Highlight
We identified C-TERMINALLY ENCODED PEPTIDE 5 (CEP5) as a novel, auxin-repressed and phloem pole-expressed signal assisting in the formation of lateral roots.
Key words: Arabidopsis, CEP5, lateral root initiation, post-translationally modified peptide, receptor kinase, XIP1.
Abstract
Roots explore the soil for water and nutrients through the continuous production of lateral roots. Lateral roots are formed at regular distances in a steadily elongating organ, but how future sites for lateral root formation become established is not yet understood. Here, we identified C-TERMINALLY ENCODED PEPTIDE 5 (CEP5) as a novel, auxin-repressed and phloem pole-expressed signal assisting in the formation of lateral roots. In addition, based on genetic and expression data, we found evidence for the involvement of its proposed receptor, XYLEM INTERMIXED WITH PHLOEM 1 (XIP1)/CEP RECEPTOR 1 (CEPR1), during the process of lateral root initiation. In conclusion, we report here on the existence of a peptide ligand−receptor kinase interaction that impacts lateral root initiation. Our results represent an important step towards the understanding of the cellular communication implicated in the early phases of lateral root formation.
Introduction
Co-ordinated positioning and development of lateral roots is central to shape root system architecture, allowing plants to adapt their below-ground organs for optimal soil exploration (De Smet, 2012; Smith and De Smet, 2012; Kong et al., 2014; Tian et al., 2014). Lateral root primordia are formed from approximately three pairs of xylem pole pericycle (XPP) cells arranged in neighbouring cell files that undergo asymmetric cell division and subsequently form a new organ (Dubrovsky et al., 2001; Kurup et al., 2005; De Smet et al., 2006, 2007; Péret et al., 2009; Lavenus et al., 2013). In the basal meristem, close to the primary root tip and before any asymmetric cell division, a periodic transcriptional mechanism specifies pre-branch sites that are competent to form lateral roots in a regular pattern (De Smet et al., 2007; Moreno-Risueno et al., 2010; Van Norman et al., 2013; Xuan et al., 2015, 2016).
Several plant hormones have been shown to affect root architecture, among which auxin has been granted a central role (Lau et al., 2008; Vanneste and Friml, 2009). In addition, a number of transcription factors and miRNAs have been shown to affect lateral root development (Satbhai et al., 2015). However, several recent studies are beginning to reveal the importance of different classes of small signalling peptides during the process of lateral root development (Ohyama et al., 2008; Delay et al., 2013; Fernandez et al., 2013, 2015; Kumpf et al., 2013; Araya et al., 2014; Bergonci et al., 2014; Czyzewicz et al., 2015). However, in Arabidopsis, very few small signalling peptides have been linked to a receptor (Murphy et al., 2012; Czyzewicz et al., 2013), and very few receptors involved in lateral root development have been identified (De Smet et al., 2008, 2009; Kumpf et al., 2013; Wierzba and Tax, 2013; Araya et al., 2014; Cho et al., 2014; Tabata et al., 2014). Recently, the leucine-rich repeat (LRR) receptor kinases XYLEM INTERMIXED WITH PHLOEM 1 (XIP1)/C-TERMINALLY ENCODED PEPTIDE (CEP) RECEPTOR 1 (CEPR1; At5g49660) and CEPR2 (At1g72180) were proposed to act as receptors for CEP1 and other members of the CEP family (Tabata et al., 2014). Both XIP1/CEPR1 and CEPR2 contain a short secretory signal peptide sequence, an N-terminal extracellular LRR receptor domain with 21 LRR repeats, a single helical transmembrane region, and a C-terminal cytoplasmic serine/threonine kinase domain. It was previously shown that a loss-of-function xip1 mutant displays anthocyanin accumulation in the leaves, xylem-like lignification of phloem in inflorescence stems, disrupted xylem vessel formation, phloem cells sometimes located adjacent to xylem cells, and shorter inflorescence stems (Bryan et al., 2012), and that the cepr1 cepr2 double mutant displays a pleiotropic phenotype, including pale green leaves, smaller rosette leaves, shorter floral stems, anthocyanin accumulation, enhanced lateral root elongation, decreased expression of nitrate transporters, and reduced nitrate uptake activity (Tabata et al., 2014). Interestingly, the Medicago truncatula compact root architecture (cra2) mutant is also affected in its root system architecture, and CRA2 was shown to be closely related to XIP1 (Huault et al., 2014).
The post-translationally modified CEP family members contain an N-terminal signal peptide sequence and a C-terminal conserved CEP domain from which the mature 15 amino acid peptide is processed (Ohyama et al., 2008; Delay et al., 2013; Roberts et al., 2013; Tabata et al., 2014). Some members of the CEP family have already been shown to regulate lateral root development (Ohyama et al., 2008; Delay et al., 2013; Mohd-Radzman et al., 2015), but in this work we functionally characterized C-TERMINALLY ENCODED PEPTIDE5 (CEP5; At5g66815) in the context of lateral root initiation. Furthermore, we explored the involvement of XIP1/CEPR1 in lateral root initiation, and could show that CEP5 and XIP1 are co-expressed during early stages of lateral root initiation, and that both affect this process.
Materials and methods
Plant materials
The following transgenic lines and mutants were described previously: pCEP5::NLS:GFP:GUS, CEP5 OE and CEP5 RNAi (Roberts et al., 2013), xip1-1 and pXIP1::GUS (Bryan et al., 2012).
Plant growth and treatment conditions
Unless mentioned otherwise, seedlings were grown at 21 °C under continuous light (110 μE m–2 s–1 photosynthetically active radiation, supplied by cool-white fluorescent tungsten tubes, Osram) on square Petri plates (12×12cm) containing 50ml of solid half-strength Murashige and Skoog (MS) growth medium supplemented with sucrose (per litre: 2.15g of MS salts, 0.1g of myo-inositol, 0.5g of MES, 10g of sucrose, and 8g of plant tissue culture agar; pH adjusted to 5.7 with KOH). For peptide treatments, medium was supplemented with CEP5pPro (DFRPTTPGHSPGIGH), CEP5pHyp (DFR{HYP}TT{HYP}GHS{HYP}GIGH), or mCEP5pHyp (DFL{HYP}HT{HYP}GHV{HYP}GISH) peptide to a concentration as indicated in the text and/or figure legends. Synthetic peptides (CEP5pPro, CEP5pHyp, and mCEP5pHyp) were obtained from GenScript (www.genscript.com/peptide-services.html?src=home), and were supplemented to growth medium with concentrations as indicated in the text and/or figure legends. For auxin treatments, medium was supplemented with indole-3-acetic acid (IAA) or 1-naphthaleneacetic acid (NAA) to a concentration as indicated in the text and/or figure legends.
Transcriptome profiling data
The naxillin treatment transcriptome data from De Rybel et al. (2012) can be searched in the Lateral Root Initiation eFP Browser (bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Lateral_Root_Initiation) (Winter et al., 2007).
Primary and lateral root phenotyping
At the indicated time, images of plates with seedlings were taken and roots were measured using ImageJ (https://imagej.nih.gov/ij/index.html) or FIJI software (Schindelin et al., 2012). For detailed staging of lateral roots, samples were cleared as described previously (Malamy and Benfey, 1997) and analysed by differential interference contrast microscopy (Olympus BX53).
Histochemical GUS assays
For GUS (β-glucuronidase) assays, plants were put overnight in 90% acetone, then transferred to a GUS-solution {1mM X-Glc, 0.5% (v/v) dimethylformamide (DMF), 0.5% (v/v) Triton X-100, 1mM EDTA (pH 8), 0.5mM potassium ferricyanide [K3Fe(CN)6], 0.5% potassium ferrocyanide [K4Fe(CN)6], 500mM phosphate buffer (pH 7)} and incubated at 37 °C for GUS staining, and finally washed in 500mM phosphate buffer (pH 7). For microscopic analysis, samples were cleared with 90% lactic acid or as described previously (Malamy and Benfey, 1997). Samples were analysed by differential interference contrast microscopy (Olympus BX53) and stereomicroscopy (Leica MZ16). For anatomical analysis (microtome transversal sectioning) of GUS-stained roots, stained samples were processed as described previously (De Smet et al., 2004).
Real-time qRT–PCR analyses
For the analysis of CEP5 expression, RNA was extracted by first performing an RNA extraction with TRI Reagent® from Sigma-Aldrich according to the manufacturer’s protocol, followed by an extra RNA extraction procedure with the Plant RNeasy Mini kit from Qiagen according to the manufacturer’s protocol to clean up the RNA further. Next, 1 μg of total RNA was used for cDNA synthesis using the iScript cDNA synthesis kit from BIORAD according to the manufacturer’s protocol. The real-time quantitative reverse transciption–PCR (qRT–PCR) was carried out on the LightCycler 480 from Roche Applied Science with the LightCycler 480 SYBR Green I Master Mix from Roche Applied Science. The expression of CEP5 (CCATGGACGAACCCTAAAAG and TGCCATCATCGTCTTGCTAT) was determined using at least three biological repeats and the reference genes EEF-1α4 (CTGGAGGTTTTGAGGCTGGTAT and CCAAGGGTGAA AGCAAGAAGA) and At2g32170 (GGACCTCTGTTGTATCA TTTTGCG and CAACCCTCTTTACATCCTCCAAAC).
SRM analysis of the CEP5 peptide
For SRM (selected reaction monitoring) experiments, the CEP5 peptide containing an isoleucine residue with heavy, stable isotopes, NH2-DFRP<hydroxy>TTP<hydroxy>GHSP<hydroxy>GI(13C6 15N)GH-COOH, was in-house synthesized by Fmoc [N-(9-fluorenyl)methoxycarbonyl] chemistry on a 433A peptide synthesizer (Applied Biosystems, Framingham, MA, USA). Frozen 5-day-old 35S::CEP5 seedlings were ground to a fine powder in liquid N2 and proteins were extracted in 50mM triethylammonium bicarbonate (TEAB) buffer containing 8M urea and the suggested amounts of protease and phosphatase inhibitors according to the manufacturer’s instructions (cOmplete protease inhibitor cocktail tablet and PhosStop phosphatase inhibitor cocktail tablet, Roche). After determining the protein concentration using the Bradford assay and diluting the protein extract twice with 50mM TEAB buffer, a total of 500 µg of protein material was filtered over a 3kDa cut-off filter (Pall Nanosep® centrifugal devices, Sigma-Aldrich) to retain only peptides with masses <3kDa in the filtrate. This peptide mixture was spiked with 10 pmol of the synthetic heavy CEP5 peptide and vacuum dried. Next, the sample was re-dissolved in 2% acetonitrile (ACN) with 0.1% trifluoroacetic acid (TFA) and used for SRM analysis. SRM analysis was performed on an Ultimate 3000 RSLC nano HPLC system (Thermo Fisher Scientific, Bremen, Germany) coupled to a TSQ Vantage (Thermo Fisher Scientific). The nano-LC system was configured with a trapping column [made in-house, 100 µm internal diameter (ID)×20mm, 5 µm beads, C18 Reprosil-HD (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany)] and an analytical column [made in-house, 75 µm ID×150mm, 3 µm beads, C18 Reprosil-HD (Dr. Maisch GmbH)]. The loading solvent consisted of 0.1% TFA in 2:98 ACN:H2O, and the nano-LC was run with 0.1% formic acid as nano-LC solvent A and 0.1% formic acid in 80:20 ACN:H2O as nano-LC solvent B. The needle voltage in the nano-ESI source was set at 1300V and the capillary temperature at 275 °C. A 5 µl aliquot of each sample was injected using a full loop injection. Injection was at 10 µl min–1 in loading solvent. After loading, the trapping column was flushed for 4min in order to pre-concentrate the components while removing buffer components, before it was put in-line with the analytical column. Compounds were eluted at 300 nl min–1 with an ACN gradient of 30min from 2% to 35% of nano-LC solvent B. The column was washed with 90% of nano-LC solvent B for 1min and equilibrated with nano-LC solvent A for 9.5min before analysis of the next sample. A dwell time of 120ms for each transition was applied. Seven transitions were monitored for both the heavy and the light form of the CEP5 peptide, with the doubly charged precursor as the first mass filter. Data analysis was performed through the Skyline software (MacLean et al., 2010).
Results and Discussion
Focused transcript profiling data identifies CEP5 as a putative regulator of lateral root development
Since the plant hormone auxin is a major regulator of primary root growth and lateral root development (Overvoorde et al., 2010; Lavenus et al., 2013), several transcript profiling studies based on auxin treatments have been performed in order to identify the molecular players involved (Himanen et al., 2004; Vanneste et al., 2005; De Smet et al., 2008). However, because of the pleiotropic effects caused by exogenous auxin application, such data sets risk compromising the spatiotemporal resolution required when looking for components specific for a single developmental process. To circumvent this, we searched for putative novel early lateral root formation regulators by screening a data set obtained through a highly focused transcript profiling analysis on seedling roots treated with the synthetic molecule naxillin. Naxillin specifically induces an auxin response in the basal meristem associated with lateral root initiation through enhancing indole-3-butyric acid (IBA) to IAA conversion in the root cap (De Rybel et al., 2012). Driven by the recurrent programmed cell death of the outermost lateral root cap cells, a periodic input of the converted auxin into the main root contributes to a fine-tuned mechanism that results in an evenly spaced lateral root distribution pattern (Xuan et al., 2016). Importantly, through its local activity, naxillin does not display the typical pleiotropic effects of exogenous application of auxin or auxin-like molecules (De Rybel et al., 2012). In order to identify novel putative early lateral root formation regulators, seedlings were grown for 72h on growth medium supplemented with the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA), which prevents lateral root initiation, followed by a transfer to growth medium supplemented with naxillin to trigger the priming event synchronously in the basal meristem. In a genome-wide transcript profiling analysis, we identified CEP5 (At5g66815) as differentially early up-regulated between non-treated and naxillin-treated seedling roots (De Rybel et al., 2012) [data not shown; see Lateral Root Initiation eFP Browser (Winter et al., 2007)]. The CEP5 gene encodes a small protein of 105 amino acids and contains a conserved 15 amino acid C-terminal CEP domain that gives rise to a small signalling peptide (Ohyama et al., 2008; Roberts et al., 2013; Tabata et al., 2014).
CEP5 expression is regulated by auxin
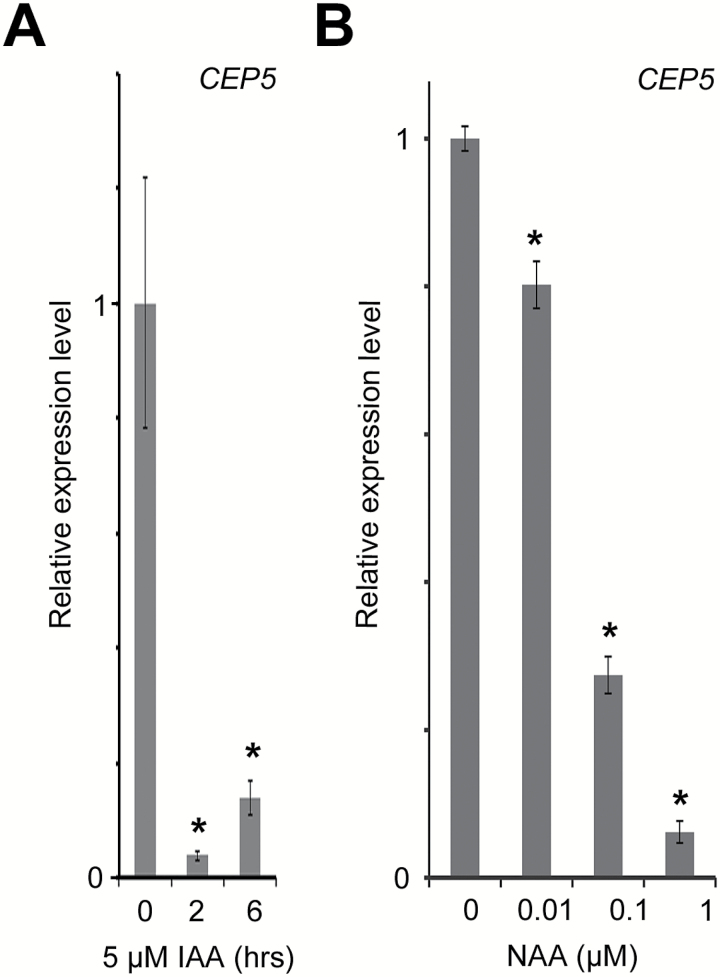
Since CEP5 is transcriptionally regulated following naxillin treatment, we subsequently checked if CEP5 expression is also auxin regulated. Treatment of wild-type roots with different concentrations of the synthetic auxin NAA or with IAA revealed that CEP5 expression was down-regulated by auxin (Fig. 1A, B). These results suggested that CEP5 expression is (directly or indirectly) regulated by auxin.
Fig. 1.
Auxin effect on CEP5 expression. (A) CEP5 expression in 7-day-old roots following the indicated hours of auxin (1 μM IAA) treatment in liquid medium. (B) CEP5 expression in 5-day-old root tips of ~5mm (including the basal meristem) following 2h of auxin (NAA) treatment at the indicated concentrations. CEP5 levels were analysed through real-time qRT–PCR. Graphs show the average ±SE of three biological replicates. *P<0.05 according to Student’s t-test compared with 0 μM NAA or IAA.
CEP5 expression is associated with early stages of lateral root development
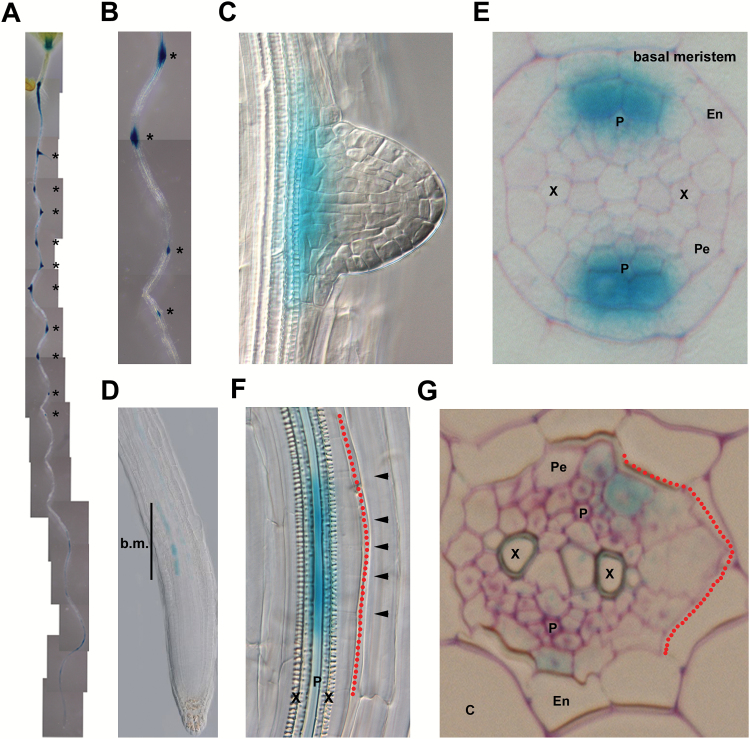
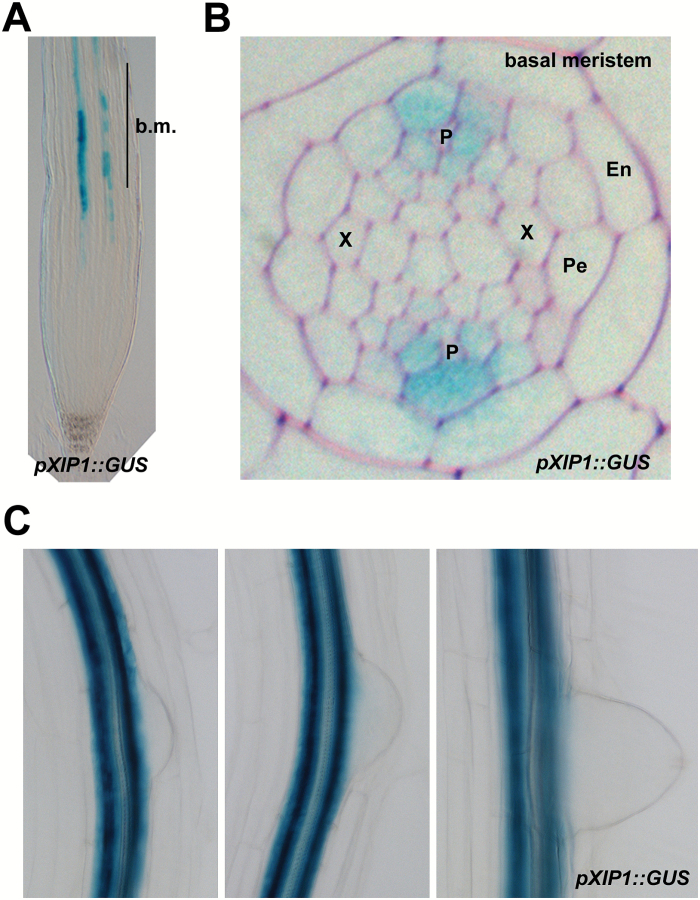
Based on its naxillin-regulated expression profile, CEP5 represents a candidate peptide to be involved in the early developmental steps toward lateral root development. Using a pCEP5::NLS:GFP:GUS reporter line (Roberts et al., 2013), we observed regularly spaced patches of CEP5 expression associated with lateral root primordia, confirming its potential involvement in this process (Fig. 2A–C). We did not detect CEP5 expression in the primary root stem cell niche; however, CEP5 was expressed in the basal meristem (Fig. 2D). The latter is important in the context of lateral root initiation as this region is defined as part of the oscillation zone where pre-branch sites are established by the input of auxin derived from the lateral root cap (De Smet et al., 2007; Moreno-Risueno et al., 2010; Xuan et al., 2016). Tissue-specific analyses showed that both in the basal meristem and during early stages of lateral root development, CEP5 was predominantly expressed in the phloem pole-associated pericycle (PPP) cells, but also—although more weakly—in the adjacent phloem (Fig. 2E–G; Supplementary Fig. S1; Supplementary Movie S1 at JXB online). This CEP5 expression pattern does not overlap with the well-documented sites of high auxin response in the primary root or during lateral root initiation, which in Arabidopsis occurs in XPP cells (De Smet et al., 2007). To check whether the expression pattern of CEP5 is perturbed under conditions of altered auxin response in the XPP cells, the pCEP5::NLS:GFP:GUS reporter line was grown on NPA. Under these conditions, we did not observe any change in the CEP5 expression pattern (such as radial expansion) compared with control conditions (Supplementary Fig. S1). Taken together, CEP5 is negatively regulated by auxin and specifically expressed in the PPP cells that are closely associated with the lateral root development process, suggesting a negative correlation with auxin activity. However, what the specific cellular threshold is, is at the moment not known.
Fig. 2.
CEP5 expression in the Arabidopsis root. Representative pictures for CEP5 expression (monitored through GUS expression in a pCEP5::NLS:GFP:GUS transgenic line) in the root: (A) in a complete seedling (overstained for illustrative reasons), (B) in a part of the root from the seedling depicted in (A), (C) at the site of a lateral root primordium, (D) at the root apex, (E) in the basal meristem on a transverse section, (F) at a site of lateral root formation with the lateral root primordium pointing to the right (outlined with the dotted red line), and (G) on a transverse section through a lateral root primordium (outlined with the dotted red line). Seedlings are 5–6 d after germination. *, Lateral root primordium; arrowheads in (F) separate individual cells; P, phloem; X, xylem; Pe, pericycle; En, endodermis; C, cortex; b.m., basal meristem.
Altering CEP5 expression levels affects root architecture
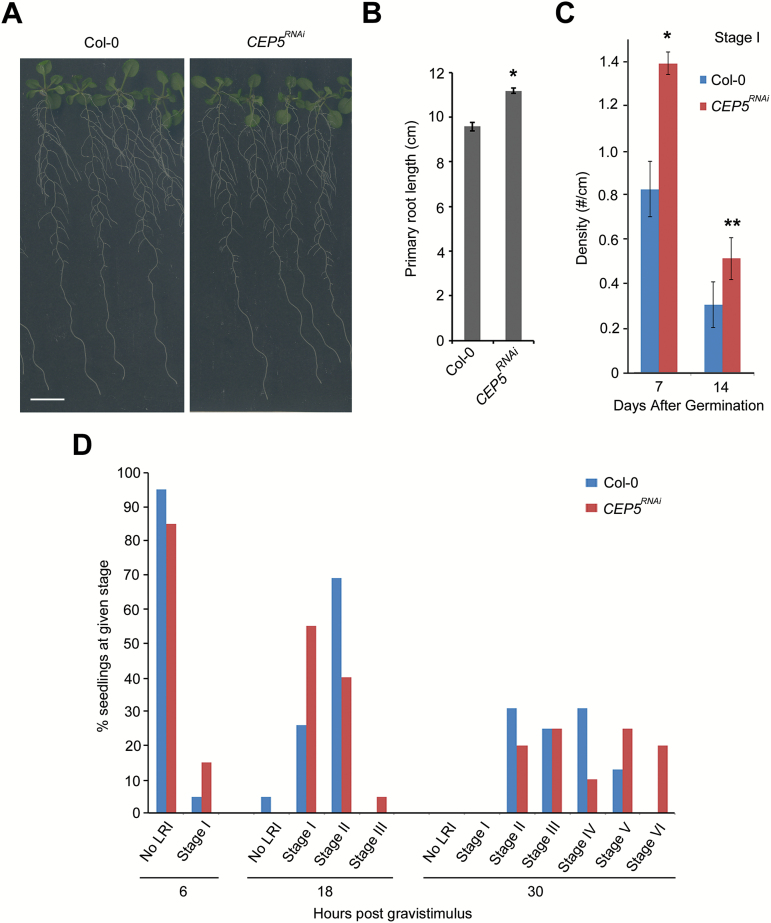
Given the spatial (appearing in common regions of the root, although not in the same cells) and temporal (being induced at the same time points) correlation of CEP5 expression with lateral root initiation and development, we assessed if CEP5 loss of function affected this process. A Cauliflower mosaic virus (CaMV) 35S promoter-driven CEP5 RNAi knockdown line (CEP5 RNAi) (Roberts et al., 2013) displayed a significant difference in primary root length compared with the control (Fig. 3A, B). In addition, detailed analyses of lateral root initiation in this CEP5 RNAi line revealed an increased number of stage I and II lateral root primordia compared with the control (Fig. 3C; Supplementary Fig' S2). Additionally, in a root bending assay (Péret et al., 2012), the CEP5 RNAi line progressed faster through lateral root developmental stages than the wild type (Fig. 3D). These loss-of-function data, together with the CEP5 expression pattern, indicate that CEP5 plays a role in early lateral root initiation events.
Fig. 3.
Effect of reduced CEP5 levels on root architecture. (A) Representative picture of the CEP5 RNAi line and Col-0 at 12 d after germination. (B) Quantification of the primary root length 12 d after germination (n ≥29). (C) Stage I lateral root primordia at the indicated seedling age (n=10). (D) Progression through lateral root stages at the indicated hours post-gravistimulus (n ≥14). Graphs in (B) and (C) show the average± SE *P<0.05 and **P<0.075 according to Student’s t-test compared with Col-0. Scale bar=1cm. (This figure is available in colour at JXB online.)
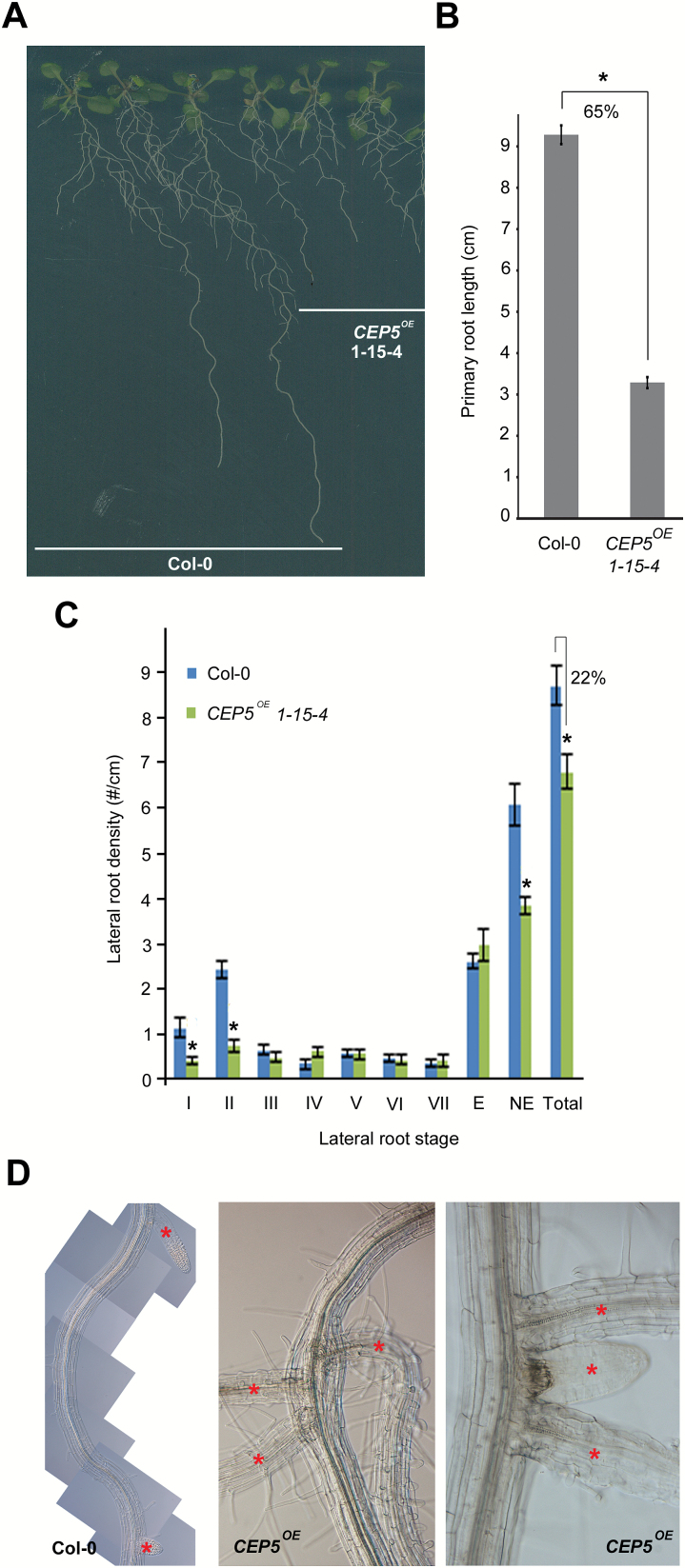
Next, we analysed a line with CaMV 35S promoter-driven constitutive overexpression of CEP5 (CEP5 OE) (Roberts et al., 2013), which displayed shorter primary roots (similar to other independent CEP5 OE lines) as compared with the wild type (Fig. 4A, B; Supplementary Fig. S2). Furthermore, the CEP5 OE line displayed a decrease in total lateral root density, with fewer non-emerged lateral roots, compared with the wild type (Fig. 4C). Detailed analyses of lateral root developmental stages showed that this was mainly due to fewer initiation events (Fig. 4C; Supplementary Fig. S3). At later stages of lateral root development, we also observed closely spaced lateral root primordia in CEP5 OE lines, which we never observed as such in wild-type roots (Fig. 4D). This gain-of-function approach further suggested that CEP5 impacts root architecture, but does not exclude that this is an indirect and/or non-specific effect due to ectopic expression.
Fig. 4.
Effect of increased CEP5 levels on primary root growth and lateral root development. (A) Representative picture of a CEP5 OE line and Col-0 at 12 d after germination. (B) Quantification of primary root length at 12 d after germination. (C) Lateral root stages I–VII (according to Malamy and Benfey, 1997) in Col-0 and a CEP5 OE line (n ≥15) at 7 d after germination. The percentage reduction in total lateral root density is indicated. E, emerged lateral roots; NE, non-emerged lateral roots; Total, sum of E and NE. (D) Regular and adjacent positioning of lateral roots in wild-type (Col-0) and CEP5 OE seedlings at 14 d after germination, respectively. Asterisks in (D) indicate lateral roots. All graphs show the average ±SE of the indicated sample numbers. *P<0.05 according to Student’s t-test compared with Col-0. (This figure is available in colour at JXB online.)
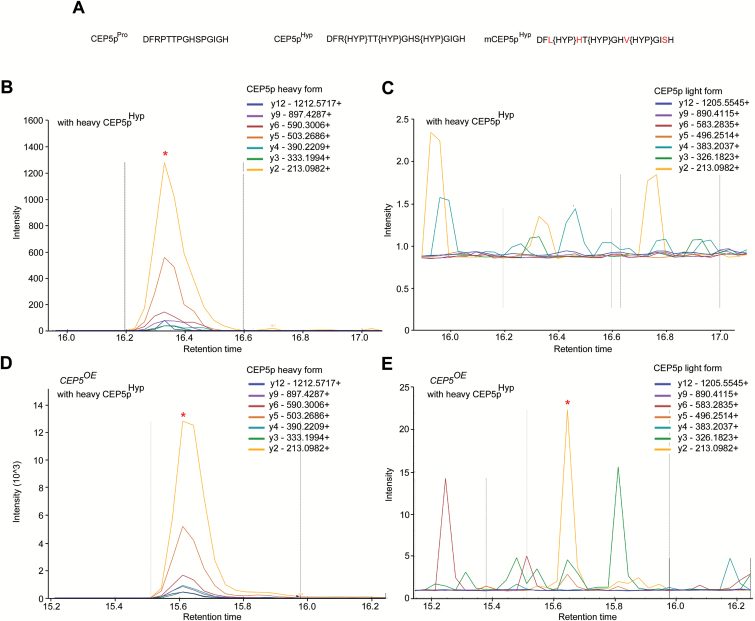
CEP5 gives rise to CEP5pHyp
CEP5 has a conserved C-terminal CEP domain, containing three proline residues and a predicted N-terminal signal peptide cleavage site that undergoes proteolytic processing to form a mature CEP5 peptide of 15 amino acids (CEP5p) (Roberts et al., 2013; Tabata et al., 2014) (Fig. 5A). However, small signalling peptides are often post-translationally modified, thereby modulating—amongst others—the ability and specificity of peptides in binding to their targets (Murphy et al., 2012). In this context, it was previously shown that members of the CEP family give rise to a peptide containing hydroxyproline (Hyp) residues (Tabata et al., 2014). To confirm that a 15 amino acid CEP5 peptide with three Hyp residues (CEP5pHyp) (Fig. 5A) is indeed present in seedlings overexpressing CEP5, we performed SRM on a CEP5 OE line. SRM is a mass spectrometry technique that allows detection and quantification of specific (low abundant) peptides in total protein preparations (Picotti and Aebersold, 2012). Indeed, in the CEP5 OE proteome spiked with a chemically synthesized version of CEP5pHyp containing an isoleucine residue with heavy, stable isotopes, transitions for both the heavy, spiked-in CEP5pHyp and the light, naturally occurring CEP5Hyp peptide could be detected (Fig. 5B–E). These results supported that a CEP5 peptide with three Hyp residues can be present in planta.
Fig. 5.
In planta CEP5 peptide. (A) Sequences for the synthetic variants of mature 15 amino acid CEP5: unmodified (CEP5pPro), with proline hydroxylation modifications on P4, P7, and P11 (CEP5pHyp), and the hydroxyprolinated mutated CEP5 sequence with four residue substitutions (R3>L, T5>H, S10>V, and G14>S; indicated in red) (mCEP5pHyp). (B–E) SRM analysis of the targeted CEP5 peptide. Characteristic y-type of fragment ions (referred to as transitions), indicated with different colours at the top of each spectrum, were monitored. As a control, the heavy CEP5pHyp alone was analysed by SRM, and the transitions of the heavy form (B) and the light form (C) were monitored. As for the latter, no transitions could be monitored, indicating the high isotopic purity of the heavy peptide. In the CEP5 OE proteome spiked with heavy CEP5pHyp, both transitions for the heavy, spiked-in peptide (D) and the light, naturally occurring peptide (E) could be detected. Red asterisk, CEP5pHyp.
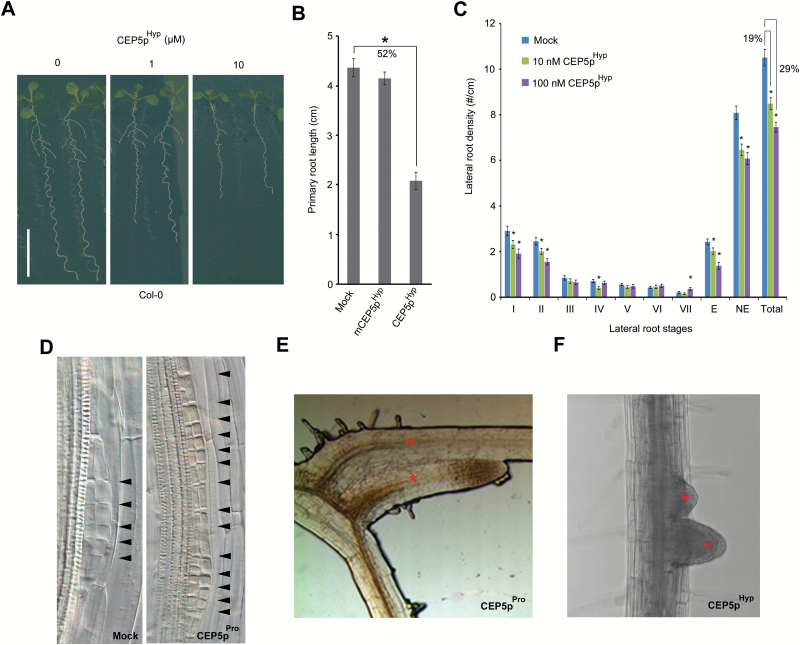
Synthetic CEP5 peptide affects root architecture
Based on previous studies (Tabata et al., 2014) and the above-described results, a synthetic CEP5pHyp peptide was generated for further analysis of CEP5 function (Fig. 5A). To assess the activity of synthesized CEP5pHyp, we first analysed its effect on primary root growth, which has previously been shown to be a straightforward, although possibly non-specific, assay to test the activity of small post-translationally modified (CEP) peptides (Delay et al., 2013). Indeed, seedlings grown in the presence of CEP5pHyp (also at low concentrations) displayed shorter roots compared with the mock-treated control and compared with a synthetic variant with four randomly positioned, but not very unlikely amino acid substitutions based on a BLOSUM62 substitution matrix within the 15 amino acid CEP5 peptide sequence, while retaining the Hyp residues at the same positions (mCEP5pHyp) (Figs 5A, 6A, B;Supplementary Fig. S4). Next, we addressed the effect of synthetic CEP5pHyp on lateral root formation. Seedlings grown in the presence of different low concentrations of CEP5pHyp displayed a decreased total lateral root density, which is mainly due to a significant reduction in lateral root initiation events (Fig. 6C; Supplementary Fig. S3). Conversely, this did not occur in mCEP5pHyp-treated seedlings (Supplementary Fig. S3). When lateral root initiation occurred, we occasionally observed regions of ectopic and/or aberrant pericycle cell divisions (observed in 10 out of 149 lateral root primordia of eight CEP5pPro-treated seedlings, while this did not occur in the untreated wild type), resulting in malformed lateral root primordia or closely spaced primordia in CEP5pPro/Hyp-treated seedlings, which differed from regularly spaced lateral roots in the wild type (Fig. 6D–F). Taken together, the similarities in primary and lateral root phenotypes between CEP5p treatment and CEP5 OE indicate that the chemically synthesized CEP5pHyp has the same bioactivity as the overexpressed CEP5. These results further support a role for CEP5pHyp in lateral root initiation.
Fig. 6.
Effect of synthetic CEP5p on primary root growth and lateral root development. (A) Representative pictures of Col-0 Arabidopsis seedlings grown on the indicated CEP5pHyp concentrations for 7 d after germination. Scale bar=1cm. (B) Quantification of primary root length of Col-0 seedlings treated with 5 µM mCEP5pHyp or 5 µM CEP5pHyp compared with mock treatment at 7 d after germination (n ≥15 per condition). The percentage reduction in primary root length is indicated. (C) Lateral root stages I–VII (according to Malamy and Benfey, 1997) upon mock or CEP5pHyp treatment at different concentrations at 9 d after germination (data from a newly grown root part of 5-day-old seedlings transferred to CEP5pHyp for 4 d, n ≥32). E, emerged lateral roots; NE, non-emerged lateral roots; Total, total lateral roots. The percentage reduction in total lateral root density is indicated. (D) Pericycle cell divisions and positioning of lateral roots in mock (left) and 1 µM CEP5pPro-treated Col-0 seedlings (right) (11 d after germination) (stage II primordia are shown) observed in 10 out of 149 lateral root primordia (n=8 seedlings), while this did not occur in the untreated wild type. (E, F) Position of lateral roots in 10 µM CEP5pPro-treated seedlings 14 d after germination (E) and in 5 µM CEP5pHyp-treated seedlings 12 d after germination (F). Scale bars=1cm. All graphs show the average ±SE of the indicated sample numbers. *P<0.05 according to Student’s t-test compared with mock. In all cases, mock refers to medium with water as used to dissolve CEP5p. Asterisk in E-F, lateral root. (This figure is available in colour at JXB online.)
The proposed CEP family receptor XIP1/CEPR1 regulates lateral root initiation
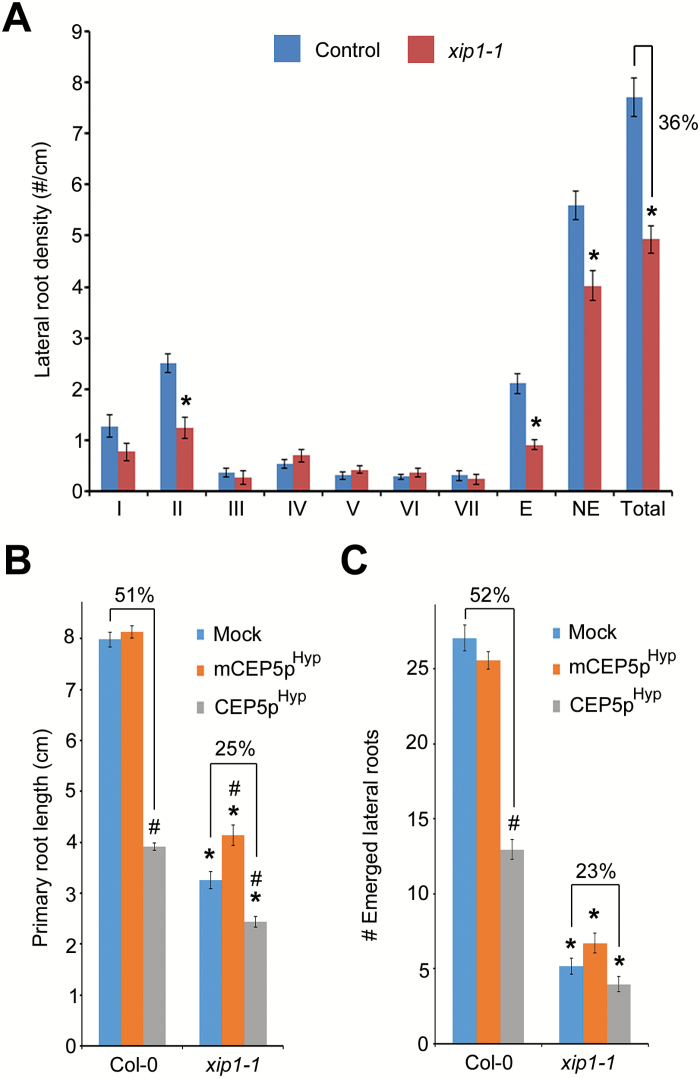
Recently XIP1/CEPR1 and CEPR2 were proposed to be the receptors for CEP peptides, including CEP5 (Tabata et al., 2014). However, a role in lateral root initiation for XIP1/CEPR1 and/or CEPR2 was not yet explored. Therefore, we performed detailed analyses of a previously described pXIP1::GUS line (Bryan et al., 2012) and we showed that XIP1/CEPR1 is expressed in the root from the basal meristem onward (Fig. 7A), a pattern that overlaps with CEP5 expression (Fig. 2D). Furthermore, tissue-specific analyses showed that XIP1/CEPR1 is expressed in the phloem pole pericycle and in the adjacent phloem (Fig. 7B), confirming the overlap with CEP5 expression (Fig. 2E), and is excluded from early stages of lateral root development (Fig. 7C), similarly to CEP5 (Fig. 2C). This expression pattern combined with the results from Tabata et al. (2014) suggested that XIP1/CEPR1 could be a receptor for CEP5 in the root and therefore might take part in lateral root initiation. To explore this further, we assessed lateral root stages and density of the previously described xip1-1 mutant (Bryan et al., 2012). This revealed a reduced total lateral root density in xip1-1 in comparison with the control, which seemed mainly due to a reduction in stage I and II lateral root primordia and—in part—to fewer emerged lateral roots (Fig. 8A; Supplementary Fig. S3), suggesting that XIP1 is a positive regulator of lateral root initiation and development.
Fig. 7.
XIP1/CEPR1 expression in the root. (A) Representative picture of XIP1 expression in the root apex. (B) Transverse section through the basal meristem in a pXIP1::GUS transgenic reporter line. P, phloem; X, xylem; Pe, pericycle; En, endodermis; b.m., basal meristem. (C) Representative pictures for XIP1 expression in different stages of lateral root development in 7-day-old seedlings. XIP1 expression was monitored through GUS expression in a pXIP1::GUS transgenic line.
Fig. 8.
Lateral root phenotype in the xip1-1 mutant. (A) Lateral root stages I–VII (according to Malamy and Benfey, 1997) in control and xip1-1 at 5 d after germination (n ≥14). (B, C) Quantification of primary root length (B) and emerged lateral root number (C) of Col-0 and xip1-1 seedlings treated with 1 µM CEP5pHyp or mCEP5pHyp compared with mock treatment at 10 d after germination (n ≥22 per condition). The percentage reduction in primary root length and lateral root number is indicated. E, emerged lateral roots; NE, non-emerged lateral roots; Total, total lateral roots. Graphs shows average ±SE. * or #, P<0.05 according to Student’s t-test compared with Col-0 or mock treatment, respectively. In all cases, mock refers to medium with water as used to dissolve CEP5p. (This figure is available in colour at JXB online.)
To evaluate further an interaction between CEP5 and XIP1, we explored to what extent xip1-1 is (in)sensitive to CEP5pHyp treatment. This revealed that, compared with the control, xip1-1 is less or not sensitive to CEP5pHyp with respect to primary root growth (Fig. 8B) or number of emerged lateral roots, respectively (Fig. 8C). These data—together with the biochemical evidence from Tabata et al. (2014)—support that CEP5 and XIP1 are a peptide ligand–receptor kinase pair in the context of lateral and primary root development. However, in general, the mutant phenotypes of the genes encoding the peptide ligand and its receptor are very similar (Butenko et al., 2009; Murphy et al., 2012; Czyzewicz et al., 2013; Kumpf et al., 2013). However, in our case, the xip1-1 root architecture phenotype is similar to that of CEP5 OE or CEP5pHyp-treated seedlings and opposite to that of CEP5 RNAi lines (Figs 4, 6), possibly suggesting that CEP5 negatively regulates XIP1 activity (e.g. by acting as an antagonist) in the context of lateral root initiation. In this context, the fact that CEP5pHyp had no strong impact on xip1-1 can also be interpreted as no further CEP5-mediated inhibitory effect if XIP1 is already absent (and hence fully inhibited). Alternatively, CEP5 does not exclusively act via the XIP1 receptor (or close homologues) in regulating root architecture. Furthermore, the observed lateral root phenotypes can be obtained through various mechanisms (e.g. the effect on lateral root initiation can impact development of nearby lateral root primordia), and further analyses will be required to unravel fully the developmental and biochemical mechanisms underlying CEP5 and XIP1 action.
Conclusion
Previously, a role for CEPs in regulating aspects of root architecture, namely nitrate-dependent lateral root elongation, was proposed. Specifically, CEPs might act as root-derived ascending N-demand signals to the shoot, where their perception by CEPRs leads to the production of a putative shoot-derived descending signal that up-regulates nitrate transporter genes in the roots (Ohyama et al., 2008; Delay et al., 2013; Tabata et al., 2014; Mohd-Radzman et al., 2015). Here, we provide evidence that CEP5 may also act (probably together with XIP1/CEPR1) during lateral root initiation. Our gain-of-function and knock-down data suggest CEP5 to be part of a lateral root inhibitory mechanism. Faster lateral root development was observed in the CEP5 RNAi line, while overexpression or treatment with the peptide resulted in fewer lateral root initiation events. The observed clustering of lateral roots in later developmental stages in the gain-of-function condition might be a secondary effect. Slowing down lateral root development can interfere with the timely development of auxin sources and therefore retard the draining of auxin from the main root. In turn, this might lead to higher auxin levels in the neighbourhood of existing primordia and induce ectopic and/or irregularly patterned primordia.
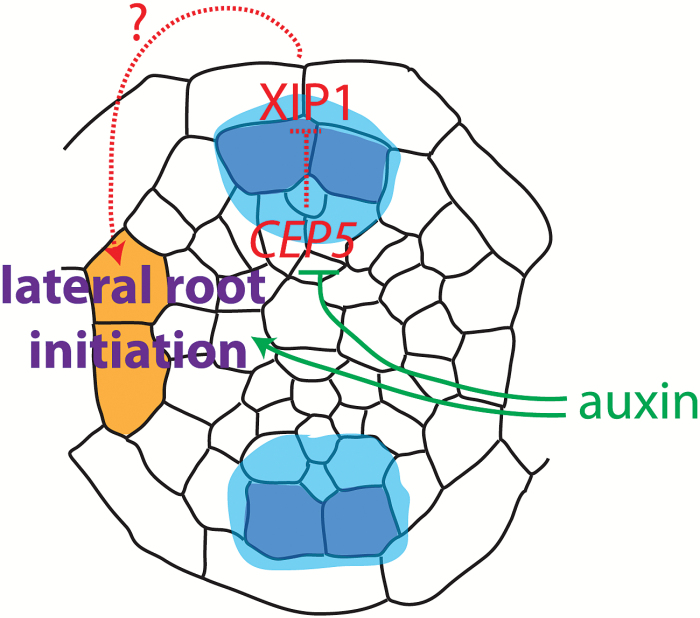
Finally, it is intriguing that a phloem-derived signal downstream of CEP5 and XIP1/CEPR1 has such an impact on lateral root initiation and development at the xylem pole (Fig. 9). So far, no mutants have been reported to show lateral root initiation at the phloem poles in Arabidopsis (and so far we have also not observed this in loss- or gain-of-function CEP5 or XIP1 lines) arguing for a strong and complex lateral root inhibition mechanism in this part of the root pericycle. Earlier, a cell cycle inhibitory mechanism, based on the pericycle-specific expression of KIP-RELATED PROTEIN2 (KRP2), a cyclin-dependent kinase inhibitor, has been proposed as essential to allow, spatially and temporally, for lateral root initiation by repressing cell division activity in the entire pericycle except for sites of lateral root initiation (Himanen et al., 2002). In the future, it will be interesting to reveal if there is any direct interaction of CEP5-dependent signalling with the control of cell cycle regulation with respect to lateral root initiation. Additionally, it will be exciting to explore alternative mechanisms on how the phloem-expressed CEP5 affects lateral root initiation in the xylem pole pericycle cells. At the moment, however, it is not yet possible to visualize CEP5 peptide reliably in planta in order to evaluate possible movement to other cells and/or tissues.
Fig. 9.
The data we have—so far—suggest that CEP5 and XIP1/CEPR1 are expressed in the phloem pole pericycle (PPP) cells (blue cells, with the highest expression in dark blue, and the domain with weaker, variable expression in light blue) associated with sites of lateral root formation and regulate lateral root initiation in the xylem pole pericycle (XPP) cells (orange cells) by a currently unknown mechanism (indicated by?). Overall, the CEP5 peptide appears to regulate XIP1 (activity) negatively. An auxin maximum in the XPP cells promotes lateral root initiation and possibly down-regulates CEP5 expression in these cells. As such, the auxin minimum in the neighbouring PPP cells probably allows CEP5 expression.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. CEP5 expression on a transverse section of the pCEP5::NLS:GFP:GUS line.
Figure S2. Analyses of CEP5 RNAi and CEP5 OE lines.
Figure S3. Lateral root phenotypes upon CEP5 perturbation and in xip1-1.
Figure S4. Bioactivity of CEP5pHyp at lower concentrations in the primary root length assay.
Movie S1. 3D reconstruction of pCEP5::NLS:GFP:GUS in the Arabidopsis root.
Acknowledgements
We thank Sarah De Cokere, Marieke Mispelaere, and Darren Wells for practical assistance, and Frans Tax for sharing materials. This work was supported by a BBSRC David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration Grant (PERG06-GA-2009-256354) (IDS), ES is a Postdoctoral Research Fellow of the Fund for Scientific Research (FWO)-Flanders (Belgium). This work was in part financed by grants from the Interuniversity Attraction Poles Programme (IAP VI/33 and IUAP P7/29 ‘MARS’) from the Belgian Federal Science Policy Office and the Research Foundation Flanders (FWO). SS received a Biotechnology and Biological Science Research Council doctoral training grant studentship. IR was supported by the Agency for Innovation by Science and Technology (IWT). BDR was funded by the Special Research Fund of Ghent University.
References
- Araya T, Miyamoto M, Wibowo J, et al. 2014. CLE–CLAVATA1 peptide–receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci T, Ribeiro B, Ceciliato PH, Guerrero-Abad JC, Silva-Filho MC, Moura DS. 2014. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. Journal of Experimental Botany 65, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AC, Obaidi A, Wierzba M, Tax FE. 2012. XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 235, 111–122. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. 2009. Plant peptides in signalling: looking for new partners. Trends in Plant Science 14, 255–263. [DOI] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, et al. 2014. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nature Cell Biology 16, 66–76. [DOI] [PubMed] [Google Scholar]
- Czyzewicz N, Shi C-L, Vu LD, Van De Cotte B, Hodgman C, Butenko MA, De Smet I. 2015. Modulation of Arabidopsis and monocot root architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 peptide. Journal of Experimental Botany 66, 5229–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewicz N, Yue K, Beeckman T, De Smet I. 2013. Message in a bottle: small signalling peptide outputs during growth and development. Journal of Experimental Botany 64, 5281–5296. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Xuan W, et al. 2012. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nature Cemical Biology 8, 798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I. 2012. Lateral root initiation: one step at a time. New Phytologist 193, 867–873. [DOI] [PubMed] [Google Scholar]
- De Smet I, Chaerle P, Vanneste S, De Rycke R, Inze D, Beeckman T. 2004. An easy and versatile embedding method for transverse sections. Journal of Microscopy 213, 76–80. [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681–690. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inze D, Beeckman T. 2006. Lateral root initiation or the birth of a new meristem. Plant Molecular Biology 60, 871–887. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. [DOI] [PubMed] [Google Scholar]
- De Smet I, Voss U, Jurgens G, Beeckman T. 2009. Receptor-like kinases shape the plant. Nature Cell Biology 11, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. Journal of Experimental Botany 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P. 2001. Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214, 30–36. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P. 2013. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiology 161, 954–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, Beeckman T, Hilson P. 2015. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. Journal of Experimental Botany 66, 5245–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. 2002. Auxin-mediated cell cycle activation during early lateral root initiation. The Plant Cell 14, 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, et al. 2004. Transcript profiling of early lateral root initiation. Proceedings of the National Academy of Sciences, USA 101, 5146–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F. 2014. Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genetics 10, e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Zhang M, De Smet I, Ding Z. 2014. Designer crops: optimal root system architecture for nutrient acquisition. Trends in Biotechnology 32, 597–598. [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Sto IM, Butenko MA, Peret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proceedings of the National Academy of Sciences, USA 110, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S, Runions J, Kohler U, Laplaze L, Hodge S, Haseloff J. 2005. Marking cell lineages in living tissues. The Plant Journal 42, 444–453. [DOI] [PubMed] [Google Scholar]
- Lau S, Jurgens G, De Smet I. 2008. The evolving complexity of the auxin pathway. The Plant Cell 20, 1738–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, et al. 2013. Lateral root development in Arabidopsis: fifty shades of auxin. Trends in Plant Science 18, 450–458 [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, et al. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Binos S, Truong TT, Imin N, Mariani M, Djordjevic MA. 2015. Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. Journal of Experimental Botany 66, 5289–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. 2012. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. The Plant Cell 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant Journal 55, 152–160. [DOI] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. 2010. Auxin control of root development. Cold Spring Harbor Perspectives in Biology 2, a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, et al. 2012. Auxin regulates aquaporin function to facilitate lateral root emergence. Nature Cell Biology 14, 991–998. [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R. 2012. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature Methods 9, 555–566. [DOI] [PubMed] [Google Scholar]
- Roberts I, Smith S, De Rybel B, Van Den Broeke J, Smet W, De Cokere S, Mispelaere M, De Smet I, Beeckman T. 2013. The CEP family in land plants: evolutionary analyses, expression studies and role in Arabidopsis shoot development. Journal of Experimental Botany 64, 5371–5381. [DOI] [PubMed] [Google Scholar]
- Satbhai SB, Ristova D, Busch W. 2015. Underground tuning: quantitative regulation of root growth. Journal of Experimental Botany 66, 1099–1112. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, De Smet I. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. [DOI] [PubMed] [Google Scholar]
- Tian H, De Smet I, Ding Z. 2014. Shaping a root system: regulating lateral versus primary root growth. Trends in Plant Science 19, 426–431. [DOI] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. The Plant Cell 17, 3035–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. 2013. To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzba MP, Tax FE. 2013. Notes from the underground: receptor-like kinases in Arabidopsis root development. Journal of Integrative Plant Biology 55, 1224–1237. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, et al. 2015. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Current Biology 25, 1381–1388. [DOI] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, et al. 2016. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351, 384–387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.