Highlight
We describe the role of RALFL34 during early events in lateral root development, and demonstrate its specific importance in orchestrating formative cell divisions in the pericycle.
Key words: Arabidopsis thaliana, ERF, GATA23, lateral root initiation, RAPID ALKALINIZATION FACTOR (RALF).
Abstract
In plants, many signalling molecules, such as phytohormones, miRNAs, transcription factors, and small signalling peptides, drive growth and development. However, very few small signalling peptides have been shown to be necessary for lateral root development. Here, we describe the role of the peptide RALFL34 during early events in lateral root development, and demonstrate its specific importance in orchestrating formative cell divisions in the pericycle. Our results further suggest that this small signalling peptide acts on the transcriptional cascade leading to a new lateral root upstream of GATA23, an important player in lateral root formation. In addition, we describe a role for ETHYLENE RESPONSE FACTORs (ERFs) in regulating RALFL34 expression. Taken together, we put forward RALFL34 as a new, important player in lateral root initiation.
Introduction
Root plasticity is one of the main adaptive traits enabling plants to cope with an ever-changing environment. Formation and positioning of lateral roots along the longitudinal primary root axis plays a vital role in nutrient acquisition and water uptake. Lateral roots are formed post-embryonically from the pericycle cells adjacent to the xylem poles (Malamy and Benfey, 1997; Dubrovsky et al., 2001; De Smet et al., 2006). Their initiation and development occur in a regular way, and depend largely on the plant hormone auxin (Laskowski et al., 1995; Malamy and Benfey, 1997; Beeckman et al., 2001; Casimiro et al., 2001; Lavenus et al., 2013). The development of lateral root primordia goes through several well-described stages, with the first stages being essential for proper lateral root primordium development (Malamy and Benfey, 1997; De Smet et al., 2008, 2010; Lucas et al., 2013; von Wangenheim et al., 2016). Typically, stage 1 comprises two rounds of asymmetric cell divisions of a small set of pericycle founder cells, forming smaller daughter cells with distinct cell fates. At stage 2, a rotation in the plane of division occurs; the cells divide periclinally toward the outer tissues forming an outer layer and an inner layer. This division normally occurs first in the two most central cells, followed by the adjacent cells. The most peripheral cells do not divide periclinally, so, as the central cells expand radially, the establishment of the lateral root primordia dome shape materializes. In stages 3–7, numerous rounds of anticlinal and periclinal cell divisions occur, establishing distinct tissue layers eventually mimicking the organization of the primary root tip. Stage 8 involves few cell divisions; however, rapid cell expansion results in penetration of the overlying tissue, emerging from the primary root. Several of the underlying genes and proteins that are involved in priming, founder cell specification or activation, and initiating and advancing lateral root development have been identified through transcript profiling and the use of gain-of-function and loss-of-function mutants, such as SOLITARY ROOT (SLR)/IAA14, AUXIN RESPONSE FACTOR7 (ARF7), ARF19, LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16), LBD29, GATA23, ARABIDOPSIS CRINKLY4 (ACR4), and several others (Fukaki et al., 2002; Tatematsu et al., 2004; Okushima et al., 2005, 2007; De Smet et al., 2008; De Rybel et al., 2010; Lavenus et al., 2013). Less information, however, is known on cellular communication during lateral root development, specifically, through the relatively recently discovered small signalling peptides.
Small signalling peptides have been shown to play a wide variety of roles in the plant, with recent evidence also showing their involvement in lateral root development (Murphy et al., 2012; Czyzewicz et al., 2013; Delay et al., 2013b ; Tavormina et al., 2015). For example, CLE-LIKE (CLEL)/GOLVEN (GLV)/ROOT GROWTH FACTOR (RGF) peptides inhibit pericycle cell divisions when overexpressed (Matsuzaki et al., 2010; Meng et al., 2012; Whitford et al., 2012; Fernandez et al., 2013, 2015). INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and its receptors HAESA (HAE) and HAESA-LIKE2 (HSL2) play a role in lateral root emergence (Kumpf et al., 2013). Various C-TERMINALLY ENCODED PEPTIDEs (CEPs) reduce (emerged) lateral root density when the synthetic peptide is exogenously applied or endogenously overexpressed (Delay et al., 2013a ; Roberts et al., 2016). CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptides inhibit lateral root emergence when overexpressed (Araya et al., 2014) and also regulate lateral root development through BIN2-mediated phosphorylation of ARFs (Cho et al., 2014). A recently discovered peptide, AUXIN-RESPONSIVE ENDOGENOUS POLYPEPTIDE 1 (AREP1), promotes lateral root organogenesis in the presence of auxin (Yang et al., 2014). Finally, RAPID ALKALINIZATION FACTOR (RALF) peptides in Arabidopsis regulate various processes predominantly through regulating cell expansion (Pearce et al., 2001; Srivastava et al., 2009; Mingossi et al., 2010; Atkinson et al., 2013; Bergonci et al., 2014; Morato do Canto et al., 2014). Some RALF peptides, such as RALF1, RALF19, and RALF23, increase emerged lateral root densities as demonstrated by RALF-silenced transgenic lines and decrease densities as shown by the use of RALF-overexpressing lines (Atkinson et al., 2013; Bergonci et al., 2014). Taken together, it is peculiar that the majority of the so far characterized small signalling peptides have a negative impact on root architecture. Speculatively, this might imply that these small signalling peptides act as specific, negative regulators of, for example, the dominant, promoting effect of auxin.
Here, we describe the role of RALF-LIKE 34 (RALFL34) in lateral root initiation, and position this small signalling peptide in the transcriptional cascade leading to a new lateral root. In addition, we describe a role for ETHYLENE RESPONSE FACTORs (ERFs) in regulating RALFL34 expression.
Materials and methods
Plant materials
We used the following lines in our research: Columbia-0 (Col-0), Landsberg erecta (Ler), ralfl34-1 (SALK_004441) (Alonso et al., 2003), ralfl34-2 (JIC_SGT4223) (Parinov et al., 1999), pGATA23::NLS:GFP (De Rybel et al., 2010), WAVE131Y (Geldner et al., 2009), pRALFL34 869bp ::n3xRFP (see below), and p35S::ERF9-GR (see below).
Growth of Arabidopsis thaliana seedlings
Seeds were surface sterilized (70% ethanol for 2min, 10% bleach for 15min, dH2O for five washes) and then stratified at 4 °C for 2 d. Seeds were then plated on half-strength Murashige and Skoog (1/2 MS) agar plates (2.154g l–1 MS, 0.1g l–1 myo-inositol, 0.5g l– MES, 10g l– bacteriological agar, pH 5.7 with 1M KOH) and germinated vertically under constant white light at 21 °C.
Genotyping of T-DNA insertion lines
The following T-DNA insertion mutant lines were used and genotyped in our investigations: ralfl34-1 (SALK_004441) using the following genotyping primers, FW primer TGACTAACCAAAAA GTCCACG; REV primer, ACGGGACCTCTAGCTCTGAAG; and T-DNA primer, ATTTTGCCGATTTCGGAAC; and ralfl34-2 (JIC_SGT4223) using the following genotyping primers, FWprimer, ATGGCAGCTTCGTCTCTC; REV primer, CTATCTCCGGCATCGAGT; and T-DNA primer, ACGGTCGGGAAACTAGCTCTAC.
Constructs
The pRALFL34::LUC was generated as follows: the 400bp RALFL34 promoter fragment was PCR amplified using the RALFL34 forward (CAACTGGACCCATCCGAA) and reverse primer (CGGCGATTGTTGGGGGA) and cloned into the pGem-T-easy vector. After sequencing confirmation, the 416bp RAFL34 promoter fragment was released from the pGem®-T Easy vector with restriction enzymes PstI and NcoI and then cloned into the LucTrap vector (Calderon-Villalobos et al., 2006; Lau et al., 2011; De Smet et al., 2013). The sequence of the final construct was confirmed. ERF9 and ERF4 were cloned into pJIT60 as follows: the ERF9 and ERF4 coding sequences were PCR amplified using ERF9 (ATGGCTCCAAGACAGGCG and CTAAACGTCCACCACCGGT) and ERF4 primers (ATGGCCAAGATGGGCTTG and TCAGGCCTGTTCCGA TGG), and were cloned into the pGem®-T-Easy vector. The sequences of the plasmids containing either ERF9 or ERF4 were confirmed, and the ERF9 and ERF4 coding sequences were released by the restriction enzyme EcoRI and cloned into the pJIT60 vector (Schwechheimer et al., 1998; Lau et al., 2011; De Smet et al., 2013). The sequences of the final constructs were confirmed. For the yeast one-hybid (Y1H) experiments, pRALFL34 fragments were cloned as follows: 416bp and 869bp RALFL34 promoter fragments were PCR amplified using RALFL34 forward (CAACTGGACCCATCCGAA or CAGTATCAGGCTTGTGTTCA) and reverse primer (CGGCGATT GTTGGGGGA or GGCGATTGTTGGGGGAAA) and then cloned into the entry vector pENTR™ 5'-TOPO (Invitrogen). The entry clones containing the 416bp or 869bp RALFL34 promoters were then recombined to yield promoter::HIS3 and promoter::LacZ reporter constructs (Deplancke et al., 2004; Brady et al., 2011) and their sequences confirmed.
Y1H
Y1H assays were performed as previously described (Gaudinier et al., 2011). Interactions were called for transcription factors that activated at least one reporter assay.
Transgenic lines
We generated pRALFL34::n3xRFP, using a previously published vector backbone (Slane et al., 2014) and a RALFL34 promoter fragment of 869bp (counting from ATG). To generate the p35S::ERF9-GR line, the coding sequence of ERF9 without a STOP codon, a glucocorticoid domain (GR), and the constitutive 35S promoter were cloned in pDONR221, pDONRP2RP3, and pDONRP4P1R, respectively, with Gateway Cloning®. A multisite LR recombination combined the entry vectors into the destination vector pK8m34GW-FAST. The sequences of both entry vectors and the expression vector were confirmed. Both pRALFL34::n3xRFP and p35S::ERF9-GR constructs were transformed into Agrobacterium sp. and then floral dipped into Col-0 plants (Clough and Bent, 1998).
Induction of lateral roots
Lateral root induction was performed through mechanical (Ditengou et al., 2008) or gravitropic bending (Péret et al., 2012; Lavenus et al., 2015; Voβ et al., 2015), as previously described. Quantification of pGATA23::NLS:GFP fluorescence was analysed on Col-0 (wild type) and ralfl34-1 seedlings. Total fluorescing nuclei (in all tissues) were counted from the quiescent centre (QC) moving up the root for ~2500 μm and density measurements were calculated as ‘fluorescing nuclei in n μm’.
DEX treatment
For dexamethasone (DEX) treatments, seedlings were grown in vitro on 1/2 MS plates (7.5g l– agar) overlaid with a nylon mesh (Prosep, 20 µm pore size). At 7 d after sowing, the mesh with seedlings was transferred to plates with 1/2 MS medium (7.5g l– agar) and plates with 1/2 MS medium containing 5 µM DEX (Sigma).
Microscopic analysis
Phenotyping was analysed under a Leica stereo dissection microscope at varying magnifications to observe and count the emerged lateral roots. Lateral roots were counted, marked, and then photographed. Root lengths were measured from the bottom of the hypocotyl to the root tip, using ImageJ software (http://imagej.nih.gov/ij/). Lateral root staging of the ralfl34-1 and ralfl34-2 mutants was performed on a Leica DMRB microsystem using differential interference contrast (DIC). Fluorescent seedlings were analysed on a Nikon confocal microscope utilizing both an Argon 488 laser and a 514 HeNe laser at ×20 and ×40 magnification.
RALFL34 auxin response quantification
Arabidopsis (ecotype Col-0) seeds were surface sterilized, stratified at 4 °C for 2 d, then plated on 1/2 MS medium and grown vertically under constant white light at 21 °C for 4 d post-germination. Seedlings were transferred to 1/2 MS liquid medium on the fourth day to acclimate overnight. Seedlings were transferred to liquid 1/2 MS containing 1-naphthaleneacetic acid (NAA) at a concentration of 10 μM (or ethanol control) and were grown for a further 0, 1, 2, 4, 6, 8, 12, and 24h; at each time point, the roots of 10 seedlings were excised, frozen in liquid nitrogen, and then total RNA extracted as below.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from lines of interest using an RNeasy kit (Qiagen) as per the manufacturer’s instructions. cDNA was generated from 1 µg or 200ng of total RNA via oligo(dT) primers using iScript (BioRad) or Superscript II (Invitrogen), respectively, as per the instructions. Quantitative PCRs were performed in at least two technical repeats, using the SYBR Green QPCR Master Mix (Quanta Biosciences), in our modified reaction protocol: Master mix, 7 μl per reaction [forward primer (100 μM), 0.1 μl per reaction; reverse primer (100 μM), 0.1 μl per reaction; SYBR (2×), 6 μl per reaction; dH2O, 0.8 μl per reaction] and template cDNA (diluted 1/100 in dH2O, 5 μl per reaction. Alternatively, the LightCycler® 480 SYBR Green I Master (2×) in Light Cycler 480 (Roche) was used, plates were filled with the JANUS Automated Workstation Perkin Elmer, primers were used at 0.5 µM, and cDNA was diluted 8× (total volume=5 µl=0.5 µl of template, 2 µl of primers, 2.5 µl; 2× mix). Gene-specific quantification was performed with intron-spanning primer pairs (where possible) designed using Primer3 software (http://bioinfo.ut.ee/primer3/). Expression was normalized using AT5G60390 (CTGGAGGTTTTGAGGCTGGTAT and CCAAGGGTGAAAGCAAGAAGA) and/or AT3G 60830 (ACTCTTCCTGATGGACAGGTG and CTCAACGATT CCATGCTCCT) and/or AT4G16100 (GGAGATTAAGC AACCTGAGGAGTG and GTGGTGGTGGTGGAGGA GAC). Primers used for qPCR: RALFL34 (CGATGCCGG AGATAGATGAAG or TGGCAGCTTCGTCTCTCAA and CAATCCACCCCTCACGACT or TGGAGAGAAATGAAA GTGAGAAGAG), GATA23 (AGTGAGAATGAAAGAAGA GAAGGG or GGGGACAATTAGGTGTTGCA and GTGGCT GCGAATAATATGAATACC or CTCCTTCGTTTCTTCA CCGC), ERF4 (TTTCTCGAGCTGAGTGACCA and GTCGGAGGAGAAGCACAGTC), and ERF9 (AGAGAGTTT CGTGGCTCCAA and CCACCGTCGTTAACCGTAGT). All qRT-PCR experiments were performed in three biological replicates (unless otherwise specified) and 2–3 technical repeats, and the data presented represent means ±SE.
In silico analyses
CORNET analyses were done using standard settings in the TF tool on https://bioinformatics.psb.ugent.be/cornet/versions/cornet3.0/main/tf (De Bodt et al., 2010, 2012; De Bodt and Inzé, 2013). We used SignalP 4.1 (Petersen et al., 2011) with standard settings to predict the presence and location of a signal peptide cleavage site in RALFL34. Genevestigator (Zimmermann et al., 2004, 2005; Hruz et al., 2008) was probed using the standard application. For the identification of transcription factor-binding sites, we used CIS-BP (Weirauch et al., 2014), and to search for the identified motif matrices in promoter sequences we used Regulatory Sequence Analysis Tools (RSAT) (Medina-Rivera et al., 2015). We used Arabidopsis eFP Browser (Winter et al., 2007) absolute or relative values to gain insight into expression patterns of selected genes.
Transient activity assays
BY-2 protoplast assays were performed as previously described (Vanden Bossche et al., 2013), using reporter (LucTrap, encoding firefly luciferase) and effector constructs (pJIT60).
Results and Discussion
RALFL34 regulates pericycle cell division patterns during lateral root initiation
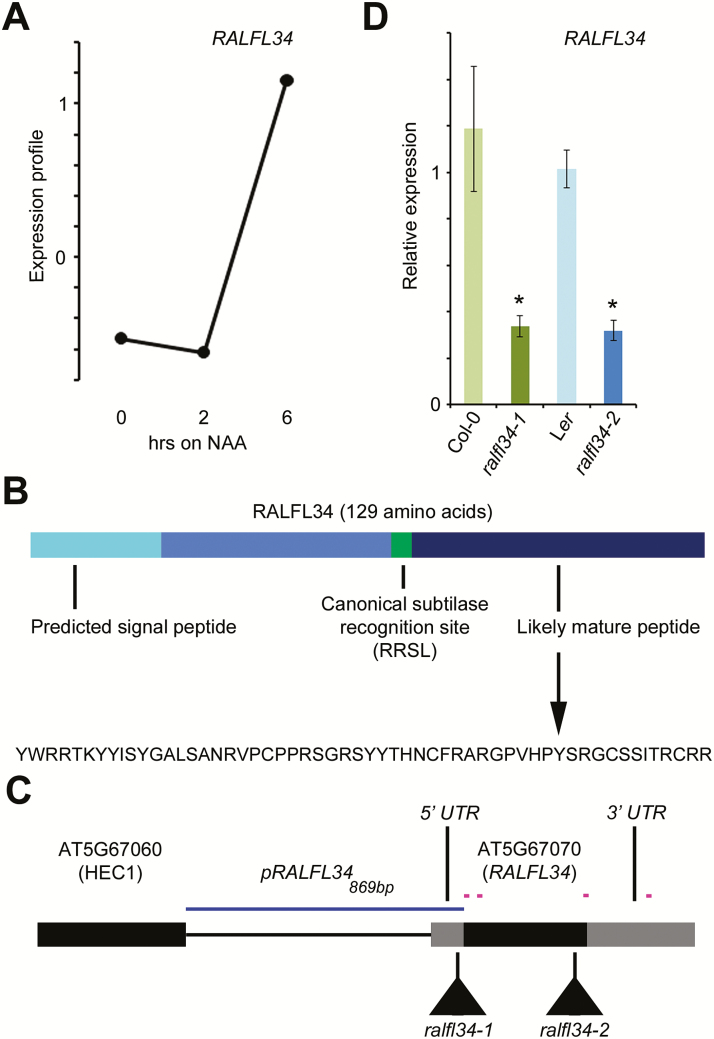
In a transcriptome analysis focusing on early stages of lateral root initiation and utilizing a lateral root inducible system based on auxin treatment leading to synchronous induction of lateral roots (Himanen et al., 2002, 2004; De Rybel et al., 2010), RALFL34 (AT5G67070) expression was up-regulated, together with a small set of 14 potential key regulatory genes for asymmetric cell division and/or cell fate specification during lateral root initiation, including ARABIDOPSIS CRINKLY4 (ACR4) (De Smet et al., 2008) (Fig. 1A). RALFL34 encodes a member of a family of cysteine-rich small signalling peptides (Murphy and De Smet, 2014) and probably gives rise to a 56 amino acid mature peptide (Fig. 1B). While members of the RALF family have been functionally characterized (Pearce et al., 2001; Olsen et al., 2002; Haruta et al., 2008, 2014; Matos et al., 2008; Srivastava et al., 2009; Atkinson et al., 2013; Bergonci et al., 2014; Morato do Canto et al., 2014; Li et al., 2015), no data are so far available for RALFs in the context of lateral root initiation.
Fig. 1.
RALFL34 expression and gene/protein topology. (A) Expression profile of RALFL34 in the pericycle of seedlings subjected to lateral root inducible system [after mixed model analysis; data from De Smet et al., (2008)]. Seedlings were grown on NPA (1-N-naphthylphthalamic acid; 0h) for 3 d. NAA, 1-naphthaleneacetic acid. (B) Schematic of RALFL34 protein, highlighting the putative signal peptide and likely mature peptide region and sequence. (C) Schematic of the RALFL34 gene, with indication of qPCR primers (pink lines), position of T-DNA insertion, and promoterregion (blue line). (D) RALFL34 expression as monitored through qPCR in ralfl34-1 and ralfl34-2 roots at 5 d after germination and their respective control lines, Col-0 and Ler. The graph depicts the average of three biological repeats (and 3–6 technical repeats) ±SE. Student’s t-test with P-value <0.01.
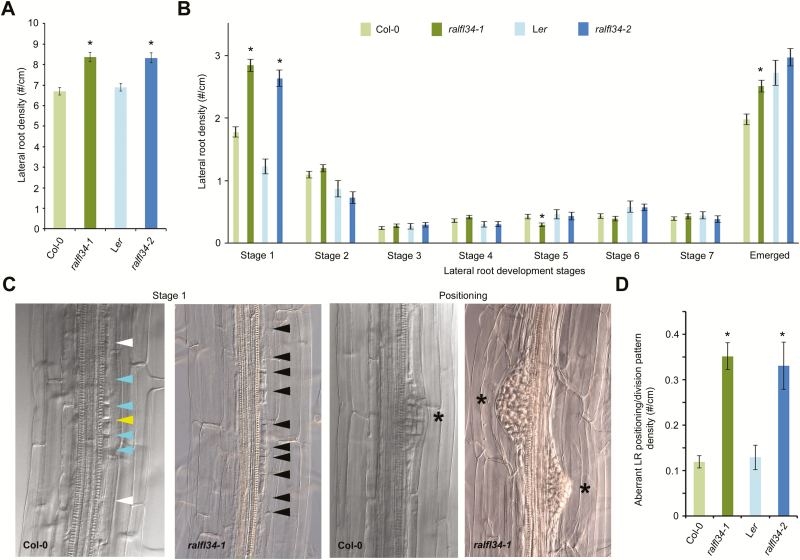
To validate a role for RALFL34 in lateral root initiation, we identified two T-DNA lines, namely ralfl34-1 (in Col-0) and ralfl34-2 (in Ler), with significantly reduced RALFL34 expression (Fig. 1C,D). Both ralfl34-1 and ralfl34-2 were analysed with respect to overall lateral root density, lateral root stage distribution, and pericycle division patterns. Overall, ralfl34-1 and ralfl34-2 displayed, respectively, a 25% and 21% increase in total lateral root density (emerged and non-emerged) compared with their control line (Fig. 2A; Supplementary Fig. S1 at JXB online). A more detailed analysis quantifying the different stages of lateral root development in ralfl34-1 and ralfl34-2 revealed that this is probably due to an enrichment of stage 1 lateral root primordia [which also included regions with divisions that did not fully resemble a typical stage 1 primordium (as shown in Fig. 2D)] compared with the control (Fig. 2B). Furthermore, we observed a 3-fold increase in ‘aberrant’ pericycle division patterns (extra cell divisions flanking the primordia) or unusually positioned (defined as lateral root primordia being closer to each other than 400 µm) in ralfl34-1 and ralfl34-2 compared with their control line (Fig. 2C, D; Supplementary Fig. S1). Based on these observations, we concluded that RALFL34 plays a role during lateral root initiation and probably acts to restrict (proliferative) cell divisions and/or mediate the spatial distribution of asymmetric founder cell divisions in the pericycle along the root’s longitudinal axis.
Fig. 2.
Lateral root phenotypes in ralfl34 mutants. (A, B) Lateral root density in ralfl34-1 (n=69) and ralfl34-2 (n=29) compared with their respective controls, Col-0 (n=79) and Ler (n=20) depicted as total lateral root density, including all stages (A) and split into lateral root stages 1–7 and emerged lateral roots (according to Malamy and Benfey, 1997) (B) at 7 d after germination. Stage 1 includes regions with extra pericycle cell divisions. (C, D) Representative DIC pictures (C) and quantification (D) of aberrant stage 1, additional pericycle divisions, or unusually positioned lateral roots in ralfl34-1 (n=69) and ralfl34-2 (n=29) compared with their respective controls, Col-0 (n=79) and Ler (n=20). An asterisk indicates a lateral root primordium. Arrowheads separate pericycle cells: central (yellow) and outer position (white) for adjacent pericycle cells undergoing two rounds of asymmetric cell divisions (blue) and extra rounds of divisions (black). All graphs show the average ±SE of the indicated sample numbers. *P<0.05 according to Student’s t-test compared with control.
RALFL34 is expressed before asymmetric pericycle cell division
To investigate at which stage during lateral root development RALFL34 is expressed, we generated a pRALFL34 869bp ::n3xRFP line (referred to as pRALFL34::n3xRFP) and combined this with a yellow fluorescent protein-NPSN12 plasma membrane marker line (WAVE131Y) (Geldner et al., 2009). RALFL34 is expressed in xylem pole pericycle cells before any visible sign of asymmetric cell division (Fig. 3A). At a later stage, RALFL34 is strongly expressed in the small central cells of the lateral root initiation site, but also in the larger flanking cells (Fig. 3B, C). In addition, we detected RALFL34 expression in the epidermis (Fig. 3A). Given that lateral root development is controlled by auxin at various levels (Lavenus et al., 2013), we explored if RALFL34 expression is regulated by auxin in the root. Treatment with the synthetic auxin NAA (1-naphthaleneacetic acid) showed minor, but significant, concentration-dependent down-regulation after 6h and minor, but significant, concentration-dependent transcriptional up-regulation following a 24h treatment (Fig. 4A). Primary auxin-responsive genes are generally significantly differentially regulated within 2h after exposure to auxin (Abel et al., 1994; Oeller and Theologis, 1995). This suggests that RALFL34 is probably not a (positively regulated) primary auxin response gene, but acts nevertheless downstream of an auxin response module. It should be noted that these results are obtained from auxin-treated seedlings grown on control medium before treatment, while the initial identification of RALFL34 (see above; Fig. 1A) was based on auxin treatment synchronously inducing asymmetric cell divisions in the pericycle following growth on the auxin transport inhibitor NPA. Furthermore, this change in RALFL34 expression is likely to be due to initiation of multiple formative asymmetric divisions, as an increased number of lateral root primordia induced by the NAA treatment are observed (Fig. 4B). Interestingly, RALFL34 was recently identified as having a mobile mRNA in a root to shoot direction (Thieme et al., 2015), suggesting that the RALFL34 domain—also in the root—is likely to be broader than reported by our pRALFL34::n3xRFP line. In this context, Arabidopsis eFP Browser data suggested the presence of RALFL34 mRNA in above-ground plant parts (Supplementary Fig. S2), but this can be derived from mobile mRNA and/or local expression.
Fig. 3.
RALFL34 expression during lateral root initiation as visualized through pRALFL34::n3xRFP (red) and WAVE 131Y (green). Arrowheads separate pericycle cells: central (yellow) and outer position (white) for adjacent pericycle cells undergoing two rounds of asymmetric cell divisions. Lateral root development stages are indicated. Ep, epidermis. Pe, pericycle (at xylem pole).
Fig. 4.
Auxin-mediated regulation of RALFL34 expression. (A) Expression of RALFL34 after 6h and 24h treatment with NAA. The graph shows the average ±SE of three biological repeats. *P<0.05 according to Student’s t-test compared with mock. (B) Representative images of pRALFL34::n3xRFP (red) expression upon 24h of NAA treatment. An asterisk indicates the position of lateral root initiation/primordium and/or dividing pericycle.
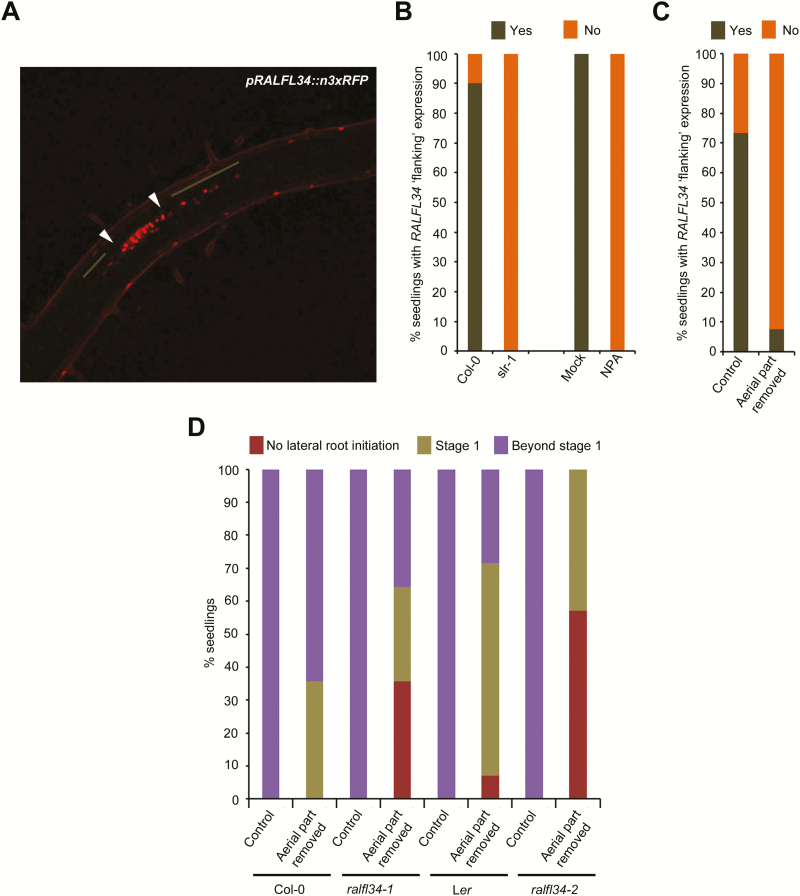
‘Flanking’ RALFL34 expression is associated with lateral root initiation
Interestingly, the pRALFL34::n3xRFP expression around the lateral root initiation site often showed a ‘flanking’ expression profile, which appears to be contained within the pericycle, that extended from the lateral root initiation site shootward and/or rootward (25/30 lateral root primordia) (Fig. 5A). We established that this ‘flanking’ expression is indeed associated with the formation of a lateral root. For this, we subjected seedlings that do not display lateral root initiation, namely grown on NPA or in the slr-1 background, to mechanical root tip bending to induce a lateral root, as was previously described (Ditengou et al., 2008). Under these experimental conditions (and 20h after the bending), we did not observe any lateral root initiation-associated (and thus probably no lateral root formation) or ‘flanking’ RALFL34 expression in the pericycle (Fig. 5B). Since RALFL34 seems to affect the number of lateral roots along the longitudinal primary root axis, we speculated that this ‘flanking’ expression might play a role in regularly positioning these lateral roots.
Fig. 5.
RALFL34 ‘flanking’ expression. (A) Representative image of pRALFL34::n3xRFP expression with expression in the lateral root primordium (between white arrowheads) and flanking the lateral root primordium (brown line) upon bending. (B, C) Percentage of seedlings showing pRALFL34::n3xRFP ‘flanking’ expression in (B) the wild type (Col-0) versus slr and mock (DMSO) versus NPA-treated seedlings upon bending and in (C) seedlings with aerial tissues removed. (D) Percentage of seedlings, with aerial tissues removed, which failed to initiate lateral roots (red), developed a stage 1 primordium (khaki), or progressed beyond stage 1 (purple), 20h post-mechanical bending.
A shoot-derived signal affects RALFL34 expression around the lateral root primordium
Since shoot-derived signals are involved in various steps of lateral root development (Reed et al., 1998; Bhalerao et al., 2002; McAdam et al., 2016), we explored the involvement of the shoot (as a source of auxin or another signal) in establishing the ‘flanking’ pRALFL34::n3xRFP expression in the root. Therefore, we removed the aerial tissues (hypocotyl and upward) and subjected pRALFL34::n3xRFP plants to the gravitropic bending assay (Péret et al., 2012; Lavenus et al., 2015; Voβ et al., 2015) to induce the formation of a lateral root. pRALFL34::n3xRFP expression was assessed for 20h after the removal of their aerial tissues and following mechanical root tip bending. Seedlings grown under control conditions (no aerial tissues removed) showed >70% pRALFL34::n3xRFP flanking expression after 20h gravistimulation (Fig. 5C). In contrast, seedlings with aerial tissues removed had only 10% with pRALFL34::n3xRFP flanking expression (Fig. 5C). These data strongly suggest that a shoot-derived signal is required for pRALFL34::n3xRFP ‘flanking’ expression.
To observe precisely the impact of removing shoot-derived signals on RALFL34-mediated lateral root formation, we utilized the gravitropic bending assay in combination with aerial tissue (hypocotyl and upward) removal on ralfl34 mutants. Without removing aerial tissues, there was no significant difference between any of the controls or mutant lines 20h post-bending, as all lateral roots progressed beyond stage 1 (Fig. 5D). However, 20h post-bending and having removed the aerial tissues, we observed a reduction in the percentage of seedlings with lateral root initiation events in ralfl34-1 (64%) and ralfl34-2 (43%), compared with the Col-0 (100%) and Ler (93%) control, respectively (Fig. 5D). In addition, we also observed a reduction in the percentage of lateral root primordia that progressed beyond stage 1 in ralfl34-1 (56%) and ralfl34-2 (0%), compared with the Col-0 (64%) and Ler (31%) control, respectively (Fig. 5D). Taken together, our results suggest that RALFL34 is required for lateral root initiation (and progression beyond this stage) and appears to interpret a shoot-derived signal that is possibly not auxin. In addition, these results and the RALFL34 ‘flanking’ expression pattern might indicate that RALFL34 is part of a lateral inhibition mechanism based on positive and negative feedback.
RALFL34 expression is negatively regulated by ERFs
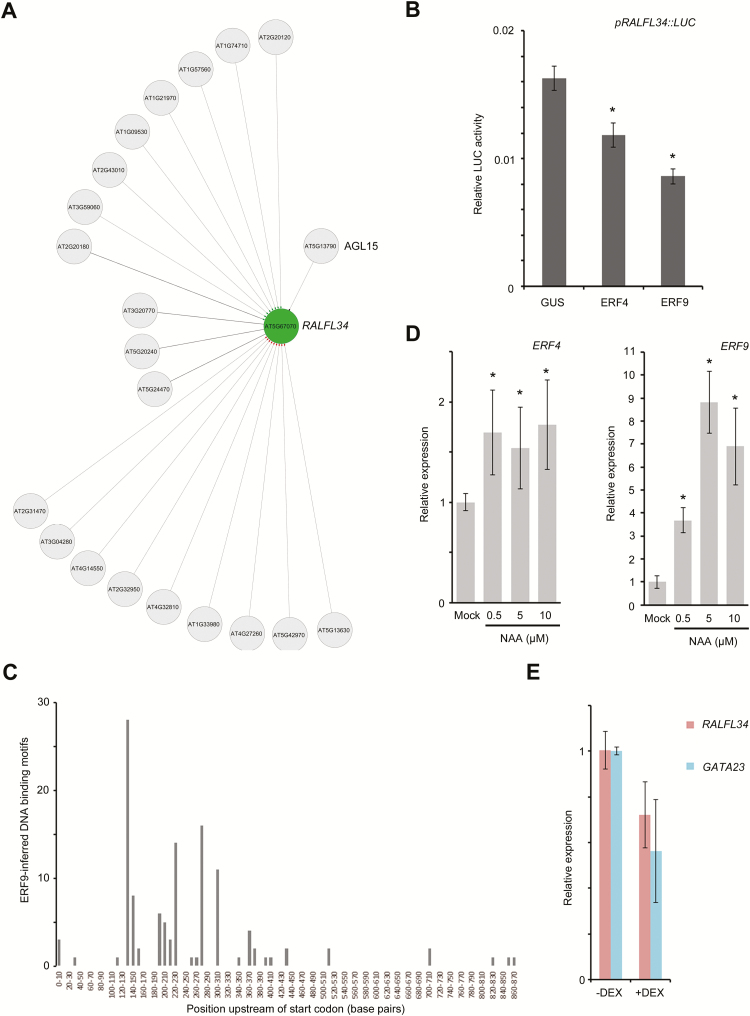
To gain insight into the regulation of RALFL34 expression (and possibly the shoot-derived signal), we initially performed an in silico analysis using CORNET, which allows the integration of regulatory interaction data sets accessible through the transcription factor tool (De Bodt et al., 2010, 2012; De Bodt and Inzé, 2013). This revealed a set of 21 transcription factors potentially regulating RALFL34 expression (Fig. 6A), of which one was also differentially expressed in the pericycle during lateral root initiation (De Smet et al., 2008) and/or all those that could be checked were to some extent differentially expressed in the Arabidopsis Lateral Root initiation eFP Browser (Supplementary Table S1). Next, we performed an enhanced yeast one-hybrid (eY1H) analysis (Gaudinier et al., 2011) on two RALFL34 upstream regulatory regions of different length (869bp or 416bp; with the original idea to narrow down the regulatory region arbitrarily), with a collection of transcription factors expressed in the root stele which includes pericycle cells (Supplementary Table S1). This revealed a set of 34 transcription factors potentially regulating RALFL34 expression, of which four were also differentially expressed in the pericycle during lateral root initiation (De Smet et al., 2008) and/or all were to some extent differentially expressed in the Arabidopsis Lateral Root initiation eFP Browser (Supplementary Table S1). This list of interacting transcription factors showed limited overlap with the CORNET data, namely only AGAMOUS-LIKE 15 (AGL15) (Supplementary Table 1). AGL15 has already been characterized extensively (Perry et al., 1999; Fernandez et al., 2000, 2014; Wang et al., 2002, 2004; Harding et al., 2003; Tang and Perry, 2003; Adamczyk et al., 2007; Hill et al., 2008; Zheng et al., 2009; Patharkar and Walker, 2015), and RALFL34 was also listed in a genome-wide identification of in vivo AGL15-binding sites (Zheng et al., 2009). To determine if AGL15 is sufficient to regulate RALFL34 expression in planta, we performed experiments using a heterologous in vivo protoplast system which monitors gene expression quantitatively by activation of a luciferase (LUC) reporter (Lau et al., 2011; Vanden Bossche et al., 2013). Based on the results (data not shown), we concluded that although AGL15 can physically bind the promoter, it was not able to reproducibly regulate the expression of pRALFL34 416bp ::LUC in this transient system. We therefore decided to focus on members of the ERF protein family, as this family was represented by seven out of 34 Y1H hits (Supplementary Table 1). Next, we specifically tested an interaction between RALFL34 expression and (the arbitrarily selected) ERF4 and ERF9 in protoplasts. This revealed a significant down-regulation of LUC expression (Fig. 6B), which is in agreement with ERF4 and ERF9 containing a repression domain (Licausi et al., 2013). In addition, we did an in silico search for ERF4 and ERF9 DNA-binding motifs. ERFs are known to bind directly to the cis-element called a GCC-box containing the core 5′-GCCGCC-3′ sequence (Ohme-Takagi and Shinshi, 1995). We could indeed find ERF4-binding motifs and ERF9 inferred binding motifs in the promoter of RALFL34. Interestingly, 92% of the identified motifs were found in the first 400bp of the promoter (Fig. 6C). We further confirmed this interaction in planta using a transgenic ERF9-GR line. This revealed that in the roots, ERF9 could indeed (mildly) down-regulate RALFL34 expression (Fig. 6E). The ERF-mediated down-regulation of RALFL34 expression possibly links RALFL34 expression to a regulation by ethylene. However, an analysis of available, general Arabidopsis Genevestigator and eFP Browser data did not reveal any significant differential regulation of RALFL34 expression upon ACC (1-aminocyclopropane-1-carboxylic acid) treatment (data not shown). However, to conclude this convincingly, a more detailed and cell- or tissue-specific expression profiling would be needed to evaluate the influence of ACC on RALFL34 expression. Interestingly, ERF4 and mainly ERF9 expression is up-regulated by auxin in the root (Fig. 6D). In addition, based on eFP Browser data, ERF4 and ERF9 appear to be ethylene inducible (Supplementary Fig. S3). In future, we will further look into this potential hormone crosstalk, but it remains possible that this ERF4– or ERF9−RALFL34 network edge is not associated with ethylene and/or lateral root development, as RALFL34 is more broadly expressed in the plant (Supplementary Fig. S2).
Fig. 6.
ERFs regulated RALFL34 expression. (A) In silico generated transcription factor network, highlighting putatively interacting proteins with RALFL34 in CORNET. Interactions are as follows: confirmed (full line), unconfirmed (dotted line), indirect (diamond), direct+unknown (disc), direct+activation (arrowhead), direct+repression [line activation (green), repression (red), and unknown (black)]. (B) Luciferase (LUC) activity upon co-expression of pRALFL34::LUC and GUS (control) or ERF4/9 in protoplasts. Luciferase assay was performed three times, each time with eight biological replicates. The graph shows the average of all 24 data points ±SE. Student’s t-test with P-value <0.01. (C) Number of ERF9-inferred DNA-binding motifs according to their position upstream of the coding sequence (start codon=position 0). (D) ERF4 and ERF9 expression upon 6h of NAA treatment at the indicated concentrations. The graph shows the average ±SE error of three biological repeats. *P<0.05 according to Student’s t-test compared with mock. (E) Expression of RALFL34 and GATA23 in an inducible ERF9 overexpression line (35S::ERF9-GR) after 4h of DEX-induced overexpression of ERF9.
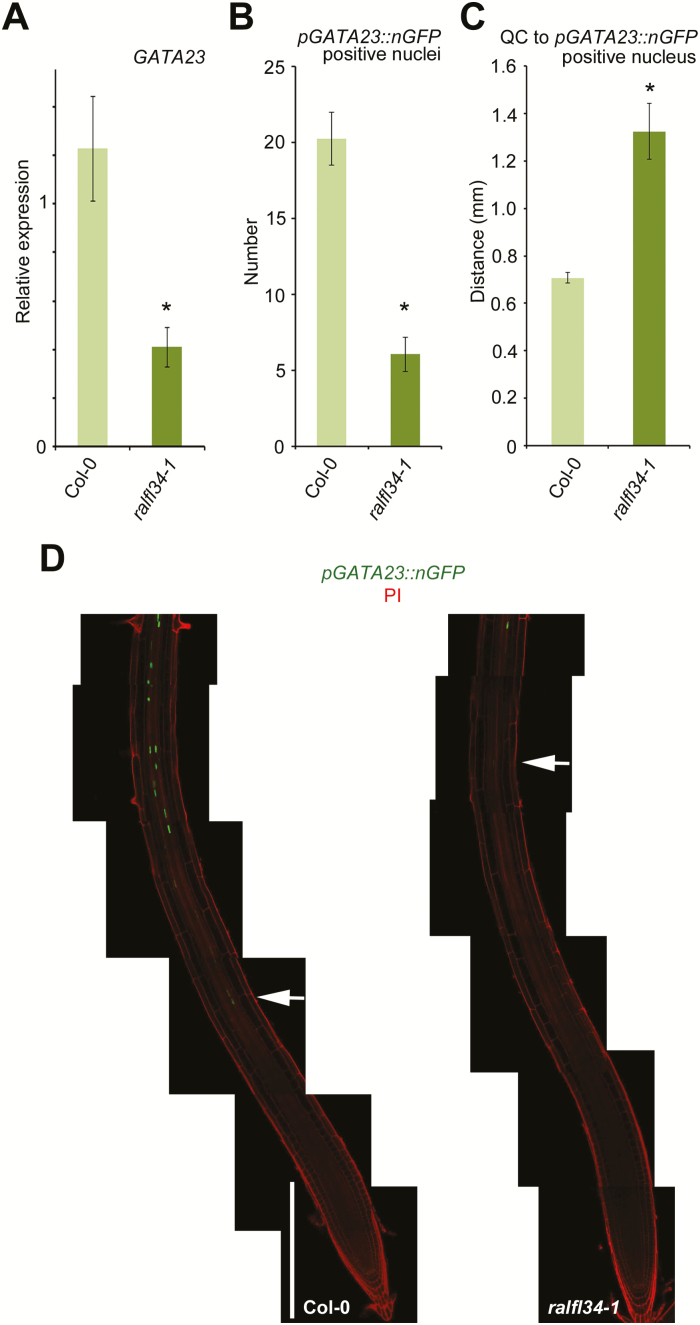
RALFL34 acts upstream of GATA23
Finally, we aimed to position RALFL34 with respect to known regulators in the cascade that leads to lateral root development (Atkinson et al., 2014; Behringer and Schwechheimer, 2015; Vermeer and Geldner, 2015). One of the earliest markers for lateral root initiation is GATA23, a transcription factor that controls founder cell identity (De Rybel et al., 2010), and we therefore analysed GATA23 expression in ralfl34-1 roots. qRT-PCR analyses on seedling roots revealed a significant down-regulation of GATA23 expression levels in ralfl34-1 compared with the control (Fig. 7A). To confirm this, we evaluated the pGATA23::NLS:GFP line (De Rybel et al., 2010) in the ralfl34-1 background. This revealed overall less green fluorescent protein (GFP)-positive nuclei in the root (Fig. 7B, C). In addition, the location of the first GFP-positive nucleus, with respect to the QC, was higher up the root (Fig. 7D). Based on these results, we conclude that RALFL34 probably acts upstream and as a positive regulator of GATA23. However, the lateral root initiation phenotypes of ralfl34-1 (this work) and a GATA23 RNAi line (fewer emerged lateral roots and fewer stage 1/2 lateral root primordia) (De Rybel et al., 2010) do not appear to match. There are potentially additional regulators of GATA23 or possibly altered expression of GATA23 in ralfl34-1 is an indirect effect, for example through a perturbed auxin balance, especially in view of GATA23 being a highly auxin-responsive gene (Supplementary Fig. S4) (De Rybel et al., 2010). Interestingly though, GATA23 expression seems also to be down-regulated upon DEX-induced ERF9 activity, suggesting that RALF34 might represent a component connecting ERF9 signalling with downstream transcriptional regulation of GATA23 (an indirect effect caused by the direct down-regulation of RALFL34) or, alternatively, ERF9 acts on GATA23 in a parallel pathway (a direct effect by the binding of ERF9 to the promoter of GATA23) (Fig. 6E).
Fig. 7.
GATA23 expression downstream of RALFL34. (A) GATA23 expression in wild-type (Col-0) and ralfl34-1 seedling roots 5 d after germination as monitored through qPCR. The graph depicts the average of three biological repeats (and 3–6 technical repeats) ±SE. Student’s t-test with P-value <0.01. (B) Representative images of pGATA23::nGFP expression in the wild type (Col-0) and ralfl34-1, white arrows indicating the first GFP expressing nucleus. White bar=300 μm. (C, D) Average number of pGATA23::nGFP-positive nuclei (C) and average length from the quiescent centre (QC) to the first nGFP-positive nucleus in the root tips of Col-0 and ralfl34-1 (D). Graphs show the average ±SE of 30 seedlings. *P<0.05 according to Student’s t-test compared with the wild type.
Conclusion
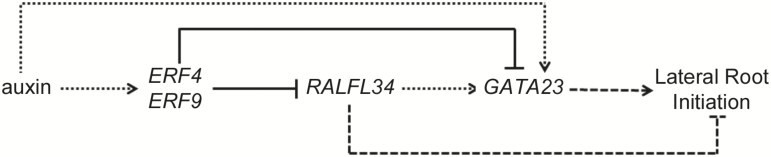
In this work, we used a candidate gene approach [based on differential expression in our transcriptomics data (De Smet et al., 2008)] to identify molecular components of lateral root initiation and associated asymmetric cell division. Our results indicate a strong developmental role for the small signalling peptide RALFL34 during the early stages of lateral root development. RALFL34 expression is down-regulated by auxin (Fig. 8). Interestingly, upstream of RALFL34 expression are auxin-inducible and ethylene-inducible ERFs that down-regulate RALFL34 expression (Fig. 8). Previously, ethylene has been shown to affect the ability of pericycle cells to undergo lateral root initiation, probably through interfering with auxin accumulation (Ivanchenko et al., 2008; Negi et al., 2008, 2010; Lewis et al., 2011). With respect to a shoot-derived signal, our data confirm that this is required for the normal progression of lateral root development from stage 1, and which acts downstream of primary auxin responsiveness (Bhalerao et al., 2002; Marchant et al., 2002; Ditengou et al., 2008). In addition, it appears that RALFL34 is interpreting a shoot-derived signal to drive the progression from founder cell to stage 1 primordia. Concurrently, RALFL34 expression in the flanking pericycle cells induced by a shoot-derived signal might be essential to restrain cell proliferation in the neighbouring pericycle cells because ralfl34 mutants are characterized by extra cell divisions in these regions. One possibility is that this shoot-derived signal is auxin, as indeed this hormone plays a role in regulating lateral root development (Lavenus et al., 2013). However, given that the primary effect of auxin is repression of RALFL34 expression, we believe other signals are likely to be involved. For example, in view of lateral root positioning, a carotenoid-derived molecule was already shown to play a role (Van Norman et al., 2014). In addition, it was recently shown that ERF109 expression is strongly up-regulated by methyl jasmonate (MeJA), and that ERF109 regulates lateral root development in response to MeJA (Cai et al., 2014). In this context, however, it is interesting to note that in the leaves ERF9 is transcriptionally down-regulated following short-term MeJA and salicylic acid (SA) treatment (Maruyama et al., 2013). Finally, we have shown that RALFL34 acts genetically upstream of GATA23 (Fig. 8), but it is not clear if this is a direct regulation or indirect due to perturbed auxin accumulation and/or responsiveness. However, GATA23 expression seems also to be regulated downstream of ERF9 (Fig. 8). Taken together, our data revealed an as yet unreported role for RALF peptides in lateral root initiation, and position RALFL34 as a possible earlier marker for founder cell identity than GATA23. In future, the spatio-temporal regulation of and genetic interactions within this small network will need to be investigated in detail in the root in order to establish its biological role further. In addition, what the underlying mechanism and targets of RALFL34 action are remain to be explored. In this respect, RALF peptides have been shown to be linked to Ca2+ release (Haruta et al., 2008, 2014; Morato do Canto et al., 2014), which—in turn—is associated with mechanical stress (Richter et al., 2009). Interestingly, mechanical stimulation of roots causes increases in Ca2+ in epidermal, cortical, endodermal, and pericycle cells of roots (potentially through stretch-activated Ca2+ channels) (Richter et al., 2009). Furthermore, lateral root initiation also requires root bending, which can be seen as a mechanical stimulus (Laskowski et al., 2008; Kircher and Schopfer, 2016; Scheres and Laskowski, 2016). In future, it will be important to determine to what extent Ca2+ levels are perturbed in ralfl34 roots under normal conditions and during (mechanical) root bending compared with wild-type roots. Another explanation might come from the recently identified RALF1 receptor, namely FERONIA (FER), a member of the highly expressed malectin receptor-like kinase family (Haruta et al., 2014). It was shown that RALF1 binds to FER, causing downstream phosphorylation on a plasma membrane-associated H+-ATPase (AHA2), resulting in an increased apoplastic pH and a decrease in cell wall elongation. AHA2 (aha2) has since been shown also to cause a decrease in lateral root density, correlating with a downstream mechanism as to why 35S::RALF1 lines have decreased lateral root density (Mlodzinska et al., 2015). Overall, RALFL34 possibly affects ion balance in the pericycle (and/or surrounding tissues) to regulate cell divisions associated with lateral root initiation.
Fig. 8.
Schematic representation of possible RALFL34-mediated regulation of lateral root initiation and regulation of ERF4, ERF9, RALFL34, and GATA23 expression in the root. Dotted lines (possibly/likely) indirect effects; dashed lines, developmental impact; full line, (likely) direct effect.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Lateral root phenotypes in ralfl34-1 and ralfl34-2.
Figure S2. Absolute expression value for RALFL34 in above-ground organs.
Figure S3. Relative expression values for ERF4 and ERF9 upon ACC treatment.
Figure S4. GATA23 expression upon NAA treatment.
Table S1. CORNET and Y1H data.
Acknowledgements
We thank Koichi Toyokura and Ute Voβ for useful suggestions and/or seeds. We thank Robin Vanden Bossche for assistance with transient protoplast assays. This work was in part funded by a Japan Society for the Promotion of Science (JSPS) overseas researcher summer fellowship, and was also supported by a Biotechnology and Biological Science Research Council David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration grant (PERG06-GA-2009-256354) to IDS. We thank the School of Biosciences and Malcolm. J. Bennett for studentship funding, and acknowledge the University of Nottingham research committee. SMB was supported by a Hellman Fellowship.
References
- Abel S, Oeller PW, Theologis A. 1994. Early auxin-induced genes encode short-lived nuclear proteins. Proceedings of the National Academy of Sciences, USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. 2007. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. The Plant Journal 50, 1007–1019. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Araya T, Miyamoto M, Wibowo J, et al. 2014. CLE–CLAVATA1 peptide–receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences, USA 11, 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson JA, Rasmussen A, Traini R, Voβ U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ. 2014. Branching out in roots: uncovering form, function, and regulation. Plant Physiology 166, 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson NJ, Lilley CJ, Urwin PE. 2013. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiology 162, 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inzé D. 2001. The peri-cell-cycle in Arabidopsis. Journal of Experimental Botany 52, 403–411. [DOI] [PubMed] [Google Scholar]
- Behringer C, Schwechheimer C. 2015. B-GATA transcription factors—insights into their structure, regulation, and role in plant development. Frontiers in Plant Science 6, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci T, Ribeiro B, Ceciliato PH, Guerrero-Abad JC, Silva-Filho MC, Moura DS. 2014. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. Journal of Experimental Botany 65, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. The Plant Journal 29, 325–332. [DOI] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, et al. 2011. A stele-enriched gene regulatory network in the Arabidopsis root. Molecular Systems Biology 7, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XT, Xu P, Zhao PX, Liu R, Yu LH, Xiang CB. 2014. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nature Communications 5, 5833. [DOI] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Kuhnle C, Li H, Rosso M, Weisshaar B, Schwechheimer C. 2006. LucTrap vectors are tools to generate luciferase fusions for the quantification of transcript and protein abundance in vivo. Plant Physiology 141, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. The Plant Cell 13, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, et al. 2014. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nature Cell Biology 16, 66–76. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Czyzewicz N, Yue K, Beeckman T, Smet ID. 2013. Message in a bottle: small signalling peptide outputs during growth and development. Journal of Experimental Botany 64, 5281–5296. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Carvajal D, Hollunder J, Van den Cruyce J, Movahedi S, Inzé D. 2010. CORNET: a user-friendly tool for data mining and integration. Plant Physiology 152, 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inzé D. 2012. CORNET 2.0: integrating plant coexpression, protein–protein interactions, regulatory interactions, gene associations and functional annotations. New Phytologist 195, 707–720. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Inzé D. 2013. A guide to CORNET for the construction of coexpression and protein–protein interaction networks. Methods in Molecular Biology 1011, 327–343. [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. a CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. Journal of Experimental Botany 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. b Regulation of Arabidopsis root development by small signaling peptides. Frontiers in Plant Science 4, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJ. 2004. A gateway-compatible yeast one-hybrid system. Genome Research 14, 2093–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, et al. 2010. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology 20, 1697–1706. [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Ehrismann JS, Axiotis I, Kolb M, Kientz M, Weijers D, Jurgens G. 2013. Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. Journal of Experimental Botany 64, 3009–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voβ U, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 107, 2705–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Vanneste S, Inzé D, Beeckman T. 2006. Lateral root initiation or the birth of a new meristem. Plant Molecular Biology 60, 871–887. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, et al. 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105, 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P. 2001. Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214, 30–36. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, Beeckman T, Hilson P. 2015. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. Journal of Experimental Botany 66, 5245–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Hilson P, Beeckman T. 2013. GOLVEN peptides as important regulatory signalling molecules of plant development. Journal of Experimental Botany 64, 5263–5268. [DOI] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC. 2000. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. The Plant Cell 12, 183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE Wang CT Zheng Y Adamczyk BJ Singhal R, Hall PK Perry SE. 2014. The MADS-domain factors AGAMOUS- LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiology 165, 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. The Plant Journal 29, 153–168. [DOI] [PubMed] [Google Scholar]
- Gaudinier A, Zhang L, Reece-Hoyes JS, et al. 2011. Enhanced Y1H assays for Arabidopsis. Nature Methods 8, 1053–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Denervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. 2009. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. The Plant Journal 59, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE. 2003. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiology 133, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Monshausen G, Gilroy S, Sussman MR. 2008. A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47, 6311–6321. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE. 2008. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. The Plant Journal 53, 172–185. [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. 2002. Auxin-mediated cell cycle activation during early lateral root initiation. The Plant Cell 14, 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, et al. 2004. Transcript profiling of early lateral root initiation. Proceedings of the National Academy of Sciences, USA 101, 5146–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG. 2008. Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. The Plant Journal 55, 335–347. [DOI] [PubMed] [Google Scholar]
- Kircher S, Schopfer P. 2016. Priming and positioning of lateral roots in Arabidopsis. An approach for an integrating concept. Journal of Experimental Botany 67, 1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Sto IM, Butenko MA, Peret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proceedings of the National Academy of Sciences, USA 110, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Maree AF, Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biology 6, e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. 1995. Formation of lateral root meristems is a two-stage process. Development 121, 3303–3310. [DOI] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jurgens G. 2011. Auxin triggers a genetic switch. Nature Cell Biology 13, 611–615. [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc’h S, et al. 2015. Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. The Plant Cell 27, 1368–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, et al. 2013. Lateral root development in Arabidopsis: fifty shades of auxin. Trends in Plant Science 18, 450–458. [DOI] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK. 2011. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138, 3485–3495. [DOI] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, et al. 2015. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, et al. 2013. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proceedings of the National Academy of Sciences, USA 110, 5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G. 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. The Plant Cell 14, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Yamoto N, Suzuki Y, Chiba Y, Yamazaki K, Sato T, Yamaguchi J. 2013. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Science 213, 79–87. [DOI] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS. 2008. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Letters 582, 3343–3347. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067. [DOI] [PubMed] [Google Scholar]
- McAdam SA, Brodribb TJ, Ross JJ. 2016. Shoot-derived abscisic acid promotes root growth. Plant, Cell and Environment 39, 652–659. [DOI] [PubMed] [Google Scholar]
- Medina-Rivera A, Defrance M, Sand O, et al. 2015. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Research 43, W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. 2012. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingossi FB, Matos JL, Rizzato AP, Medeiros AH, Falco MC, Silva-Filho MC, Moura DS. 2010. SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Molecular Biology 73, 271–281. [DOI] [PubMed] [Google Scholar]
- Mlodzinska E, Klobus G, Christensen MD, Fuglsang AT. 2015. The plasma membrane H(+)-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiologia Plantarum 154, 270–282. [DOI] [PubMed] [Google Scholar]
- Morato do Canto A, Ceciliato PH, Ribeiro B, Ortiz Morea FA, Franco Garcia AA, Silva-Filho MC, Moura DS. 2014. Biological activity of nine recombinant AtRALF peptides: implications for their perception and function in Arabidopsis. Plant Physiology and Biochemistry 75, 45–54. [DOI] [PubMed] [Google Scholar]
- Murphy E, De Smet I. 2014. Understanding the RALF family: a tale of many species. Trends in Plant Science 19, 664–671. [DOI] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. 2012. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. The Plant Cell 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. 2008. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. The Plant Journal 55, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Sukumar P, Liu X, Cohen JD, Muday GK. 2010. Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. The Plant Journal 61, 3–15. [DOI] [PubMed] [Google Scholar]
- Oeller PW, Theologis A. 1995. Induction kinetics of the nuclear proteins encoded by the early indoleacetic acid-inducible genes, PS-IAA4/5 and PS-IAA6, in pea (Pisum sativum L.). The Plant Journal 7, 37–48. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell 19, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Mundy J, Skriver K. 2002. Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biology 2, 441–451. [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V. 1999. Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. The Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Walker JC. 2015. Floral organ abscission is regulated by a positive feedback loop. Proceedings of the National Academy of Sciences, USA 112, 2906–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA., Jr 2001. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proceedings of the National Academy of Sciences, USA 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, et al. 2012. Auxin regulates aquaporin function to facilitate lateral root emergence. Nature Cell Biology 14, 991–998. [DOI] [PubMed] [Google Scholar]
- Perry SE, Lehti MD, Fernandez DE. 1999. The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiology 120, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. 1998. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiology 118, 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. 2009. Mechanical stimuli modulate lateral root organogenesis. Plant Physiology 151, 1855–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I, Smith S, Stes E, et al. 2016. CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. Journal of Experimental Botany (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Laskowski M. 2016. Root patterning: it takes two to tangle. Journal of Experimental Botany 67, 1201–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Smith C, Bevan MW. 1998. The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Molecular Biology 36, 195–204. [DOI] [PubMed] [Google Scholar]
- Slane D, Kong J, Berendzen KW, et al. 2014. Cell type-specific transcriptome analysis in the early Arabidopsis thaliana embryo. Development 141, 4831–4840. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH. 2009. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. The Plant Journal 59, 930–939. [DOI] [PubMed] [Google Scholar]
- Tang W, Perry SE. 2003. Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. Journal of Biological Chemistry 278, 28154–28159. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. 2004. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. The Plant Cell 16, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. 2015. The plant peptidome: an expanding repertoire of structural features and biological functions. The Plant Cell 27, 2095–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, et al. 2015. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nature Plants 1, 15025. [DOI] [PubMed] [Google Scholar]
- Vanden Bossche R, Demedts B, Vanderhaeghen R, Goossens A. 2013. Transient expression assays in tobacco protoplasts. Methods in Molecular Biology 1011, 227–239. [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Zhang J, Cazzonelli CI, Pogson BJ, Harrison PJ, Bugg TD, Chan KX, Thompson AJ, Benfey PN. 2014. Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proceedings of the National Academy of Sciences, USA 111, E1300–E1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, Geldner N. 2015. Lateral root initiation in Arabidopsis thaliana: a force awakens. F1000Prime Reports 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, Maizel A. 2016. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Current Biology 26, 439–449. [DOI] [PubMed] [Google Scholar]
- Voβ U, Wilson MH, Kenobi K, et al. 2015. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nature Communications 6, 7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Caruso LV, Downie AB, Perry SE. 2004. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. The Plant Cell 16, 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang W, Zhu C, Perry SE. 2002. A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. The Plant Journal 32, 831–843. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, et al. 2012. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Developmental Cell 22, 678–685. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Song Y, Yang H, Liu Z, Zhu G, Yang Y. 2014. An auxin-responsive endogenous peptide regulates root development in Arabidopsis. Journal of Integrative Plant Biology 56, 635–647. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. 2009. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. The Plant Cell 21, 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W. 2005. Gene-expression analysis and network discovery using Genevestigator. Trends in Plant Science 10, 407–409. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.