KOSR was correlated with data from 2C embryos, but not oocytes (Figure 3A, z values of 5.1 and 2.4, respectively). However, these correlations were not as strong as those to later stages of development (Figure 2G). Conversely, serum-cultured ESCs showed a correlation with oocytes (z = 5.6) but not 2C embryos (z = −5.6) (Figure 3A). This may be a result of the correlation in gene expression between the late blastocyst and oocytes that we found when re-analyzing published datasets (Figure S3A). To further assess the significance of a potential 2C-like population, we asked whether culture in different conditions could induce the expression of a reporter for the 2C-specific marker (Falco et al., 2007, Hirata et al., 2012), Zscan4. An induction of Zscan4-Emerald was observed in both 2i and KOSR (Figures 3B and S3B).
Figure 3.
2i and KOSR ESCs Can Form 100% ESC-Derived Mice When Injected into 2C Embryos
ESCs were derived (D) in and cultured (C) under various conditions; D and C in serum, D in serum and C in 2i, D and C in 2i, D in 2i and C in serum, and D and C in KOSR. Single ESCs from each of these conditions were injected into 2C embryos.
(A) Graph showing the correlation of serum-, 2i-, and KOSR-cultured ESCs with published microarray data of oocytes and 2C embryos (Macfarlan et al., 2012).
(B) Flow cytometry profiles for Zscan4-Emerald ESCs, immunostained for the marker of undifferentiated ESCs PECAM-1, cultured in serum, 2i, and KOSR.
(C) Representative 2C embryos immediately after injection with a single ESC expressing a constitutive H2B-Tomato marker.
(D) Confocal optical sections of an immunostained late blastocyst resulting from the injection of a single 2i-cultured ESC, expressing a constitutive H2B-Tomato marker, into a 2C embryo.
(E) Representative images of litters containing 100% ESC-derived agouti mice generated from the injection of a single 2i- or KOSR-cultured cell into a 2C embryo, along wild-type C57BL/6 littermates. Asterisks indicate mice with ESC contribution.
(F and G) Single ESCs were injected into 2C embryos and their contribution to full-term chimeras assessed according to coat color. (F) Graph showing the percentage of chimeras born out of the total number of pups and quantification of coat color chimerism. n Values are provided above bars and refer to the number of independent ESC clones that were tested for each condition. Error bars indicate SEM. (G) Representative images of full-term mice generated from the injection of single-cell ESCs, D and C in serum (upper panel), or D and C in 2i (lower panel), into 2C embryos. Asterisks indicate mice with ESC contribution.
(H) Graph showing the percentage of chimeras born out of the total number of pups and quantification of coat color chimerism. n Values are provided above bars and refer to the number of independent ESC clones tested for each condition.
(I) ESCs were pre-cultured in serum, 2i, or KOSR for three passages and then switched to XEN cell medium for two passages. Images show representative colonies.
(J) Quantification of the average proportion of colonies under each condition expressing GATA6. A distinction was made between colonies with >20 GATA6+ cells and those with <20 GATA6+ cells. Error bars indicate SD of the mean of five randomly selected fields of view.
See also Figure S3.
Based on the capacity of 2i- and KOSR-cultured ESCs to stimulate extraembryonic and 2C gene expression, we asked whether they were capable of contributing to the earliest stages of embryonic development. We injected single 2i-cultured ESCs labeled with H2B-Tomato into 2C embryos (Figure 3C). In the majority of cases (n = 8/9), 100% of NANOG+ Epi cells were generated from the single injected ESC (Figures 3D, S3C, and S3D). We also observed a contribution of ESC progeny to the extraembryonic trophoblast (Figure S3C). While 2i-cultured ESCs introduced at the 2C stage divided and populated the ICM, the majority of serum-cultured cells were excluded during pre-implantation development (Figures S3E and S3F).
Despite the high levels of chimerism observed in single-cell injections as late as mid-gestation (Figure S3D) (Morgani et al., 2013), no live-born chimeras were obtained from 129/Ola lines injected as single cells. In contrast, full-term ESC-derived mice were obtained from the injection of single F1 129S2;C57BL/6N hybrid 2i- or KOSR-cultured ESCs, but not from those cultured in serum (Figures 3G–3I and Table 1). As with morula injections, these mice showed a high, virtually homogeneous contribution to ectoderm, judged by coat color and internal organs (Figures S3G and S3I). All tested ESC-derived mice contributed to the germline (n = 5/5, Table 2). Similar to the single-cell morula injections (Figures 1G and 1I), transfer of serum-derived ESCs to 2i culture endowed these cell lines with the capacity to contribute to 2C embryos (Figures 3F and 3G; Table 1). In addition, when 2i-derived ESCs were switched to serum-containing medium, either directly or by gradual adaptation, these cells lost this capacity (Figure 3F).
We then asked whether the inability of serum-cultured ESCs to contribute to 2C embryos was due to their previously observed reduced clonogenicity (Figure 1G). We injected five ESCs into 2C embryos but no full-term chimeras were obtained (Figure 3H and Table 1). When these serum-derived ESCs were switched to 2i culture, the injection of multiple ESCs into 2C embryos resulted in six out of seven 100% ESC-derived mice (Figure 3H and Table 1). This suggests that, as well as exhibiting a lower level of clonogenicity in morula injection, serum-cultured ESCs cannot contribute to early 2C embryos, even when injected as multiple cells.
We recently showed that a totipotent fraction of 2i-cultured cells is characterized by the expression of an HV transgene and the coexpression of embryonic and extraembryonic determinants within individual cells (Morgani et al., 2013). These cells also demonstrated an enhanced capacity to differentiate toward the extraembryonic lineages. As KOSR-cultured ESCs coexpressed embryonic and extraembryonic markers and demonstrated a similar clonal capacity to contribute to early pre-implantation development, we asked whether KOSR-cultured cells could generate extraembryonic cell types, both trophoblast and extraembryonic endoderm (XEN). We have previously shown that 2i-cultured ESCs generate CDX2+ trophoblast-like cells when cultured in trophoblast stem cell (TSC) medium (Morgani et al., 2013). ESCs cultured in either serum or KOSR did not have this capacity (Figure S3K). Serum-cultured ESCs died after two passages while KOSR-cultured cells survived but did not express CDX2. However, when ESCs were transferred to XEN cell media, in the absence of lineage-promoting cytokines, both KOSR and 2i-cultured ESCs displayed a XEN-like morphology, downregulated the pluripotency marker NANOG, and expressed the endoderm marker GATA6 (Figures 3I and 3J). While the majority of serum-cultured ESCs downregulated NANOG, they did not express GATA6. Preliminary data from injections of KOSR-cultured ESCs into morulae suggests that these cells may also to contribute to both embryonic and extraembryonic lineages in vivo (Figure S3J). Taken together, these data indicate that ESCs cultured in 2i or KOSR have the capacity to generate extraembryonic lineages and this correlates with their gene-expression profiles, in addition to their capacity to colonize 2C embryos.
Discussion
In this paper we have shown that the culture conditions in which ESCs are grown can be used to re-program their potency irrespective of the protocol by which they were derived. Single ESCs cultured in 2i and KOSR culture conditions demonstrated the capacity to contribute effectively to embryonic development from both the morula and 2C stage. Moreover, this enhanced capacity to contribute to embryonic development correlated with the expression of extraembryonic-associated RNAs and 2C determinants. Thus, while pluripotency factors were expressed in all three culture conditions, ESCs showed distinct gene-expression profiles, and these profiles appear to be correlated with cell behavior in vivo.
Canonical ESC derivation protocols yield variable efficiencies depending on the genetic background of the mice used (Auerbach et al., 2000, Brook and Gardner, 1997). Using improved ESC derivation protocols, it is now possible to generate ESCs from a variety of backgrounds (Nichols et al., 2009a, Nichols and Ying, 2006, Yang et al., 2009, Ying et al., 2008). Although we analyzed inbred (129) and outbred (F1) genetic backgrounds in this study, to compare different derivation approaches we started with embryos of an F1 genetic background known to generate high-quality ESC lines based on hybrid vigor (Eggan et al., 2001, Rideout et al., 2000). The ability of single ESCs to contribute to 2C or morula stages was independent of background, but the capacity to generate live-born entirely ESC-derived mice appeared to be linked to an F1 genetic background. However, as the 129 ESCs used in this study have been in culture for longer periods, cultural adaptation may contribute to this reduced efficiency. Nevertheless, previous data show that F1 hybrid ESC lines produce viable offspring with increased efficiency and suggest that genetic heterozygosity is a requirement to support post-natal development of entirely ESC-derived mice (Eggan et al., 2001, Kirchain et al., 2008).
Previous studies suggest that approximately one out of ten serum-cultured ESCs contribute to embryonic development (Wang and Jaenisch, 2004). If 2i and KOSR cultures generated more single-cell chimeras by increasing the proportion of functionally potent ESCs, we would also expect to observe some chimeras from serum-cultured ESCs (n = 87 injected embryos). As a result, it is not merely that serum-cultured ESCs contain fewer “pluripotent” cells, but that their clonogenic capacity to self-renew is limited. The increased functional potential of individual 2i-cultured (Morgani et al., 2013) and KOSR-cultured ESCs to generate embryonic and extraembryonic lineages could be a feature of their increased clonogenicity. It has previously been shown that ESCs cultured in the presence of the 2i inhibitors as well as an FGF receptor inhibitor (3i) exhibit increased clonogenicity compared with serum-cultured cells in vitro (Ying et al., 2008), and single 2i-cultured cells show an increased capacity to generate multiple cell types in differentiation assays (Morgani et al., 2013). If 2i- and KOSR-cultured ESCs undergo increased numbers of self-renewing cell divisions, even in the presence of signals that promote lineage restriction, they may generate a critical mass to effectively contribute to both Epi and extraembryonic lineages. In support of this hypothesis, ESC clones exhibiting a greater rate of proliferation generate a higher number of chimeras than those with slower proliferation rates (Wang and Jaenisch, 2004).
Although individual clonogenic ESCs can appear totipotent, the fraction of these cells in different culture conditions may vary, and this could explain why aggregate gene-expression data indicate that 2i cells represent distinct developmental stages (Boroviak et al., 2014). Here we find two clear signatures in the 2i dataset, early pre-implantation development and Epi, perhaps representing this heterogeneity.
We also observed that ESC culture in KOSR enhances their functional potency. At a transcriptional and functional level, these cells are biased toward endoderm. This is distinct from cells cultured in 2i that can differentiate into both trophoblast and extraembryonic endoderm in vitro and in vivo (Morgani et al., 2013). However, ESCs grown and derived in either condition exhibit enhanced potency on a single-cell level. Thus ESC culture conditions may enhance potency by promoting competence for extraembryonic lineages. Moreover, a number of recent reports on ESC totipotency use KOSR in their culture medium (Abad et al., 2013, Macfarlan et al., 2012). As a result, it is intriguing to speculate that this media condition can enhance the reprogramming of normal ESCs to a totipotent, clonogenic state.
Experimental Procedures
Mouse Line Maintenance
Mouse lines were maintained under 12/12-hr light/dark cycles in the designated facilities at the University of Copenhagen. Mice were checked for copulation plugs each morning and embryos were considered embryonic day 0.5 (E0.5) on the day of plug detection. All animal work was carried in accordance with European legislation. All work was authorized by and carried out under Project License 2012-15-2934-00743 issued by the Danish Regulatory Authority.
ESC Line Derivation
ESCs derived in this study were F1 hybrid 129S2;C57BL/6N, and 129S6;C57BL/6N lines. Embryos were obtained from mice supplied by commercial vendors with their corresponding certificates (129S6, Taconic; 129S2, Charles River; C57BL/6, Harlan). Females were superovulated at 4 (C57BL/6) or 7 (129) weeks old and mated with fertile males (8–24 weeks old). For derivation in 2i, E2.5 embryos were flushed from oviducts in M2 medium and cultured in KSOM (Millipore) for approximately 8 hr until the evening where they were transferred to KSOM + 2i (1 μM PD0325901, Stemgent; 3 μM CHIR99021, Biovision). Embryos were cultured overnight until E3.5 blastocysts, then transferred to individual wells of gelatinized 96-well plates containing 2i/LIF ESC culture medium (2i inhibitors in N2B27 medium). Embryos were cultured for 5–8 days at 37°C, 5% CO2, and 90% humidity, until a cluster of cells generated from the ICM emerged. ICM outgrowths were picked and dissociated into smaller clusters in Accutase (Sigma) for 3–5 min at 37°C before being plated into fresh medium for expansion. The same protocol was used to derive ESCs in serum and KOSR conditions but with pre-culture of embryos in KSOM alone and then transfer to 96-well plates containing either serum/LIF or KOSR/LIF. Derivation of ESC lines in serum/LIF and KOSR/LIF was only possible on mouse embryonic fibroblasts (MEFs), although in 2i/LIF conditions ESCs could be derived on gelatin.
ESC Culture
ESC lines were maintained in serum/LIF (Canham et al., 2010), 2i/LIF (Ying et al., 2008) as previously described or KOSR/LIF medium (80% Knockout DMEM [Gibco], 20% Knockout Serum Replacement [Gibco], non-essential amino acids [Gibco], glutamine and sodium pyruvate [Gibco], 0.1 mM β-mercaptoethanol [Sigma]). H2B-Tomato ESCs were generated using E14 ESCs from the 129/Ola background (Morgani et al., 2013). Newly derived ESC lines described are F1 hybrid 129S6;C57BL/6N lines. F1 hybrid lines are named as follows. 2i-derived lines: TCF2.2, TCF2.5, TCF2.6; serum-derived lines: TCF3.4, TCF3.5; KOSR-derived lines: TCF5.30, TCF5.45. ESC lines were switched from their original derivation medium to alternative medium conditions. In the case of serum-cultured cells, ESCs were maintained in the serum for a minimum of 12 passages before being switched to 2i for three passages prior to injection. F1 ESC lines were also electroporated with a constitutive pCAG Kozak-Venus marker (a kind gift from Dr. Heiko Lickert). Zscan4-Emerald ESCs were obtained from the Ko laboratory (Zalzman et al., 2010).
XEN and TSC Differentiation Assays
For XEN cell differentiation, ESCs were cultured for three passages in serum, 2i, or KOSR and transferred to XEN cell medium (RPMI [Sigma], 15% fetal bovine serum, glutamine and sodium pyruvate, 0.1 mM β-mercaptoethanol) for two passages (6 days total). Cells were immunostained for GATA6 and NANOG and the average proportion of GATA6+ colonies was quantified using five random fields of view per condition. As there was variation in the number of GATA6+ cells/colony, we distinguished between colonies with <20 GATA6+ cells or >20 GATA6+ cells. Overall colony size did not significantly differ. For TSC differentiation, ESCs were cultured for three passages in serum, 2i, or KOSR, then transferred to TSC medium for three passages (total 6 days) (Tanaka et al., 1998). Resultant cultures were stained for CDX2.
Alkaline Phosphatase Staining
ESCs were plated at clonal density and cultured for 9–10 days. Alkaline phosphatase staining was carried out as per manufacturer’s instructions. Colonies were imaged using a Nikon AZ-100 microscope.
Chimera Generation and Embryo Culture
Embryos were obtained as for ESC derivation. Single or multiple (5) ESCs were injected into 2C or 8-cell embryos. Inbred cell lines were sorted by flow cytometry prior to injection (Figure S3F) as previously described (Morgani et al., 2013). For 2C embryo injection, E1.5 wild-type C57BL/6N embryos were flushed from oviducts. Embryos were de-compacted in PB1 medium without calcium and magnesium for 20 min at room temperature and a single cell introduced by microinjection. The same process was used for injection of E2.5 morula. Resultant embryos were cultured ex vivo in KSOM microdrops covered with mineral oil (Sigma) or transferred to pseudopregnant CD1 females (8–13 weeks old) and allowed to develop to term.
Microarray Analysis
Unsorted ESCs were maintained in serum/LIF, 2i/LIF, or KOSR/LIF. Samples were collected from three independent ESC clones from the 129/Ola background: E14, HVJu5.1, and HVJu9.3 (Morgani et al., 2013). For 2i/LIF, five samples were collected with two samples each for HVJu5.1 and HVJu9.3 cell lines; for serum/LIF and KOSR/LIF conditions, one sample was collected for each cell line. Each condition is represented at least as biological triplicates. RNA isolation was performed using Qiagen RNeasy kits according to the manufacturer's instructions. RNA pellets were re-suspended in Nuclease Free Water (Ambion) and RNA concentration was measured using the Nanodrop system. 260/280 values for all samples were in the range of 1.8–2.0. RNA quality was additionally assessed using the Agilent 2100 BioAnalyzer with all samples having a RIN of ≥9.7. Transcriptome analysis was carried out in-house with one-color 8 × 60K Mouse Gene Expression Microarrays (Agilent Technologies, G4852A) as per manufacturer's instructions. In brief, 150 ng of total RNA was labeled using the Low Input Quick Amp Labeling Kit (Agilent Technologies, 5190-2305). Labeled samples were hybridized overnight, then washed and scanned using the high-sensitivity protocol (AgilentG3_HiSen_GX_1color) on a SureScan microarray scanner (Agilent Technologies). Probe intensities were obtained by taking the gProcessedSignal from the output of Agilent feature extraction software using default settings. Probe intensities were analyzed using the NIA Array Analysis tool (http://lgsun.grc.nia.nih.gov/ANOVA/).
Analysis of microarray data was carried out using ExAtlas online tool for meta-analysis of gene-expression data developed by the NIA (http://lgsun.grc.nia.nih.gov/exatlas/) (Sharov et al., 2015). Pearson correlation between datasets was measured using the following parameters: false discovery rate (FDR) ≤0.05, fold change ≥2, angle 0.8, expected proportion of false positives (EPFP) 0.5. To obtain correlating gene lists (Table S10), the following parameters were used: FDR 0.1, fold change ≥1.5, angle 0.8, EPFP 0.5. Gene ontology analysis was performed using the Gene Ontology Consortium online tool (http://geneontology.org).
Immunostaining of ESCs
ESCs were plated in ibiTreat 15 μ-Slide 8-well dishes (Ibidi). ESCs were immunostained as described by Canham et al. (2010). Primary antibodies were used at the following concentrations: Gata6 (R&D, AF1700 [Morgani et al., 2013]) 1:100, Gata6 D61 × 104 XP (Cell Signaling, 5851 [Lim et al., 2014]) 1:200, Oct4 (C-10 Santa Cruz Biotechnology, sc5279 [Hall et al., 2009]) 1:100, Sox2 (Santa Cruz, sc17320 [Huang et al., 2012]) 1:100, Nanog (eBioscience, 14-5761 [Morgani et al., 2013]) 1:200, Cdx2 (MU392A, Biogenex [Nichols et al., 2009b]) 1:200. Alexa Fluor secondary antibodies (Invitrogen) were diluted 1:500. When immunostaining for HV, a conjugated anti-GFP-Alexa 488 antibody (1:200, Molecular Probes, A21311 [Morgani et al., 2013]) was added at the same time as the secondary antibodies. Cells were imaged on a Leica TCS SP8.
Immunostaining of Embryos
Immunostaining of embryos was carried out as described by Nichols et al. (2009b). The primary antibodies were used at the following dilutions: Nanog (eBioscience, 14-5761 [Le Bin et al., 2014]) 1:200, Cdx2 (Biogenex, MU392A-UC [Nichols et al., 2009b]) 1:200. Alexa Fluor secondary antibodies were used at 1:500. Embryos were imaged in an Attofluor chamber (Life Technologies) on a 25-mm glass coverslip using a Leica TCS SP8 confocal.
Organ Disaggregation and Flow Cytometry
Internal organs were dissected in PB1 medium and imaged using a Nikon AZ100. A section of organ was manually disaggregated with scissors in PBS. Dissociated tissue was disaggregated in trypsin with 20 μL/mL DNase (Qiagen) while shaking at 37°C for 15 min. Disaggregated Kozak-Venus chimeric organs were re-suspended in PBS with 100 ng/mL DAPI and analyzed. The disaggregated spleen of chimeric mice generated from unlabeled 129 ESCs was re-suspended in PBS containing 1:50 anti-CD229.1-APC 30C7 conjugated antibody (560188, BD Pharmingen [Tawara et al., 2012]). Cells were incubated on ice for 15 min, then washed three times with PBS before being re-suspended in PBS with 100 ng/mL DAPI. Data were collected using a BD LSR Fortessa. Flow cytometry analysis was carried out using FlowJo software (Tree Star) first by gating on forward and side scatter to identify a cell population and eliminate debris, then to gate DAPI-negative, viable cells before assessing the level of Venus or APC. PDGFRα flow cytometry was carried out as described by Rugg-Gunn et al. (2012).
Antibody Staining and Flow Cytometry of ESCs
ESCs were collected by trypsinization, and antibody staining for PECAM-1 (BD Pharmingen, 551262 [Morgani et al., 2013]) was carried out as described by Canham et al. (2010). Data were collected using a BD LSR Fortessa. Flow cytometry analysis was carried out using FlowJo software (Tree Star) first by gating on forward and side scatter to identify a cell population and eliminate debris, then to gate DAPI-negative, viable cells and PECAM-1-APC-positive cells, before assessing the level of Venus or GFP. For Zscan4-Emerald analysis, gating was based on E14 wild-type ESCs.
qRT-PCR
Three independent ESC lines (E14, HVJu5.1, and HVJu9.3) were cultured in serum/LIF, 2i/LIF, or KOSR/LIF for three passages. RNA was isolated from cells (RNeasy, Qiagen). cDNA was synthesized from 1 μg of RNA using Superscript III according to the manufacturer's guidelines (Invitrogen). qRT-PCR was carried out on a Lightcycler480 (Roche) using UPL technology (Roche) utilizing primer sequences specified in Supplemental Experimental Procedures. Primers were used at a concentration of 1 μM. The geometric mean of the housekeeping genes Tbp, Gapdh, and Pgk1 was used for normalization of Ct values detected for each sample.
Statistical Analysis
Statistical analysis was performed using the QuickCalc GraphPad (http://www.graphpad.com/quickcalcs/index.cfm). For non-parametric data, two-tailed chi-square tests were performed. For parametric data comparing two samples, an unpaired Student's t test was used. A p value is shown when the difference between compared groups is significant. The statistical significance of the overlap of gene lists was assessed using http://nemates.org/MA/progs/overlap_stats.cgi, using a genome size of 25,000. The representation factor is the number of overlapping genes divided by the expected number of overlapping genes drawn from two random, independent groups. A representation factor >1 indicates a greater overlap than expected by chance.
Author Contributions
J.M.G. conceived of and executed experiments. S.M.M. conceived of and executed experiments and wrote the paper with J.M.B. R.A.B., K.B., and S.A. executed experiments. C.B. designed and supervised experiments. J.M.B. designed and supervised experiments and wrote the paper with S.M.M.
Acknowledgments
We thank the entire Brickman laboratory for comments and critical discussion; Gelo de la Cruz and Alexei Sharov for technical assistance; and Heiko Lickert and Minoru Ko for reagents. This work was funded by the Novo Nordisk Foundation Section for Basic Stem Cell Biology and the Faculty of Health Sciences at the University of Copenhagen. S.M.M. was supported by a University of Copenhagen Studentship.
Published: August 9, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and ten tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.07.009.
Accession Numbers
The microarray data have been deposited under accession number GEO: GSE72445.
Supplemental Information
References
- Abad M., Mosteiro L., Pantoja C., Canamero M., Rayon T., Ors I., Grana O., Megias D., Dominguez O., Martinez D. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- Auerbach W., Dunmore J.H., Fairchild-Huntress V., Fang Q., Auerbach A.B., Huszar D., Joyner A.L. Establishment and chimera analysis of 129/SvEv- and C57BL/6-derived mouse embryonic stem cell lines. Biotechniques. 2000;29:1024–1028. doi: 10.2144/00295st04. 1030, 1032. [DOI] [PubMed] [Google Scholar]
- Beddington R.S., Robertson E.J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A., Evans M., Kaufman M.H., Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- Brook F.A., Gardner R.L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham M.A., Sharov A.A., Ko M.S., Brickman J.M. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Eggan K., Akutsu H., Loring J., Jackson-Grusby L., Klemm M., Rideout W.M., 3rd, Yanagimachi R., Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Falco G., Lee S.L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A., Doetschman T., Korn R., Serfling E., Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc. Natl. Acad. Sci. USA. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarek J.B., Zyzynska K., Saiz N., Piliszek A., Frankenberg S., Nichols J., Hadjantonakis A.K., Plusa B. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139:129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T., Amano T., Nakatake Y., Amano M., Piao Y., Hoang H.G., Ko M.S. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Osorno R., Tsakiridis A., Wilson V. Vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012;2:1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Kirchain S.M., Hayward A.M., Mkandawire J.M., Qi P., Burds A.A. Comparison of tetraploid blastocyst microinjection of outbred Crl:CD1(ICR), hybrid B6D2F1/Tac, and inbred C57BL/6NTac embryos for generation of mice derived from embryonic stem cells. Comp. Med. 2008;58:145–150. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand Y., Brulet P. An in situ assessment of the routes and extents of colonisation of the mouse embryo by embryonic stem cells and their descendants. Development. 1990;110:1241–1248. doi: 10.1242/dev.110.4.1241. [DOI] [PubMed] [Google Scholar]
- Le Bin G.C., Munoz-Descalzo S., Kurowski A., Leitch H., Lou X., Mansfield W., Etienne-Dumeau C., Grabole N., Mulas C., Niwa H. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development. 2014;141:1001–1010. doi: 10.1242/dev.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.L., Ooi B.N., Jungebluth P., Sjoqvist S., Hultman I., Lemon G., Gustafsson Y., Asmundsson J., Baiguera S., Douagi I. Characterization of stem-like cells in mucoepidermoid tracheal paediatric tumor. PLoS One. 2014;9:e107712. doi: 10.1371/journal.pone.0107712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S.M., Canham M.A., Nichols J., Sharov A.A., Migueles R.P., Ko M.S., Brickman J.M. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 2013;3:1945–1957. doi: 10.1016/j.celrep.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Ying Q.L. Derivation and propagation of embryonic stem cells in serum- and feeder-free culture. Methods Mol. Biol. 2006;329:91–98. doi: 10.1385/1-59745-037-5:91. [DOI] [PubMed] [Google Scholar]
- Nichols J., Jones K., Phillips J.M., Newland S.A., Roode M., Mansfield W., Smith A., Cooke A. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 2009;15:814–818. doi: 10.1038/nm.1996. [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y., Huber W., Tsumura A., Kang M., Xenopoulos P., Kurimoto K., Oles A.K., Arauzo-Bravo M.J., Saitou M., Hadjantonakis A.K. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 2014;16:27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymirou W.T., Auerbach W., Frendewey D., Hickey J.F., Escaravage J.M., Esau L., Dore A.T., Stevens S., Adams N.C., Dominguez M.G. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat. Biotechnol. 2007;25:91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- Rideout W.M., 3rd, Wakayama T., Wutz A., Eggan K., Jackson-Grusby L., Dausman J., Yanagimachi R., Jaenisch R. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat. Genet. 2000;24:109–110. doi: 10.1038/72753. [DOI] [PubMed] [Google Scholar]
- Robertson E., Bradley A., Kuehn M., Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- Rossant J., Lis W.T. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev. Biol. 1979;70:255–261. doi: 10.1016/0012-1606(79)90022-8. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Cox B.J., Lanner F., Sharma P., Ignatchenko V., McDonald A.C., Garner J., Gramolini A.O., Rossant J., Kislinger T. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev. Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburi S., Azuma S., Sato E., Toyoda Y., Tachi C. Developmental fate of single embryonic stem cells microinjected into 8-cell-stage mouse embryos. Differentiation. 1997;62:1–11. doi: 10.1046/j.1432-0436.1997.6210001.x. [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Schlessinger D., Ko M.S. ExAtlas: an interactive online tool for meta-analysis of gene expression data. J. Bioinform. Comput. Biol. 2015;13:1550019. doi: 10.1142/S0219720015500195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Hamazaki T., Hankowski K.E., Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Smith A.G., Heath J.K., Donaldson D.D., Wong G.G., Moreau J., Stahl M., Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Suemori H., Kadodawa Y., Goto K., Araki I., Kondoh H., Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous beta-galactosidase expression. Cell Differ. Dev. 1990;29:181–186. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A.K., Nagy A., Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawara I., Sun Y., Lewis E.C., Toubai T., Evers R., Nieves E., Azam T., Dinarello C.A., Reddy P. Alpha-1-antitrypsin monotherapy reduces graft-versus-host disease after experimental allogeneic bone marrow transplantation. Proc. Natl. Acad. Sci. USA. 2012;109:564–569. doi: 10.1073/pnas.1117665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Wang Z., Jaenisch R. At most three ES cells contribute to the somatic lineages of chimeric mice and of mice produced by ES-tetraploid complementation. Dev. Biol. 2004;275:192–201. doi: 10.1016/j.ydbio.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Ward C.M., Stern P., Willington M.A., Flenniken A.M. Efficient germline transmission of mouse embryonic stem cells grown in synthetic serum in the absence of a fibroblast feeder layer. Lab. Invest. 2002;82:1765–1767. doi: 10.1097/01.lab.0000043123.37057.f6. [DOI] [PubMed] [Google Scholar]
- Williams R.L., Hilton D.J., Pease S., Willson T.A., Stewart C.L., Gearing D.P., Wagner E.F., Metcalf D., Nicola N.A., Gough N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Yang W., Wei W., Shi C., Zhu J., Ying W., Shen Y., Ye X., Fang L., Duo S., Che J. Pluripotin combined with leukemia inhibitory factor greatly promotes the derivation of embryonic stem cell lines from refractory strains. Stem Cells. 2009;27:383–389. doi: 10.1634/stemcells.2008-0974. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M., Falco G., Sharova L.V., Nishiyama A., Thomas M., Lee S.L., Stagg C.A., Hoang H.G., Yang H.T., Indig F.E. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.