Abstract
The 2014 i2b2 natural language processing shared task focused on identifying cardiovascular risk factors such as high blood pressure, high cholesterol levels, obesity and smoking status among other factors found in health records of diabetic patients. In addition, the task involved detecting medications, and time information associated with the extracted data. This paper presents the development and evaluation of a natural language processing (NLP) application conceived for this i2b2 shared task. For increased efficiency, the application main components were adapted from two existing NLP tools implemented in the Apache UIMA framework: Textractor (for dictionary-based lookup) and cTAKES (for preprocessing and smoking status detection). The application achieved a final (micro-averaged) F1-measure of 87.5% on the final evaluation test set. Our attempt was mostly based on existing tools adapted with minimal changes and allowed for satisfying performance with limited development efforts.
Graphical abstract
1. Introduction
The 2014 i2b2 (Informatics for Integrating Biology and the Bedside) challenge proposed several different tasks: clinical text de-identification, cardiovascular risk factors identification, software usability assessment, and novel data uses. Our efforts focused on the second track, identifying risk factors for heart disease based on the automated analysis of narrative clinical records of diabetic patients [1]. The annotation guidelines for the task defined eight categories of information associated with increased risk for heart disease: 1) Diabetes, 2) Coronary Artery Disease (CAD), 3) Hyperlipidemia, 4) Hypertension, 5) Obesity, 6) Family history of CAD, 7) Smoking and 8) Medications associated with the aforementioned chronic diseases. Each category of information (except family history of CAD and smoking status) had to be described with indicator and time attributes. The indicator attribute captures indications of the risk factor in the clinical text. For instance, Diabetes could be identified using a mention of the disease (i.e. “patient has h/o DMII”), or a hemoglobin A1c value above 6.5 mg/dL (i.e. “7/18: A1c: 7.3”) while CAD could be identified using a mention (i.e. “PMH: significant for CAD”), or an event (i.e. “CABG in 1999”). The time attribute specifies the temporal relation to the Document Creation Time (DCT). It could take any one of the following values: before DCT, during DCT or after DCT. We refer the reader to [2] for a complete description of the annotation guidelines. For this challenge, we built a natural language processing (NLP) application based on the Apache UIMA (Unstructured Information Management Architecture) [3] and reusing existing tools previously developed to address similar tasks in previous i2b2 challenges. In this paper, we present our approach to extract relevant information from clinical notes, discuss performance results, and conclude with remarks about our experience adapting existing NLP tools.
2. Background
Extracting information from clinical notes has been the focus of a growing body of research these past years [4]. Common characteristics of narrative text used by physicians in electronic health records (e.g., telegraphic style, ambiguous abbreviations) make it difficult to access such information automatically. Natural Language Processing (NLP) techniques are needed to convert information from the unstructured text to a structured form readily processable by computers [5, 6]. This structured information can then be used to extract meaning and enable Clinical Decision Support (CDS) systems that assist healthcare professionals and improve health outcomes [7]. Among the earliest attempts to develop NLP applications in the medical domain, the LSP (Linguistic String Project) [8], and MedLEE (Medical Language Extraction and Encoding system) [9] were prominent examples. More recent applications include MetaMap [10] developed by the National Library of Medicine to map terms in biomedical text with concepts in the UMLS (Unified Medical Language System) Metathesaurus [11]. cTAKES [12] was developed at the Mayo Clinic and is described as “large-scale, comprehensive, modular, extensible, robust, open-source” application based on Apache UIMA. It can be used to preprocess clinical text, find named entities and perform additional advanced NLP tasks such as coreference resolution. Textractor [13] is another UIMA-based application that was originally developed at the University of Utah to extract medications, their attributes, and reasons for their prescription from clinical notes.
When extracting information from clinical notes, NLP applications must take local contextual and temporal information into account for improved accuracy. Contextual information is important to determine if concepts are affirmed or negated (e.g., ‘denies any chest pain’), or if the subject of the information is the patient or someone else (e.g., ‘mother has diabetes’). Popular algorithms for negation detection in clinical notes include NegExpander [14] and NegEx [15]. Temporal information is critical to establish chronological order of events described in patient notes and to resolve mentions of procedures or laboratory results to specific time points for accurate analysis [16, 17]. The ConText algorithm [18] proposed by Chapman et. al. is an extension of NegEx that allows analysis of contextual information like negation (negated, affirmed), temporality (historical, recent, hypothetical), and experiencer (patient, other). The development of NLP applications typically requires significant efforts and relies on annotated clinical text for training and testing. Widely accessible and shared annotated corpora in the medical domain are still rare, mainly because of strict patient privacy rules. This scarcity has been an obstacle to developing state-of-the-art NLP approaches for clinical text [19]. To address this obstacle and enable direct comparison of NLP approaches in the clinical domain, i2b2 shared NLP tasks have been organized almost annually since 2006. The challenges started with an automated de-identification [20] and smoking status detection [21] challenges. In 2008, the i2b2 challenge focused on identifying information about obesity and 15 co-morbidities [22]. In 2009, the third i2b2 challenge [23] was focused on identifying medications and associated information such as dosage and frequency. This was followed by challenges for medical concept extraction, assertion and relations classification in 2010 [24], followed by coreference resolution tasks in 2011 [25] and a temporal relations classification in 2012 [26].
To reduce development efforts, many authors have reused NLP tools or resources such as ConText, sentence boundary detectors and part-of-speech taggers from OpenNLP project [27], the Stanford parser [28], or the Weka machine learning framework [29], but the majority of their applications were still new developments. Reusing larger components or even existing NLP applications could allow for further development effort reduction. A good example was the application developed by Wellner et al. [30] for the 2006 i2b2 de-identification task. It was based on the adaptation of two applications originally designed for recognizing named entities in newswire text. The process involved running two applications out-of-the-box as a baseline and then gradually introducing a few task-specific features, using bias parameters to control feature weights, and adding lists of common English words during development to improve performance. With minimal effort, they were able to obtain very high performance for the task. Although their attempt used applications out-of-the-box as baselines, they had to re-train the models with new task-specific features to achieve high performance. Our attempt focused on adapting existing tools that were developed to solve similar tasks in the past, and do it without feature engineering and re-training of machine learning models.
3. Methods
3.1. Datasets
The i2b2 NLP shared task organizers distributed two annotated datasets (SET1 and SET2) to be used for development and training. These sets were released separately, with a few weeks interval. SET1 was composed of 521 de-identified clinical notes and SET2 was composed of 269 de-identified notes; therefore, a total of 790 documents were available for training. The test set was released three days before final submission and consisted of a total of 514 de-identified clinical notes.
3.2. NLP Application Overview
As already mentioned, our application was based on the Apache UIMA framework, with components adapted from two existing applications. Because of the various nature of information to be extracted in this task, we experimented with different approaches for different categories of information. For example, Textractor’s dictionary-based lookup component was used to detect mentions of chronic diseases, in addition to mentions of CAD events as defined in the annotation guidelines. The results of the lookup module were then filtered using lists of UMLS Metathesaurus concept identifiers CUIs for disease and risk factor concepts defined for the task. Smoking status was identified using the existing classifier available from cTAKES. Medications and the various test results (hemoglobin A1c, glucose, blood pressure, cholesterol, etc) were identified using pattern matching with regular expressions. Family history of CAD was detected by modifying the contextual analysis of the detected CAD mentions using ConText’s ‘experiencer’ analysis.
The application pipeline is depicted in Figure 1 and described below. The analysis of clinical text begins with a preprocessing stage that consists in segmenting the text into sections, splitting it into sentences, tokenizing and assigning part-of-speech tags to the input text with cTAKES. This is followed by running the smoking status classifier from cTAKES “out-of-box” to classify each patient record to a smoking status category: CURRENT, PAST, EVER, NEVER, UNKNOWN. The existing cTAKES SMOKER label was changed to EVER, as defined for this i2b2 task.
Figure 1.
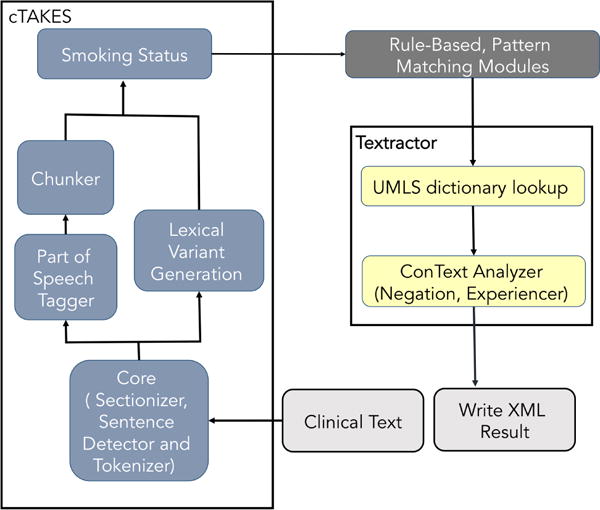
Overview of NLP application pipeline with adapted components from cTAKE and Textractor
The text analysis then continues with rule-based pattern matching modules for detecting medications and laboratory test results. Medications were detected with a manually curated terminology of synonymous terms and abbreviations linked to each medications category. These lists were compiled using UMLS Metathesaurus terminologies and lists of common abbreviations found in clinical narratives (manually built by local domain experts); and then manually grouping the concepts into medication categories. The number of terms used for each medications varied widely, ranging from as few as 3 (e.g. for metformin) to more than 50 (e.g. for beta blockers and aspirin). Laboratory test results and vital signs were detected using regular expressions and the associated values were compared with abnormality thresholds defined in the guidelines. For instance, the phrase “Cholesterol-LDL 08/26/2091 148” indicates an LDL cholesterol concentration of 148 mg/dL, which is above the normal concentration of 100 mg/dL and should therefore be included as a risk factor. Special attention was paid to avoid incorrect values that were part of other numeric expressions (e.g., dates) by restricting regular expression matches to reasonable value ranges and imposing specific conditions on number boundaries (see examples in Table 1). Two regular expressions were used for each relevant laboratory test or vital sign indicator; one for capturing the term and the other for numerical value associated with the laboratory test or vital sign.
Table 1.
Examples of regular expressions used for matching test mentions and values.
| Laboratory/Test | Regular expression for mention | Regular expression for value |
|---|---|---|
| Glucose (for Diabetes mellitus) | (fasting)?(blood)?(glucose|\bGLU (−poc)?\b|\bBG\b|(blood)sugar(s)?| \bFS\b|\bBS\b|fingerstick|\bFG\b) | (?<!/|\d)(\d\d\d?)(−\d\d\d)?(?!/|\d|\w) |
| Blood Pressure (for Hypertension) | (?<!\w)((s)?BP[s]?|b/p|((blood| systolic)[ ]+pressure[s]?)| hypertensive)[:]?(?!\w) | (?<!/|\d)(\d\d\d)/(\d\d\d?)(?!/\d|\d) |
The application then proceeded with the UMLS Metathesaurus lookup module from Textractor. This module uses Apache Lucene-based [31] dictionary indexes to detect disease and risk factor terms. Before the dictionary lookup, acronyms were expanded and tokens normalized by removing unwanted stopwords. The lookup module then matched terms that belonged to one of the predefined UMLS semantic types for diseases (i.e., T019, T033, T046, T047 and T061). Matching was performed at the token level first, and then expanded to match at the noun phrase chunk level. All detected concepts were then filtered based on their CUIs to only include concepts belonging to one of the five disease and risk factor categories identified in the guidelines: CAD, Diabetes mellitus, Obesity, Hyperlipidemia, and Hypertension.
Finally, the application performed contextual analysis of all extracted and filtered information to exclude negated concepts, verify that the patient was the experiencer, and produce time attributes for each concept in relation to the DCT. Negation and experiencer analysis was performed using a local implementation of the ConText algorithm, as available in Textractor. Detection of family history of CAD was handled by considering all extracted CAD concepts with an experiencer other than the patient (e.g., “mother has history of CAD”) as a present family history of CAD. If all CAD concepts were identified as belonging to the patient, or if no CAD concepts were found in the clinical note, then family history of CAD was set to not present.
We experimented with various uses of ConText’s temporal analysis (i.e., concepts classified as recent, historical or hypothetical) in order to map them to the corresponding time attribute values (i.e., before DCT, during DCT or after DCT). However, initial results on the training data using this approach were not satisfying. As an alternative approach, we used the most common time value found for each category of information in the training data. For example, chronic diseases such as CAD and most medications were continuing (i.e., existed before, during, and after the hospital stay or visit) and therefore annotated with all three time attribute values in the reference standard. As another example, laboratory test results varied with examples like hemoglobin A1c and glucose tests that were mostly ‘before DCT’, and others like hypertension that were mostly ‘during DCT’.
4. Results
After development and refinement based on the training corpus (SET1 and SET2), the NLP application processed the testing corpus when made available, and the application output was sent to the shared task organizers for analysis. The application output was compared with the reference standard using the evaluation script provided by the shared task organizers and all extracted information classified as true positive (i.e., output matches with the reference standard), false positive, or false negative. Metrics used included recall, precision, and the F1-measure (details in [1]). The results for each class of information are presented in Table 2. For overall averages, both macro- and micro-averages are included. Each separate class-indicator combination is reported using micro-averages only. The evaluation script contained an option to calculate results separately for each class of information using the –filter option. It also allowed computing specific class and indicator attribute values such as the class DIABETES and indicator attribute value of mention using the option –conjunctive. Results for each disease category are presented for mention and each disease-specific indicators separately as in the annotation guideline. The SMOKING category results are presented as status only, and MEDICATION results are aggregated for all the categories correctly identified in the clinical records. All results in the table were computed for all three values of time attribute for each class and no attempt made to separate ‘before DCT’, ‘during DCT’ and ‘after DCT’ results for each class.
Table 2.
Macro- and micro-averaged overall results including the micro-averaged breakdown of final results for every class of information given in terms of Precision, Recall and F1-measure.
| Indicator | Precision | Recall | F1-measure | |
|---|---|---|---|---|
| CAD | mention | 0.883 | 0.9651 | 0.9222 |
| symptom | 0.2095 | 0.4429 | 0.2844 | |
| event | 0.6457 | 0.5899 | 0.6165 | |
| test | 0.4557 | 0.6102 | 0.5217 | |
| DIABETES | mention | 0.9512 | 0.9887 | 0.9696 |
| A1C | 0.8611 | 0.7561 | 0.8052 | |
| glucose | 0.1486 | 0.3333 | 0.2056 | |
| HYPERLIPIDEMIA | mention | 0.9899 | 0.827 | 0.9011 |
| high cholesterol | 0.5714 | 0.3636 | 0.4444 | |
| high LDL | 0.84 | 0.7241 | 0.7778 | |
| HYPERTENSION | mention | 0.9918 | 0.9891 | 0.9904 |
| high BP | 0.8571 | 0.5231 | 0.6497 | |
| OBESITY | mention | 0.7562 | 1.0 | 0.8612 |
| BMI | 0.9231 | 0.7059 | 0.8 | |
| SMOKING | 0.8638 | 0.8672 | 0.8655 | |
| MEDICATION | 0.8282 | 0.8911 | 0.8585 | |
| FAM. HIST. of CAD | 0.9494 | 0.9494 | 0.9494 | |
| Macro-average | 0.8494 | 0.8914 | 0.8699 | |
| Micro-average | 0.8552 | 0.8951 | 0.8747 | |
As shown in Table 2, the application achieved an overall micro-averaged F1-measure of 87.47% and a macro-averaged F1-measure of 86.99%. In most disease categories, accuracy was highest for mentions of disease with micro-averaged F1-measures of 92.22%, 94.94%, 96.96%, 90.11%, and 99.04% for CAD, family history of CAD, Diabetes, Hyperlipidemia, and Hypertension, respectively. Medications, mentions of Obesity and Smoking status identification accuracy reached micro-averaged F1-measures of 85.85%, 86.12% and 86.55%, respectively. Accuracy was lower with other information categories such as laboratory tests, CAD events and symptoms with F1-measures ranging from 20.56% to %80.
5. Discussion
As presented above, the application accuracy for mentions of the various diseases, smoking status, medications and family history was higher than accuracy for any other indicator type defined in the annotation guidelines (e.g., laboratory tests, CAD events and symptoms). The dictionary lookup approach with terminological content from the UMLS Metathesaurus for detecting disease mentions was successful for this task. Similarly, the smoking status classifier from cTAKES successfully identified and classified smoking status information (F1-measure of about 87%) despite the fact that the model was used out-of-the-box, without any training on the new corpus for the current i2b2 NLP task. The identification of medications and their attributes reached an F1-measure of about 86% when using regular expressions and manually curated lists of terms, demonstrating the feasibility of this approach for the type of narrative notes used in this shared task. The precision obtained for medications was lower (83%) than recall (89%) and hence affected the final F1-measure. This is mainly due to the way we chose to generate the time attribute by using the continuing times scenario (i.e., generating ‘before DCT’, ‘during DCT’ and ‘after DCT’ temporal information tags for every medication detected in the notes). Obviously, there will be false positives associated with this approach when medications strictly occur for either one or two of the time values in the clinical notes. In addition, since the medication term lists were created manually, some spelling variations and terms could have been missed, therefore producing some false negatives and affecting overall recall. An example of spelling variation is the term ‘nitroglycerine’ in the nitrate group category, which appeared in both corpora as ‘nitroglycerin’. The latter was not in the nitrate list used by our application and hence caused some false negatives. An example of completely missed terms was sublingual nitroglycerin mentioned as ‘SL NTG’. Among disease mentions, the Hyperlipidemia class had the lowest recall (83%) and Obesity had the lowest precision (76%). The former was mostly due to some clinical reports containing annotations for Hyperlipidemia mentions appearing as ‘elevated serum cholesterol’, ‘elevated lipids’ and ‘high cholesterol’ cholesterol? that were missed by our application because of inaccurate chunking. In addition, we did not have the corresponding CUI codes for some of them in our dictionary lookup module. There were at least two cases in the testing corpus where Hyperlipidemia was mentioned directly following a word with no space in between such as ‘hemodialysis Hyperlipidemia’ which our application missed also. The low precision with Obesity was caused by including the UMLS concept ‘overweight’ in our list of CUIs for Obesity. Although ‘overweight’ was used as indicator for obesity in one record in the reference standard corpora, its use produced many false positives since ‘overweight’ often does not indicate obesity. There were also false positive mentions of Obesity produced by our application in cases where ‘obese’ was mentioned without indicating Obesity (e.g., “abdomen is slightly obese” and “Abdomen: Moderately obese”). The other indicators for diseases and risk factors were quite challenging and our approach using regular expressions at the lexical level was not always effective. With the exception of hemoglobin A1c laboratory tests (for Diabetes), BMI (for Obesity), and cholesterol LDL (for Hyperlipidemia), the application performance was modest with an F1-measure ranging from 21% for the blood glucose indicator up to 65% for the blood pressure indicator. Some of the challenges with these indicators are summarized below,
Lexical and spelling variations: Some laboratory indicators for diseases are mentioned with many lexical variations and acronyms. Table 1 shows the regular expressions used to capture blood glucose for diabetes and blood pressure for hypertension. As shown, glucose can be described with a variety of terms like BG, BS, FS and FG; and blood pressure can be described with terms like BP and b/p. This is an example of some of the limitations with our approach. and a comprehensive strategy to deal with this issue to enable better accuracy would be needed.
Extracting laboratory numerical results accurately: When the application finds matching terms for laboratory or test indicators, it must proceed with extracting associated numerical values and compare them to threshold levels for abnormality. Extracting numerical values may be straightforward when they immediately follow the term and are expressed as single units such as in the phrase “FSBG was 353”. However, other phrases can be more challenging like “FG 120–199; now 68–172, although 172 = outlier, mostly in the 70–130”. In this case, ranges of values are expressed with ‘–’, and multiple units are expressed with temporal and frequency modifiers (i.e. ‘now’ and ‘mostly’).
Training data sparseness: The number of training examples available was sometimes too low to allow for the variety needed for adequate application generalization. For instance, in the case of cholesterol indicator for Hyperlipidemia, the total number of available annotations was only 9 in the whole set of 790 training documents. In contrast, there were about 33 annotations available for the LDL indicator.
Complex time analysis. Test and laboratory indicators require more sophisticated time attribute analysis and this is another limitation of our approach. Unlike chronic disease mention annotations which were mostly characterized with ‘continuing’ time attribute (i.e. before, during and after DCT), most of the laboratory and vital sign annotations were characterized by a variety of time attribute values. For instance, hemoglobin A1c and glucose tests were usually conducted in a prior visit and hence mostly annotated with ’before DCT’ while blood pressure (BP) was mostly measured during the patient visit and hence had mostly ’during DCT’ time value. To examine the impact of time attributes on performance of our application, we followed the “fixed” evaluation procedure described in [32] and produced results for some indicators after replacing the value of time attribute with ‘before DCT’ in all annotations from our application output and in the testing reference standard (see Table 3). This evaluation considers true positives, false positives and false negatives for each individual annotation while ignoring the time attribute (i.e. application output is not penalized for incorrect time values). As shown in table 3, the performance of our application improved when the time component was ignored in the evaluation (compare with results from Table 2). Our decision to use the most common time attribute values for each of these indicators caused a loss in precision and recall contributing to lower overall F1-measure score.
Table 3.
Results for Medications and some disease indicators after fixing the time attribute to the same value in both application output and testing reference standard.
| Precision | Recall | F1-measure | |
|---|---|---|---|
| Glucose | 0.2568 | 0.6129 | 0.3619 |
| High Cholesterol | 0.7143 | 0.5 | 0.5882 |
| High BP | 0.8908 | 0.5792 | 0.702 |
| MEDICATIONS | 0.8791 | 0.8826 | 0.8808 |
6. Conclusion
Our rapid approach, adapting resources from existing applications for the 2014 i2b2 challenge, allowed for performance similar to other more sophisticated application developed for this task which used additional manual annotations or multiple machine learning classifiers [1]. We think that existing NLP resources should be reused, and most can be adapted and used at least as baseline for future tasks in the clinical domain. Improvements for future attempts shall focus on a comprehensive strategy to tackle spelling errors and variations, acronyms disambiguation, and more refined temporal analysis. Use of standard terminologies, as available in the UMLS Metathesaurus, should be the basis for these clinical information extraction tasks as they already contain well-defined concepts associated with multiple terms. Finally, regular expressions and pattern matching can be useful for extracting information such as name-value pairs from short phrases (e.g. ‘Cholesterol- LDL 08/26/2091 148’). However, longer phrases containing complex syntactic structures require the use of advanced parsing techniques to identify constituents and relations between them. In the future, we plan to explore advanced techniques such as dependency parsing or semantic role labeling to reduce errors appearing with long phrases requiring deeper contextual analysis to be accurately extracted. For instance, in the following sentence: “Prior to her bypass surgery on the right leg, she underwent a Persantine MIBI which showed only 1 mm ST depressions and was considered not diagnostic”; it is important for an application to link the negated phrase “was considered not diagotstic” with the noun phrase “Persantine MIBI” to conclude that although the patient had the MIBI test performed, the result was not diagnostic and therefore the test indicator (i.e. ‘MIBI’) ruled out CAD.
Highlights.
We used natural language processing (NLP) to extract heart disease risk factors
Components were adapted from two existing NLP applications
We used existing tools without feature engineering or re-training of models
Our system achieved an overall micro-averaged F1-measure of 87.47%
Adapting existing tools allowed for performance comparable to sophisticated systems
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Abdulrahman Khalifa, Email: abdulrahman.aal@utah.edu.
Stéphane Meystre, Email: stephane.meystre@utah.edu.
References
- 1.Stubbs A, Kotfila C, Xu H, Uzuner Ö. Practical Applications for NLP in Clinical Research: the 2014 i2b2/UTHealth Shared Tasks. Proceedings of the i2b2 2014 Shared Task and Workshop Challenges in Natural Language Processing for Clinical Data. 2015 (in press) [Google Scholar]
- 2.Stubbs A, Uzuner Ö. Annotating Risk Factors for Heart Disease in Clinical Narratives for Diabetic Patients. Proceedings of the i2b2 2014 Shared Task and Workshop Challenges in Natural Language Processing for Clinical Data. 2015 doi: 10.1016/j.jbi.2015.05.009. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrucci D, Lally A. Uima: an architectural approach to unstructured information processing in the corporate research environment. Natural Language Engineering. 2004;10(3–4):327–348. [Google Scholar]
- 4.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;35:128–44. [PubMed] [Google Scholar]
- 5.Pratt A. Medicine computers and linguistics. Biomed Eng. 1973:87–140. [Google Scholar]
- 6.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. Journal of the American Medical Informatics Association. 2011;18(5):544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? Journal of biomedical informatics. 2009;42(5):760–772. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi E, Lyman M, Sager N, Friedman C, Macleod C. A database of computer-structured narrative: methods of computing complex relations, in: Proceedings of the Annual Symposium on Computer Application in Medical Care. American Medical Informatics Association. 1985:221. [Google Scholar]
- 9.Friedman C, Johnson SB, Forman B, Starren J. Architectural requirements for a multipurpose natural language processor in the clinical environment. Proceedings of the Annual Symposium on Computer Application in Medical Care, American Medical Informatics Association. 1995:347. [PMC free article] [PubMed] [Google Scholar]
- 10.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the AMIA Symposium, American Medical Informatics Association. 2001:17. [PMC free article] [PubMed] [Google Scholar]
- 11.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meystre SM, Thibault J, Shen S, Hurdle JF, South BR. Textractor: a hybrid system for medications and reason for their prescription extraction from clinical text documents. Journal of the American Medical Informatics Association. 2010;17(5):559–562. doi: 10.1136/jamia.2010.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aronow DB, Fangfang F, Croft WB. Ad hoc classification of radiology reports. Journal of the American Medical Informatics Association. 1999;6(5):393–411. doi: 10.1136/jamia.1999.0060393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Melton GB, Parsons S, Hripcsak G. A temporal constraint structure for extracting temporal information from clinical narrative. Journal of biomedical informatics. 2006;39(4):424–439. doi: 10.1016/j.jbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Bramsen P, Deshpande P, Lee YK, Barzilay R. Finding temporal order in discharge summaries. AMIA annual symposium proceedings, Vol. 2006, American Medical Informatics Association. 2006:81. [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman WW, Chu D, Dowling JN. ConText: An algorithm for identifying contextual features from clinical text. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing, Association for Computational Linguistics. 2007:81–88. [Google Scholar]
- 19.Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. Journal of the American Medical Informatics Association. 2011;18(5):540–543. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. Journal of the American Medical Informatics Association. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. Journal of the American Medical Informatics Association. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzuner O. Second i2b2 workshop on natural language processing challenges for clinical records. AMIA… Annual Symposium proceedings/AMIA Symposium, AMIA Symposium. 2007:1252–1253. [PubMed] [Google Scholar]
- 23.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010;17(5):514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. Journal of the American Medical Informatics Association. 2012 doi: 10.1136/amiajnl-2011-000784. amiajnl–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. Journal of the American Medical Informatics Association. 2013 doi: 10.1136/amiajnl-2013-001628. amiajnl–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton T, Kottmann J, Baldridge J, Bierner G. Opennlp: A java-based nlp toolkit. 2005 [Google Scholar]
- 28.De Marneffe MC, MacCartney B, Manning CD, et al. Generating typed dependency parses from phrase structure parses. Proceedings of LREC. 2006;6:449–454. [Google Scholar]
- 29.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The weka data mining software: an update. ACM SIGKDD explorations newsletter. 2009;11(1):10–18. [Google Scholar]
- 30.Wellner B, Huyck M, Mardis S, Aberdeen J, Morgan A, Peshkin L, Yeh A, Hitzeman J, Hirschman L. Rapidly retargetable approaches to de-identification in medical records. Journal of the American Medical Informatics Association. 2007;14(5):564–573. doi: 10.1197/jamia.M2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bialecki A, Muir R, Ingersoll G. Apache lucene 4. SIGIR 2012 workshop on open source information retrieval. 2012:17–24. [Google Scholar]
- 32.Grouin C, Moriceau V, Zweigenbaum P. Combining glass box and black box evaluations in the identification of heart disease risk factors and their temporal relations from clinical records. Journal of biomedical informatics. doi: 10.1016/j.jbi.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


