Abstract
Liquid chromatography-mass spectrometry (LC-MS)-based metabolomics studies require highly selective and efficient chromatographic techniques. Typically employed reversed-phase (RP) methods fail to target polar metabolites but the introduction of hydrophilic interaction liquid chromatography (HILIC) is slow due to perceived issues of reproducibility and ruggedness and a limited understanding of the complex retention mechanisms.
In this study, we present a comparison of the chromatographic performance of a traditional RP-C18 column with zwitterionic, amide-, alkyl diol- and aminoalkyl-based HILIC and mixed-mode columns. Our metabolite library represents one of the largest analyte sets available and consists of 764 authentic metabolite standards including amino acids, nucleotides, sugars and other metabolites representing all major biological pathways and commonly observed exogenous metabolites (drugs).
Coverage, retention patterns and selectivity of the individual methods are highly diverse even between conceptually related HILIC methods. Furthermore, we show that HILIC sorbents having highly orthogonal selectivity and specificity enhance the coverage of major metabolite groups in (semi-) targeted applications compared to RP. Finally, we discuss issues encountered in the analysis of biological samples based on results obtained with human plasma extracts. Our results demonstrate that fast and highly reproducible separations on zwitterionic columns are feasible but knowledge of analyte properties is essential to avoid chromatographic bias and exclusion of key analytes in metabolomics studies.
Keywords: High-performance liquid chromatography, hydrophilic interaction liquid chromatography, reversed phase, mass spectrometry, metabolomics, selectivity
Graphical Abstract
1. Introduction
Metabolomics investigates the low-molecular weight intermediates and products of biological processes in tissue, biofluids or cell culture extracts (1–3). It is a relatively recent addition to the bioanalytical “omics” fields located downstream of the more established genomics, transcriptomics and proteomics. Common challenges arise not only from the complexity and large dynamic range of metabolites in biological samples but also from the chemical diversity of the metabolome (4).
Liquid chromatography coupled to mass spectrometry (LC-MS) dominates the metabolomics field owing to the superior detection limits and large dynamic ranges of modern MS techniques. Inadequate or poorly adjusted chromatographic performance, on the other hand, is currently believed to be responsible for significant limitations of metabolomics studies ranging from insufficient metabolome coverage to poor reproducibility (5, 6). For example, widely used reversed-phase (RP) sorbents in superficially porous particle format are efficient chromatographically but they fail to effectively retain many polar compounds. Many essential metabolites such as carbohydrates or amino acids (AAs) have polar-ionic characteristics and a derivatization step is necessary to render them hydrophobic enough for RP separation. Derivatization is typically limited to specific functional groups such as amino or hydroxyl, and can therefore only address a fraction of the total polar metabolome (7–9).
Another approach to increase coverage of the polar metabolome is the introduction of hydrophilic interaction liquid chromatography (HILIC). HILIC utilizes highly polar stationary phases (SPs) capable of forming a water-rich layer on their surface when operated under hydro-organic mobile phase (MP) conditions. Hydrophilic solutes are retained based on their partitioning between the water-poor bulk MP and the water-rich layer on the SP (10, 11). Polar and ionic interactions, hydrogen bonding and even dispersive interactions can also contribute to retention in HILIC (12). Many different HILIC-compatible sorbents exist, among them bare silica as well as polymers or silica gel bearing charged or “neutral” modifications (13–15). Different modifications can give rise to different sorbent-solute interactions and, consequently, different chromatographic results. Hence, there is no unified retention model for HILIC separations. Furthermore, seemingly small changes in mobile phase composition can have a larger impact on HILIC separations than on RP, which has led to a widespread perception of HILIC as being less “rugged” than RP (12).
Recent efforts to elucidate HILIC retention mechanisms, characterize columns and predict separation performance include the application of quantitative structure-retention relationships (QSRRs) and linear solvation energy relationship (LSER) calculations (16, 17). LSER approaches utilize solute properties to establish system constants and characterize HILIC columns, for example by relating retention to hydrophobicity, charge state and H bond donor/acceptor properties (18–20). However, such models have limited predictive power and are often impractical or inaccessible outside of dedicated chromatography research laboratories. HILIC method development can become labor-intensive and time-consuming, especially to inexperienced chromatography users. Consequently, there is a need to provide more information on the applicability of HILIC methods for metabolomics studies in order to utilize the optimization potential residing in the chromatographic separation.
In this work, we discuss chromatographic issues relevant to metabolomics study design and address some of the limitations attributed to HILIC when compared to RP. Selectivity differences between RP and HILIC columns as well as among HILIC columns are assessed using straightforward strategies appropriate for the early stages of method development.
To this end, we determined the chromatographic parameters of a library of 764 commercially available endo- and exogenous metabolite standards. RP and neutral (diol, amide), zwitterionic (sulfobetaine, phosphorylcholine) and weak anion exchanger (WAX)-type aminoalkyl HILIC columns were evaluated for their retention and separation capabilities towards a highly diverse set of representative metabolites. We investigated the total library coverage and retention distributions of the individual columns. These quantitative performance parameters are important in untargeted metabolomics, where large numbers of unknown “features” (m/z signals at a given retention time) are detected. Furthermore, we assessed the selectivity and specificity of the individual columns towards selected sub-groups of the metabolite library. The respective sections are particularly relevant for targeted applications. Finally, we determined applicability of HILIC in metabolomics analysis of human plasma extracts (HPE). These studies lay the groundwork for incorporation of HILIC in standard metabolomics analysis of plasma.
2. Materials and Methods
2.1 Reagents and chemicals, library of standards
LC-MS grade water, methanol, acetone and acetonitrile were purchased from Fluka via Sigma-Aldrich (St. Louis, MO). Mobile phase additives ammonium formate, ammonium acetate and formic acid (99.9% or LC-MS purity) were also from Fluka or Sigma-Aldrich.
The 764 metabolite library standards (Fig. 1, full list: ESM II) were of analytical or higher grade and were obtained from Fluka/Sigma Aldrich, Acros Organics (Fisher Scientific, Pittsburgh, PA), Alfa Aesar (Ward Hill, MA), Avanti Polar Lipids, Inc. (Alabaster, AL), Cayman Chemical (Ann Arbor, MI), Chem-Impex International, Inc. (Wood Dale, IL), MP Biomedicals (Santa Ana, CA), Santa Cruz Biotechnology (Dallas, TX), Supelco (Bellefonte, PA), TCI America (Cambridge, MA), Toronto Research Chemicals (Toronto, Canada) and other providers. Equal volumes of ca. 1mg/mL stock solutions were combined to yield 20 library mixtures containing about 40 non-isobaric compounds each. The library solutions were diluted in MeOH/H2O (50:50, v/v) for RP measurements and in ACN/H2O (80:20) for HILIC injections. Injection volume was 3 μL.
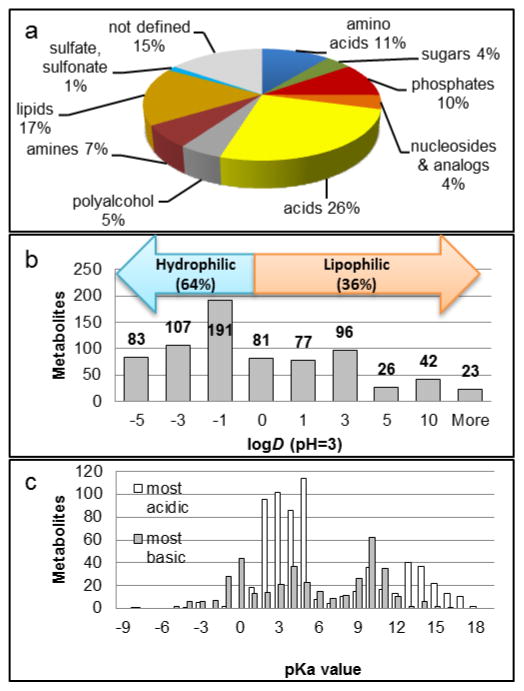
Fig. 1. Properties of metabolites in the analyte set.
a: Sub-groups of metabolites. Percentage based on total library (764 compounds). b: Octanol-water partition coefficients logD of 726 ionizable compounds at pH=3. (Missing values: Data not available.) c: Acid dissociation constants pKa for 652 acidic and 381 basic moieties. Note: Some metabolites contain >1 ionizable group.
2.2 Biological samples (human plasma)
Pooled human plasma was obtained from Innovative Research (Novi, MI). The frozen sample was thawed for 1 h on ice, vortexed briefly and separated into aliquots of 100 μL. Metabolite extraction was initiated by addition of 400 μL of methanol/acetone/acetonitrile (1:1:1 v/v/v). After 1 min of vortexing, the samples were incubated at 4 °C for 30 min. The samples were centrifuged at 14,000 rpm at 4 °C and the supernatant including the lipid layer was collected, transferred to microtubes and dried under N2. The metabolites were reconstituted in 50 μL ACN/H2O (80:20 v/v), vortexed briefly and transferred to autosampler vials. Spiked plasma extracts for the estimation of detection limits were prepared by adding 1, 2 or 5 μL of standard mixtures containing known concentrations (millimolar range) of metabolites to plasma extract before drying. 60 metabolites were selected based on their known or expected presence in blood according to data from the human metabolome database (21) (http://hmdb.ca, accessed November 2015) and each mixture contained 10 different, non-isobaric compounds.
2.3 HPLC instrumentation, columns and chromatographic conditions
All measurements were conducted using an Agilent 1200 HPLC system that included a binary pump, degasser, thermostated autosampler and column compartment coupled to an Agilent 6520 quadrupole-time of flight (QTOF) hybrid mass spectrometer (Agilent Technologies, Foster City, CA) via a dual electrospray ionization (ESI) source. Purine and HP-921 (Agilent Technologies) were continually infused as reference masses. MS detection was carried out in positive ion mode except for the high pH method on column B (Table S1, see Electronic Supplementary Material, ESM I). ESI evaporation/ionization settings were adjusted for different column dimensions and LC flow rates. For detailed information on LC gradient conditions and ESI settings the reader is referred to Table S1 (see ESM I). Table S1 provides details on the columns and chromatographic conditions used in this study.
Theoretical void times t0 were calculated from column length Lc and mobile phase velocity u according to Equations 1 and 2. F represents the mobile phase flow rate, dc the column diameter and εtot the total porosity of the packing material (estimated here to be 0.8).
| Equation 1 |
| Equation 2 |
Void times calculated using Equations 1 and 2 were found to be in excellent agreement with the experimental elution times of unretained compounds as determined from the respective extracted ion chromatograms. Retention factors k were calculated from the analyte retention times tR and theoretical void times t0.
2.4 Software and online databases
The LC-MS system was controlled with ChemStation software (Agilent Technologies). Data evaluation was performed using the “Find by formula” feature in Agilent MassHunter Qual, version 7.00, MassHunter Quant, version 7.00, and Microsoft Excel 2010. Predicted logP, logD and pK values for reference compounds were retrieved from the CAS repository, calculated by Advanced Chemistry Development (ACD/Labs, Software V11.02). Metabolite plasma levels were retrieved from the HMDB (accessed November 2015) (21). The HMDB was also consulted for assignment of metabolites to metabolite classes in case of ambiguity (see 2.1).
3. Results and Discussion
Different characteristics of the chromatographic separation may become critical in targeted or untargeted metabolomics (3, 22, 23). For example, untargeted metabolomics requires high peak capacities, near-ideal peakshapes and excellent retention time stability in order to facilitate reliable MS detection of a large number of features and the alignment of peaks across samples and groups. Both approaches are affected by the inherent bias of chromatographic separations which can cause exclusion, poor retention or incomplete separation of critical analytes. Consequences include increased time demand for data analysis or irreproducible and unreliable results diminishing the impact of bioanalytical studies. It is therefore essential to select appropriate chromatographic sorbents for each metabolomics study. Currently, a common approach in targeted metabolomics is “RP for non-polar and HILIC for the rest”, which fails to take into account that HILIC columns are heterogeneous and their separation performance is less predictable than that of RP materials (6). The complex, multi-increment retention processes occurring in HILIC render this technique flexible but also more challenging in terms of method development and selectivity prediction (18–20, 24, 25)..
The purpose of this study was to reduce the gap between mechanistic studies of HILIC systems and the metabolomics applications of LC. To this end, we investigated the performance of six mixed-mode (MM) and HILIC methods towards an in-house library of 764 authentic endogenous and exogenous metabolite standards (Fig. 1). LC parameters including retention factors, peak distribution and peak shapes are compared to a typical RP-C18 separation. Selectivity ranges and complementarity (“orthogonality”) of HILIC and RP separations as well as the performance of tested columns towards specific metabolite groups is discussed.
A full list of the retention times and retention factors obtained for the library compounds with all 7 methods is available in ESM II.
3.1 LC columns and metabolite library
For our study, 6 mixed-mode, ionic and neutral HILIC columns (B–G, Table S1, see ESM I) were compared to an RP-C-18 column frequently employed in bioanalytical studies (A). With the exception of B (polymer-based), all materials are silica-based. For details on the individual sorbents, the reader is referred to the Supporting Information (ESM I, Table S1) and materials provided by column manufacturers.
The following relationships between methods A–G are important for the discussion of results:
Methods A (also referred to as “RP-C18”) and C (“MM-RP”) both have RP character.
C (MM-RP) and D (“MM-HILIC”) are carried out on the same (diol) column with mobile phases promoting RP-type (C) and HILIC-type (D) interactions, respectively.
B (sulfobetaine, “pHILIC”) and E (phosphorylcholine “cHILIC”) are zwitterionic (“neutral”) HILIC methods.
D (diol) and F (amide) are “neutral” HILIC methods.
E and G (“TSK-NH2”, alkylamine) have weak anion exchanger characteristics due to a terminal amino group.
G (amide) and F (amine) target similar analytes but have different hydrogen bond donor/acceptor properties.
In order to identify trends for metabolite groups, the library compounds were divided into 10 metabolite classes based on their chemical properties (Fig. 1a). 20 mixtures containing ca. 40 non-isobaric metabolites each were created so that every metabolite group was represented in every mixture.
As shown in Fig. 1b, 2/3 of the reference compounds are polar-hydrophilic with octanol-water distribution coefficients logD<1. 89% carry acidic or basic moieties as indicated by their acid dissociation constants pKa (Fig. 1c). Lipids and related compounds such as steroids make up 17% of the library, This class of largely hydrophobic compounds is the subject of “lipidomics” and will not be discussed in detail in this contribution (27–29).
The major part of the library – as well as a substantial fraction of the metabolome – is comprised of compounds that are appropriate targets for hydrophilic interaction LC. HILIC materials retain polar analytes by promoting their partition from a water-poor bulk mobile phase into a water-rich layer on the stationary phase surface. However, depending on the type of surface modification, hydrogen bonding and electrostatic interactions between polar and ionic groups can become dominant, alter elution orders and produce issues such as peak-tailing (12, 24). Our observations are described in the following sections.
3.2 Library coverage and retention profiles of HILIC methods
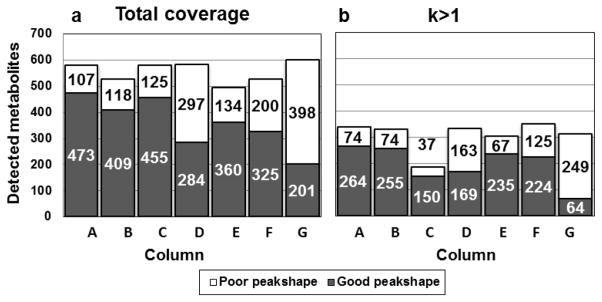
The first characteristic assessed in this study was the library coverage, i.e. the percentage of library metabolites that produced an identifiable MS peak after separation with methods A–G., The benchmark RP method A facilitated the detection of 75% of the metabolite standards contained in the library regardless of peakshape (column height in Fig. 2a). Methods B–G afforded an average total coverage of 71 ± 5.4% with the highest total number of metabolites being detected by method G (77%).
Fig. 2. Library coverage of RP column A and mixed-mode and HILIC columns B–G.
a: Chromatographic peak-shape for all 764 compounds. b: Well-retained compounds with retention factors k ≥ 1.
We frequently observed split, excessively broad, double or multiple peaks and tailing or fronting phenomena which were characterized as “poor peakshape” in the chromatographic evaluation (white segments, Fig. 2. See also ESM II: SI2_Metabolite_librarydate.xlsx for definitions of poor peak shapes). Non-ideal peaks are common in HILIC due to the complex retention mechanisms. Most importantly, strong electrostatic interactions can lead to broad or tailing peaks (26, 30). Similarly slow mass exchange kinetics can result in asymmetric peaks at high flow rates (24, 30). Poor peakshapes may also originate from unsuitable injection solvent having high elution strength. The 80:20 (v/v) ACN/H2O mixture used in this study is believed to minimize these effects (note that methods A and C utilize a H2O/MeOH mixture). Poor peakshapes are particularly undesirable in untargeted studies because they resist automated peak alignment, integration and MSn analysis and significantly increase the time required for data analysis. Metabolites producing “good”, ideally Gaussian peakshapes on the individual columns is shown as dark segments in Fig. 2. RP method A produced good peaks for 65 % of the library while the individual HILIC methods covered between 29 % (G) and 59 % (B). Chemical properties are not evenly distributed among the library compounds (Fig. 1b). Consequently, there are major discrepancies between total coverage and coverage of specific subgroups by the individual methods (see 3.4).
For 50 compounds, among them lipophilic bile acids and fatty acid esters and easily degradable small organic acids, none of the methods tested provided acceptable peakshapes and the respective compounds were excluded from the following evaluation.. The following results are based on a total of 714 detectable library compounds.
In addition to high coverage, a uniform distribution of peaks across the entire chromatogram is desirable in untargeted metabolomics. Optimal use of the available separation space minimizes ionization issues originating from co-eluting and overlapping peaks.
In order to account for differences in column diameter, particle size and flow rate of methods A–G. retention times were transformed into retention factors k. Fig. S1 (see ESM I) displays retention profiles of the tested columns based on the retention factor distributions of the metabolites detected with each method. In addition to many poorly retained metabolites (see below), method A shows extremely strong retention (k>10) of 15 % of the library. The strongly retained fraction is largely identical with the metabolite subgroup “lipids and related compounds” and increases analysis time significantly, which can become a disadvantage in large studies.
The distribution of analyte peaks across the separation space appears more uniform in HILIC, especially on the neutral amide column F and the zwitterionic columns B and E compared to RP-type methods A and C. In part, this can be attributed to the absence of strong ionic or hydrophobic interactions which would promote irreversible adsorption. Importantly, the HILIC columns exhibiting favorable, balanced retention profiles also show the highest “good” coverage, which makes them particularly attractive for untargeted separations (Fig. 2b).
44% and 67% of the compounds producing “good peaks” were poorly retained with k<1 on columns A and C. The respective numbers for HILIC methods were 38 (B), 41 (D), 35 (E) 31 (F) and 69 % (G). Poorly retained solutes interact with the stationary phase in an unspecific, often irreproducible way due to the absence of suitable interaction sites. This makes them difficult to target during method optimization. Co-eluting, unretained compounds cause ionization suppression in the ESI source in complex biological samples. Shifting retention times and “loss” of peaks may be observed in crowded chromatogram areas For realistic representation of the chromatographic performance, we further adjusted the “successfully detected” metabolites to include only well-retained compounds with k>1. Fig. 2b depicts the library coverage of methods A–G. Successfully addressed metabolites, characterized by good retention and good peak-shape, are represented by dark segments. Most HILIC methods and RP-C18 show similar recognition potential. White segments show the fraction of metabolites with good retention but poor peak-shapes. This group has significant optimization potential.
It was observed that the chromatographic results of each of the 20 metabolite mixtures reflected the total library coverage closely. This shows that while a large metabolite library is beneficial for metabolite identity confirmation, meaningful column tests can be carried out with a small set of representative compounds, greatly reducing the time required for method development.
3.3 Selectivity and orthogonality of RP and HILIC methods
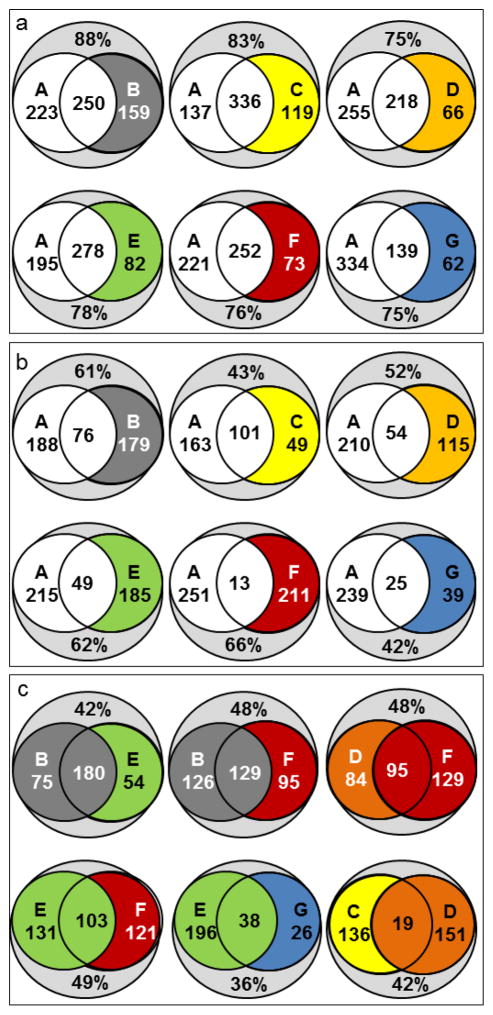
Having access to a large compound library provides an opportunity to test the presumption of orthogonality of RP and HILIC methods and elucidate the selectivity differences between different HILIC columns on a broad scale. Fig. 3 illustrates that the library coverages of methods B–G overlap with that of RP-C18 method A. The top panel (Fig. 3a) displays the data for all metabolites detected with good peakshape, the middle panel (Fig. 3b) only for those with k>1 on the respective column. As discussed in 3.1, method A captures 65% of the analyte set and methods B–G capture 28–63 %. Combining method A with any of the methods B–G affords total coverages of 75–85% of the library. 139 (method G) to 336 (method C) metabolites or 20–45% of the compounds are detected by both methods (redundancy). The combined library coverage for metabolites with k>1 was determined to be between 42% and 66% (Fig. 3b). Only 37% of the library produce k>1 on RP-C18 method A. The lowest redundancy was observed at 2% between methods A and F. Combining method A with any HILIC method yielded total library coverages of up to 66% (A & F). Fig. 3c shows the redundancy and increase in coverages for metabolites with k>1 on zwitterionic B and E (top left), neutral diol-HILIC D with amide F (top right), B and E with neutral F (top center, bottom left), zwitterionic E with WAX-type G (bottom center) and mixed-mode methods C and D. The combined coverages of 36–49 % tend to be lower than those of HILIC methods with RP method A due to the closer relationship between the HILIC columns. Nonetheless, there is considerable diversity in the metabolites targeted by the individual HILIC methods. Even conceptually related zwitterionic columns B and E, whose capture 75 and 50 unique metabolites, respectively. For neutral HILIC methods D and G, the respective numbers are 74 and 129. The same column operated under RP (C) and HILIC (D) conditions captures only 19 metabolites in both modes but ca. 140 uniquely with each method. These comparisons illustrate that HILIC methods, although commonly considered similar, are not necessarily interchangeable. Furthermore, the choice of column can introduce significant methodological bias. In multi-dimensional separations the largest potential gain in information can be obtained by coupling highly orthogonal methods. To estimate the degree of orthogonality, the logarithmic retention factors logk of metabolites on two columns are plotted against each other (Fig. S2, see ESM I). With methods A and B, many metabolites show poor retention on one and strong retention on the other column, resulting in a deviation from the diagonal indicative of high orthogonality. In contrast, methods B and E exhibit similar selectivity with detected metabolites clustering near the diagonal. Obviously, the opposite orientation of ionic interaction sites has only a limited influence on the selectivity of the zwitterionic columns.. Zwitterionic and neutral HILIC materials B and F, on the other hand, give rise to different selectivity profiles due to their different capabilities of engaging in electrostatic and H bond interactions with solutes. The comparison between F (amide-modified, an H-bond acceptor) and G (amino-modified, an H-bond donor) corroborates this notion. As expected, the two RP-type methods A and C display similar selectivity. Hence, their combination would result in a limited gain in information. High complementarity is observed between methods A G (RP vs. WAX-type HILIC), between A and F (RP vs. amide), between HILIC methods B and F (zwitterionic vs. neutral) and between methods E and G (phosphorylcholine vs. amino).
Fig. 3. Coverage comparison of RP-C18, mixed-mode and HILIC methods.
Numbers inside circles: Metabolites exclusively detected with each method. Center: compounds detected with both. Percentages relative to detectable compounds in library (714). a: Metabolites with good peakshapes. b: Well-retained compounds with good peakshapes and retention factors k ≥ 1. c: Overlapping selectivity ranges for zwitterionic HILIC methods (B vs. E), zwitterionic and neutral SPs (E and B vs. F), SPs with terminal amino groups (E vs. G), neutral HILIC (D vs. F) and for a mixed-mode column run in RP and HILIC modes (C vs. D).
3.4 HILIC specificity: Targeting individual metabolite groups
Assessing the recognition potential of individual HILIC methods towards defined metabolite groups or pairs provides important starting information for the development of targeted assays. As shown in Fig. 1a, the main polar groups in the library included acids, carbohydrates, amino acids, and phosphates. Selected aspects of their chromatographic behavior are discussed in detail below. The metabolite groups are color-coded in the supplementary data file (ESM II).
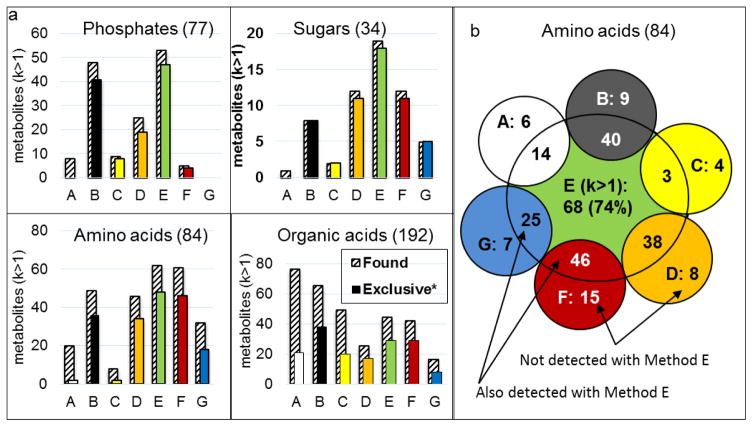
Fig. 4a depicts the coverage of methods A–G for phosphates, sugars, amino acids and organic acids as shaded bars. Colored bars illustrate the superior recognition of the respective metabolite subgroup with HILIC methods B–G compared RP-C18 method A. Except for organic acids, all subsets are more successfully targeted by HILIC methods. Fig. S3 (see ESM I) depicts the coverage of methods A–G for all metabolite subgroups. It is evident that selectivity varies significantly between RP and HILIC but, more importantly, also between the HILIC methods. For example, the zwitterionic HILIC method E successfully retains a large portion of the amino acid subgroup ((Fig. 4b), presumably through double ion pairing with the phosphorylcholine moiety. Despite the high coverage achieved with method E, each one of the other methods captured metabolites not detected with method E. Similar observations were made for carbohydrates (not shown).
Fig. 4. Coverage of metabolite groups.
Shaded bars: retention factor>1, good peak-shape, colored bars: number of metabolites not captured by RP-C18 method A. For method A, “exclusive” compounds are not detected by B–F. b. Complementarity of phosphorylcholine-based column E and other HILIC columns for amino acids subgroup.
Sugars/carbohydrates are difficult to address chromatographically due to the presence of multiple highly polar hydroxyl groups. As expected, underivatized sugars were not successfully targeted by RP methods (Fig. 4a). Only one of the 34 carbohydrates and hexose amines in the library was well retained on RP-C18 column A and only two on MM-RP column C. In contrast, all sugars were captured by at least one HILIC method with the exception of arabinose. Individual HILIC columns retained up to 90% of the carbohydrates but we frequently observed multiple peaks. Anomerization of reducing carbohydrates occurs on shape-selective HILIC sorbents with strong H bond contributions to retention. Low selectivity was observed for isobaric sugars such as hexoses and slightly higher selectivity for homologues (pent- vs. hexoses). Column E retained carbohydrates more strongly than column B and distributed peaks more evenly. With this method, 19 out of 34 carbohydrates (58% of the library) produced good peakshapes. The reason for this is likely found in the terminal amino group of sorbent E which addresses the sugar hydroxyl moieties more favorably than the negatively charged terminal sulfonate of sorbent B Column G covered only 21% of the sugar subset despite its terminal amino groups, with 15% showing good retention and peakshape. It is conceivable that the mobile phase lacked the elution strength to interrupt the electrostatic interactions that are likely to occur between the partially charged hydroxyl and amino groups. In extreme cases, Schiff base formation of the terminal amines with reducing sugars may lead to permanent retention (15, 18, 24).
The neutral HILIC methods F and D produced sharp peaks for 35% of the carbohydrate library. Sugars eluted early from diol-modified column D which has H bond donor properties similar to those of sugars. Column F acts primarily as an H bond acceptor. Consequently, it retained sugars well and showed favorable peak distribution. For both neutral HILIC methods, sugar retention seemed to be strongly influenced by hydrophilic partitioning (relatively high correlation between logk and logD, data not shown).
In addition to the 23 proteinogenic (20 DNA-encoded) α-amino acids (AAs), the library contained 61 β-AAs, unusual AAs and small peptides whose common feature is the presence of chargeable amino and carboxylate groups. Highly polar functionalities, an amphoteric nature and the pH-dependent charge state make them challenging targets for RP separations.
Our study showed that native AAs are most successfully targeted by neutral or zwitterionic HILIC methods. For example, compared to only 25% of AAs covered method A under acidic conditions, method E successfully retained 75% at near-neutral pH where the zwitterionic form is prevalent. Combining methods E and F afforded almost complete coverage (99%) of the entire AA library. With method B, AA coverage was reduced to 56%. The high pH of 9.2 utilized in method B disrupts attraction of the amino terminus by the terminal sulfonate and results in loss of retention compared to method E’s neutral pH.
Despite the lack of long-range ionic interactions, the neutral amide column F covered 72% of the AA library. Its retention profile showed two clusters of weakly (k≤3) and strongly (k≥6) retained AAs. Interestingly, the early eluting compounds included the lipophilic thyroid hormones thyroxine (T4), triiodothyronine (T3) and their precursor diiodotyrosine, all of which were well retained on RP-C18 column A and belonged to the few very strongly retained AAs on MM-RP column C. The latter captured the overall lowest number of amino acids (less than 10%). Overall, amino acids are more successfully addressed by stationary phases having H bond acceptor properties (F) than by H bond donors such as C/D.95% of amino acids showed moderate to strong retention on column G but most of them were detected as split or broad peaks. That only 38 % produced symmetric, narrow peaks is due to the simultaneous presence of the zwitterionic, the positively and the negatively charged form and their differential affinity by the stationary phase. Modifying the mobile phase pH and additive type would afford better control of electrostatic interactions, especially repulsion between the amino groups of sorbent and analytes.
The nucleoside subgroup contained 31 nucleobases and derivatives, including the purines adenine (A) and guanine (G) and the pyrimidines cytosine (C), thymine (T) and uracil (U), as well as their ribo- and deoxyribonucleosides. A, T and U are capable of forming three H bonds whereas G and C can form a maximum of two. U, G and T have relatively moderate basicity (pKa values of ca. 9.5) and their neutral and basic forms can coexist at neutral or slightly acidic pH(24).
In our study, 90% of the nucleoside library were covered by RP method A but only 45% showed k>1 and acceptable peakshapes. In contrast, the zwitterionic sorbent B retained more than 70% and produced an advantageous distribution of nucleoside peaks. Interestingly, the second zwitterionic method E covered only 31 % of the library. In addition, overall retention was reduced, which could be due to either the different packing materials (B: polymer, E: silica) or the absence of a terminal acid moiety on the surface of E.
On amide column F, we observed inconsistent elution sequences of the nucleobases (U<T≪G~A<C) compared to the respective nucleosides (U<A<C, G not detected) and deoxynucleosides (T<dU<dC<dG~dA). This indicates that the sugar moiety modulates selectivity for the latter two, possibly through H bonding, which has been shown to be a significant contributor to retention on amide columns (24). Comparing retention of the purine and pyrimidine series on the different HILIC columns, purine derivatives A and G tend to be more strongly retained than pyrimidines U and T. The retention of C and its derivatives, however, resembles that of purines.
RP-type separations A and C gave rise to identical elution sequences for the nucleobase series C<A<G<U<T. In contrast, no two HILIC columns yielded the same elution orders except for E and G (T<U<A<C<G). The latter two also provided identical elution sequences for the nucleosides (A~U<G<C, all with poor peakshapes on column G) and deoxynucleosides (T<U<A<C<G). It is likely that the terminal amino group, the common feature of columns E and G, is crucial for their selectivity towards nucleosides. The nucleobase elution order was conserved for the deoxynucleosides (T<dU<dA<dC<dG), which suggests that recognition of these metabolites by E and G is determined by interactions of the SP with the nucleobase moiety rather than the deoxyribose. Retention factors of nucleobase, nucleoside and deoxynucleoside (e.g., adenine, adenosine and deoxyadenosine) were typically similar, whereas higher selectivity was observed between the purine and pyrimidine series. We specifically noted differences between columns G and E as a result of the dissimilar hydrogen bond acidity of the two sorbents (primary vs. quaternary amine) and between B and E (inverse orientation of ionic sites).
We observed good peakshapes for pyrimidine bases U and T on column E but not on column B. The reverse was true for the purine bases. The elution orders of (T)<A<U<C<G for nucleosides and T<dU<dA<dC<dG for deoxynucleosides on polymer-based column B at high pH were in accordance with those reported in the literature for sulfobetaine-modified silica under acidic conditions (24), signifying a negligible influence of the packing material on the selectivity in this case.
On RP-type columns A and C, the nucleosides A, G and U eluted before their respective deoxynucleosides. Reversal of this sequence was observed on zwitterionic columns B and E, where dA, dG and dU eluted earlier than A, G and U, respectively MM-HILIC method D was not capable of resolving the two species, and F and G produced poor peakshapes.
Phosphate-containing compounds included sugar phosphates and nucleotides having one to three phosphoryl groups. Only 10% of the 77 phosphates in the analyte set were captured with k>1 on column A, which is not surprising considering the strongly acidic nature of these metabolites. On the other hand, WAX column G did not produce acceptable peaks for any phosphate and many phosphates could not even be detected with this method. We concluded that the 0.1% formate gradient utilized in method G was not sufficient to facilitate the displacement of the strong acids from the amino AX sites.
Zwitterionic columns, which display more moderate ion exchanger effects, successfully targeted 70 % of the phosphates. The high pH of 9.2 employed in method B facilitates quantitative deprotonation of the surface sulfonate (26). The resulting electrostatic repulsion of phosphates counterbalances their attraction by the positively charged betaine and facilitates timely elution. Interestingly, method E afforded similar results at neutral pH. Both zwitterionic methods provided a favorable, relatively uniform peak distribution across the run time But the absence of ionic interactions in the neutral MM and amide methods C/D and F reduced coverage to 32% (D), 10% (C) and even <7% for method F.
On zwitterionic columns, retention times increased in the order cXMP < XMP < XDP < XTP (X … nucleobase, c… cyclic). Clearly, electrostatic interactions play a crucial role in the selector-solute interactions. The selectivity of B and E for nucleobases was lower by comparison. Cyclic, deoxy, mono-, di- and triphosphates showed similar retention regardless of the base. More precisely, coelution was observed for guanosine and uridine monophosphates, and all detected diphosphates on B and E. All investigated columns showed only marginal selectivity toward isomeric sugar phosphates such as fructose-, glucose- and mannose-6-phosphate. However, retention was significantly improved on HILIC columns compared to method A. The most promising candidates for mobile phase optimization that could facilitate baseline resolution of sugar phosphates are C and E.
The library subgroup of organic acids included 194 small carboxylic acids such as lactate and tartrate, short-chain fatty acids, N-protected AAs and pharmaceutically relevant compounds such as non-steroidal anti-inflammatory drugs (NSAIDs). RP-C18 method A covered a total of 60% of the acid library with 40% of good peakshapes and k>1, which represents the highest coverage of all methods for the acid subset. Weak retention was frequently observed for small acids such as lactate or heterocycles such as quinate or oxaloacetate due to their limited capacity for hydrophobic interaction. 13 acids were detected by RP-C18 exclusively but not by any HILIC method. Overall distribution of acids, however, was more favorable on zwitterionic column B (34 % coverage). The latter was the only column that produced good peaks for both isocitrate and citrate (partial separation, α=1.03). As shown in Fig. 4a, maximum acid coverage (59%) was achieved by combination of A and B. On the other hand, both methods had high rates of poorly retained compounds (20% and 25%, respectively). The second zwitterionic column E yielded a relatively low coverage of 25%. Under neutral conditions, there exists presumably a strong counterion effect from the internal phosphate moiety on the carboxylates. Method E did not yield detectable peaks for short- and medium-chain fatty acids, method E did not yield detectable peaks. On sorbent B, these acids were barely retained (k<1) but still produced sharp, clearly identifiable peaks. In contrast, medium-length dicarboxylic acids (C-5 through C-10) were well retained on both zwitterionic sorbents. The retention time of dicarboxylic acids increased with hydrocarbon chain length on RP column A in accordance with a retention mechanism driven by dispersive interactions. On HILIC columns B and E, retention was found to be inversely proportional to chain length Unexpectedly, WAX-type column G also under performed in the separation of acids and covered only 8% of the library. Among the well-retained compounds were N-protected moderately polar and even basic amino acids, which was unexpected as they are not primary targets for anion exchangers. α- and β-ketoacids such as 3-hydroxypyruvate and acetoacetate were excluded (k<0). Dicarboxylic acids (glutarate, dihydroxyfumarate and others), aromatic and heterocyclic acids and α-hydroxycarboxylic acids also eluted ahead of or close to t0, which hints at electrostatic repulsion phenomena. Irreversible adsorption did not seem to be an issue but the high rate of poor peakshapes in this group (75 out of 91 acids with k>1) was indicative of poorly controlled ionic interactions. This illustrates how crucial method development including optimization of ionic additive type and content is for HILIC separations, even when a highly specific method is employed.
3.5 HILIC for biological samples: Selected issues
3.5.1 With biological samples, column conditioning is essential for stable retention times
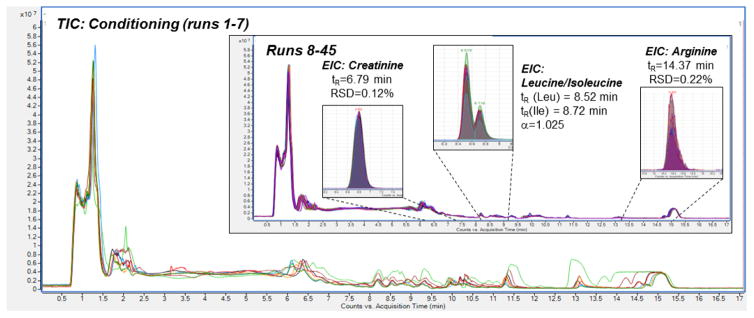
In untargeted metabolomics, retention time stability is essential for efficient and accurate peak alignment and feature identification, both of which are very time-consuming and error-prone tasks. Compound libraries containing the retention times and MS/MS data of authentic standards are important tools in the final data analysis steps. We determined that that retention times for the library compounds remained stable in spiked human plasma extracts (HPE) compared to matrix-free library mixtures with retention time deviation typically < 0.1 min for method B (data not shown). Next, 45 consecutive HPE injections were performed to investigate the reproducibility. Method B was chosen due to its broad applicability and large coverage range which was deemed most appropriate for future untargeted studies.
We determined that 3–5 consecutive matrix-free blank gradient runs were required to saturate the ionic interaction sites and establish a stable water layer in the HILIC system. Nonetheless, poor total ion chromatogram (TIC) reproducibility was observed during the first 7 injections of HPE (Fig. 5). This is indicative of increasing saturation of non-specific adsorption sites in the column with sample components over time. When analyzing complex biological samples such as plasma using HILIC, it is conceivable that irreversible adsorption of some components occurs through strong electrostatic/ionic or covalent bonds involving the surface modification or residual silanol groups. Column B is designed to minimize both phenomena due to its polymer-based support material and the zwitterionic modification. The latter reduces irreversible adsorption by exerting a “counterion effect”, i.e., partial repulsion on charged compounds.
Fig. 5. Shifting peaks during HILIC column conditioning with human plasma extract.
During the first runs, reproducibility of the total ion chromatogram (TIC) is poor. After completed conditioning, retention times are stable with an RSD of <2%. Chromatograms recorded with method B.
After conditioning was complete, the TIC trace as well as individual extracted ion chromatograms showed satisfactory reproducibility (Fig. 5, Insert). Extracted ion chromatograms were investigated and confirmed the initial assessment. For example, the relative standard deviation (RSD) of the retention time of well-retained amino acid Arg (mean tR=14.4 min, k=9.4) was 0.12%. Less strongly retained creatinine (mean tR =6.8 min, k=3.9) gave an RSD of 0.2 %. All investigated metabolites (k>1 for all of them) showed retention time RSDs below 2% and no correlation was found between retention time and injection number, suggesting that the fluctuations were random. Both column bleeding and build-up of irreversibly retained material would likely have resulted in decreased retention over time. It must be mentioned that peak area reproducibility was significantly lower. RSDs ranged from 2–30% for selected, representative analytes and were higher for low-abundance compounds. No trends regarding the peak area vs. the number of injections were observed. Inconsistent peak areas might indicate MS-related issues rather than represent a result of inadequate chromatographic separation. Due to the rather slow scan speed of high accuracy MS instruments such as the Q-TOF (2–3 scans/s in full scan mode), narrow peaks of less than 30 s width might not be accurately represented. Furthermore, detection limits are relatively high. Poor peak area reproducibility in “crowded” areas of the chromatogram where multiple peaks co-elute and create a high TIC signal could hint at competitive ionization issues but we did not observe this for compounds with k>1.
3.5.2 Spiked samples on columns B and E
The metabolite library contained 345 compounds which have been quantified in blood with concentrations ranging from 8*10−6 to 6*103 μmol/L according to data from the Human Metabolome Database. At least another 107 are expected to be present in blood according to an HMDB search performed in Nov. 2015. Disregarding detection limits, the establishment of which is a task typically performed in targeted methods, the percentage of metabolites expected in plasma and known to produce good peaks is 245 out of 452 (54%) for method B. Based on retention times collected using the library and accurate m/z measurements, we were able to unequivocally identify 110 metabolites in human plasma extracts (HPE) in a straightforward fashion. 25 more were identified with the help of HPE samples spiked with increasing concentrations of the metabolites in question. Among the latter were isobaric compounds which eluted close to each other and whose retention time deviated from the one determined for the matrix-free library standard such as Ile and Leu (Fig. 5, insert).
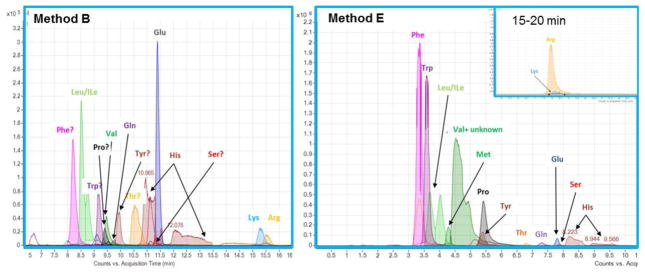
Fig. 6 shows extracted ion chromatograms obtained with methods B and E for proteinogenic AAs. With both methods, the analytes are spread out over the gradient run time whereas they exhibit weak retention on RP columns such as A (see ESM II for respective data). The zwitterionic HILIC methods show high selectivity for AA side chains. For example, hydrophobic AAs (Phe, Leu/Ile, Val) elute early and basic amino acids Lys and Arg exhibit the strongest retention. Poor peakshapes (broad, split peaks) are observed for His on both columns, and Val co-elutes with an unidentified compound on column E. Differences in signal intensity are related to the detection mode (ESI negative for method B, ESI positive for method E), which represents an aspect of method development not considered in this study. However, both HILIC methods are more successful in detecting non-protected AAs in a biological sample than the standard RP method.
Fig. 6. Extracted ion chromatograms of proteinogenic amino acids obtained with methods B (left panel) and E (right panel).
Hydrophobic amino acids elute earlier and highest retention times are observed for basic amino acids lysine (Lys) and arginine (Arg).
4. Conclusions
In this study, we report on the chromatographic performance of an RP-C18, 2 mixed-mode and 5 HILIC columns towards a library of 764 metabolites. We investigated their coverage, retention capabilities, selectivities and specificity towards moderately and strongly hydrophilic metabolites to gauge the suitability of the individual methods for targeted and untargeted metabolomics.
The applicability of our results for the analysis of samples of human plasma extract is demonstrated by the identification of more than 130 compounds in a straightforward fashion using a zwitterionic HILIC method. Our qualitative study emphasized the importance of appropriate equilibration in HILIC applications to overcome the perceived issue of poor reproducibility in HILIC.
We found that the most successful methods are capable of addressing about 70% of the metabolite library and attributed the missed fraction to chromatographic bias, i.e. the failure to retain certain metabolites due to a lack of compatible interaction sites. For example, RP and MM-RP typically missed highly polar compounds such as phosphates whereas HILIC methods performed poorly for lipids and related compounds. This fraction of poorly and un-retained compounds is unlikely to be addressed by optimization efforts. A “glass ceiling” may exist for the type and number of detectable and quantifiable compounds in metabolomics studies that utilize one-dimensional separations. Consequently, future efforts towards improved metabolomics methodology must involve the advancement of broadly applicable derivatization techniques and multidimensional separations coupling highly orthogonal LC techniques in addition to high-efficiency and high-selectivity approaches. At this point it should be noted that this work did not involve extensive method development. In particular, for methods B–G no mobile phase optimization was carried out in order to avoid selectivity bias towards a particular metabolite (group). Based on the critical role of the mobile phase in HILIC separations, considerable selectivity variation is expected upon changes to solvent composition and ionic additive type and content.
Nonetheless, our studies show that HILIC methods markedly improve the coverage of polar metabolite groups such as phosphates or carbohydrates compared to RP and therefore represent a worthy alternative to conventional separations. Zwitterionic sorbents such as sulfobetaine were found to have particularly broad application ranges, suggesting large potential for untargeted applications. Selectivity was highly diverse among the HILIC methods investigated. For example, the amide-HILIC column was particularly successful in retaining nucleosides while the phosphorylcholine sorbent was found to be most appropriate for the separation of carbohydrates.
In order to choose an appropriate sorbent, the most important questions to consider are the nature of the study (targeted, semi- or untargeted) and the properties of key analytes (most importantly pKa, logD and H bond characteristics) as well as ion exchange and hydrogen bond donor and acceptor characteristics of the sorbents. Our studies corroborate earlier findings of zwitterionic and H bond acceptor sorbents having broad applicability ranges and high stability. Zwitterionic sorbents in particular produce excellent chromatographic results for charged compounds, whereas hydrogen bond acceptors such as amide perform well for moderately polar analytes.
The retention factors for the library compounds as determined in our column screening study are made available as a searchable MS Excel file in the Electronic Supplementary Material (ESM II) and can assist researchers in choosing a suitable HILIC column based on the results for representative target analytes.
In conclusion, there is great potential for the implementation of HILIC methods in the metabolomics field due to their selectivity and specificity being complementary and in some cases even orthogonal to more established RP methods. Based on our evaluation of 764 authentic metabolite standards, we expect the biological information accessible via HILIC to be highly relevant in the ongoing search for biomarkers of health and disease as well as metabolic phenotyping.
Supplementary Material
Fig. S1 Retention profiles for RP (A, C) and HILIC (B, D–G) methods based on retention factors of detected metabolite library compounds. Chromatographic conditions according to Table S1
Fig. S2 Estimation of method orthogonality for RP and HILIC methods from plots of logarithmic retention factors. The deviation from the diagonal is larger for methods having high orthogonality (e.g., A/B or B/F, top row) compared to methods with similar selectivity (B/E, A/C, bottom row). Note that this comparison includes only compounds detected with both of the two methods being compared
Fig. S3 Metabolite subgroups and their retention on columns A–G. The numbers on top of each panel denote total number of identified metabolites from the library of 714. Dark blue segments signify good and light blue segments signify poor peakshape (double/multiple, split, broad or tailing peaks). Missing fraction is shown as red segments. See Table S1 for method description and Supporting Information II (xlsx file) for full list of library compound classification and results
Table S1 Columns and mobile phase conditions for methods A–G. * Note that methods C and D utilize the same column but different mobile phase modes
Acknowledgments
The authors would like to thank Prof. Wolfgang Lindner (University of Vienna) for the generous loan of HILIC columns used in this study. Research reported in this publication was supported in part by grants from the National Institutes of Health: DK094292, DK089503, DK082841, DK081943, U2C-ES-026553, DK097153 and UL1TR000433.
Abbreviations
- A
adenine/adenosine
- Arg
arginine
- C
cytosine/cytidine
- ESI
electrospray ionization
- G
guanine/guanosine
- HILIC
hydrophilic interaction liquid chromatography
- His
histidine
- HMDB
human metabolome database
- HPE
human plasma extract
- Ile
isoleucine
- LC-MS
liquid chromatography-mass spectrometry
- Leu
leucine
- LSER
linear solvation energy relationships
- Lys
lysine
- MM
mixed-mode (stationary phase)
- MP
mobile phase
- m/z
mass-to-charge ratio
- Phe
phenylalanine
- QSSR
quantitative structure-selectivity relationships
- QTOF
quadrupole-time of flight (mass spectrometer)
- RP
reversed phase
- RSD
relative standard deviation
- SP
stationary phase
- T
thymine/thymidine
- Val
valine
- WAX
weak anion exchanger
Footnotes
Compliance with Ethical Standards
No experiments requiring Institutional review board (IRB) approval were carried out. The biological samples in this study were obtained from commercial sources and therefore IRB exempt.
The authors declare that no conflict of interest exists.
References
- 1.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1(2):153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 2.Gika HG, Theodoridis GA, Plumb RS, Wilson ID. Current practice of liquid chromatography–mass spectrometry in metabolomics and metabonomics. Journal of Pharmaceutical and Biomedical Analysis. 2014;87(0):12–25. doi: 10.1016/j.jpba.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 3.Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and Diabetes: Analytical and Computational Approaches. Diabetes. 2015;64(3):718–32. doi: 10.2337/db14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS. Advances in metabolite identification. Bioanalysis. 2011;3(15):1769–82. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- 5.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protocols. 2011;6(7):1060–83. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 6.Gika HG, Wilson ID, Theodoridis GA. LC–MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. Journal of Chromatography B. 2014;966:1–6. doi: 10.1016/j.jchromb.2014.01.054. [DOI] [PubMed] [Google Scholar]
- 7.Liu R, Jia Y, Cheng W, Ling J, Liu L, Bi K, et al. Determination of polyamines in human urine by precolumn derivatization with benzoyl chloride and high-performance liquid chromatography coupled with Q-time-of-flight mass spectrometry. Talanta. 2011;83(3):751–6. doi: 10.1016/j.talanta.2010.10.039. [DOI] [PubMed] [Google Scholar]
- 8.Song P, Mabrouk OS, Hershey ND, Kennedy RT. In Vivo Neurochemical Monitoring Using Benzoyl Chloride Derivatization and Liquid Chromatography–Mass Spectrometry. Analytical Chemistry. 2011;84(1):412–9. doi: 10.1021/ac202794q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenz MA, El Azzouny MA, Kennedy RT, Burant CF. Metabolome Response to Glucose in the β-Cell Line INS-1 832/13. Journal of Biological Chemistry. 2013;288(15):10923–35. doi: 10.1074/jbc.M112.414961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alpert AJ. HILIC at 21: Reflections and perspective. Journal of Chromatography A. 2011;1218(35):5879. doi: 10.1016/j.chroma.2011.05.071. [DOI] [PubMed] [Google Scholar]
- 11.Alpert AJ. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. Journal of Chromatography A. 1990;499(C):177–96. doi: 10.1016/s0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- 12.Greco G, Letzel T. Main Interactions and Influences of the Chromatographic Parameters in HILIC Separations. Journal of Chromatographic Science. 2013;51(7):684–93. doi: 10.1093/chromsci/bmt015. [DOI] [PubMed] [Google Scholar]
- 13.Buszewski B, Noga S. Hydrophilic interaction liquid chromatography (HILIC)—a powerful separation technique. Anal Bioanal Chem. 2012;402(1):231–47. doi: 10.1007/s00216-011-5308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jandera P. Stationary and mobile phases in hydrophilic interaction chromatography: a review. Analytica Chimica Acta. 2011;692(1–2):1–25. doi: 10.1016/j.aca.2011.02.047. [DOI] [PubMed] [Google Scholar]
- 15.Jandera P. Stationary phases for hydrophilic interaction chromatography, their characterization and implementation into multidimensional chromatography concepts. Journal of Separation Science. 2008;31(9):1421–37. doi: 10.1002/jssc.200800051. [DOI] [PubMed] [Google Scholar]
- 16.Al-Haj MA. Quantitative Structure-Retention Relationships with Model Analytes as a Means of an Objective Evaluation of Chromatographic Columns. Journal of chromatographic science. 2001;39(1):29–38. doi: 10.1093/chromsci/39.1.29. [DOI] [PubMed] [Google Scholar]
- 17.Chirita R-I, West C, Zubrzycki S, Finaru A-L, Elfakir C. Investigations on the chromatographic behaviour of zwitterionic stationary phases used in hydrophilic interaction chromatography. Journal of Chromatography A. 2011;1218(35):5939–63. doi: 10.1016/j.chroma.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Schuster G, Lindner W. Comparative characterization of hydrophilic interaction liquid chromatography columns by linear solvation energy relationships. Journal of Chromatography A. 2013;1273(0):73–94. doi: 10.1016/j.chroma.2012.11.075. [DOI] [PubMed] [Google Scholar]
- 19.Schuster G, Lindner W. Additional investigations into the retention mechanism of hydrophilic interaction liquid chromatography by linear solvation energy relationships. Journal of Chromatography A. 2013;1301:98–110. doi: 10.1016/j.chroma.2013.05.065. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Guo Z, Shen A, Yu L, Xiao Y, Xue X, et al. Hydrophilic-subtraction model for the characterization and comparison of hydrophilic interaction liquid chromatography columns. Journal of Chromatography A. 2015;1398:29–46. doi: 10.1016/j.chroma.2015.03.065. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Research. 2013;41(D1):D801–D7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC–MS-based targeted metabolomics. Journal of Chromatography B. 2008;871(2):236–42. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theodoridis GA, Gika HG, Wilson ID. Metabolomics in Practice. Wiley-VCH Verlag GmbH & Co. KGaA; 2013. LC-MS-Based Nontargeted Metabolomics; pp. 93–115. [Google Scholar]
- 24.Kawachi Y, Ikegami T, Takubo H, Ikegami Y, Miyamoto M, Tanaka N. Chromatographic characterization of hydrophilic interaction liquid chromatography–stationary phases: Hydrophilicity, charge effects, structural selectivity, and separation efficiency. Journal of Chromatography A. 2011;1218(35):5903–19. doi: 10.1016/j.chroma.2011.06.048. [DOI] [PubMed] [Google Scholar]
- 25.Sampsonidis I, Witting M, Koch W, Virgiliou C, Gika HG, Schmitt-Kopplin P, et al. Computational analysis and ratiometric comparison approaches aimed to assist column selection in hydrophilic interaction liquid chromatography tandem mass spectrometry targeted metabolomics. Journal of Chromatography A. 2015;1406:145–55. doi: 10.1016/j.chroma.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, Watson DG, Wang L, Westrop GD, Coombs GH, Zhang T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. Journal of Chromatography A. 2014;1362:168–79. doi: 10.1016/j.chroma.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 27.Hyötyläinen T, Orešič M. Analytical Lipidomics in Metabolic and Clinical Research. Trends in Endocrinology & Metabolism. doi: 10.1016/j.tem.2015.08.006. [DOI] [PubMed]
- 28.Wang M, Han X. Multidimensional Mass Spectrometry-Based Shotgun Lipidomics. In: Raftery D, editor. Mass Spectrometry in Metabolomics. Methods in Molecular Biology. Vol. 1198. Springer; New York: 2014. pp. 203–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Astarita G. Lipidomics: An Evolving Discipline in Molecular Sciences. International journal of molecular sciences. 2015;16(4):7748–52. [Google Scholar]
- 30.Heaton JC, McCalley DV. Some factors that can lead to poor peak shape in hydrophilic interaction chromatography, and possibilities for their remediation. Journal of Chromatography A. doi: 10.1016/j.chroma.2015.10.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Retention profiles for RP (A, C) and HILIC (B, D–G) methods based on retention factors of detected metabolite library compounds. Chromatographic conditions according to Table S1
Fig. S2 Estimation of method orthogonality for RP and HILIC methods from plots of logarithmic retention factors. The deviation from the diagonal is larger for methods having high orthogonality (e.g., A/B or B/F, top row) compared to methods with similar selectivity (B/E, A/C, bottom row). Note that this comparison includes only compounds detected with both of the two methods being compared
Fig. S3 Metabolite subgroups and their retention on columns A–G. The numbers on top of each panel denote total number of identified metabolites from the library of 714. Dark blue segments signify good and light blue segments signify poor peakshape (double/multiple, split, broad or tailing peaks). Missing fraction is shown as red segments. See Table S1 for method description and Supporting Information II (xlsx file) for full list of library compound classification and results
Table S1 Columns and mobile phase conditions for methods A–G. * Note that methods C and D utilize the same column but different mobile phase modes