Abstract
The embryonic brain and spinal cord initially form through the process of neural tube closure (NTC). NTC is thought to be highly similar between rodents and humans, and studies of mouse genetic mutants have greatly increased our understanding of the molecular basis of NTC with relevance for human neural tube defects. In addition, studies using amphibian and chick embryos have shed light into the cellular and tissue dynamics underlying NTC. However, the dynamics of mammalian NTC has been difficult to study due to in utero development until recently when advances in mouse embryo ex vivo culture techniques along with confocal microscopy have allowed for imaging of mouse NTC in real time. Here, we have performed live imaging of mouse embryos with a particular focus on the non-neural ectoderm (NNE). Previous studies in multiple model systems have found that the NNE is important for proper NTC, but little is known about the behavior of these cells during mammalian NTC. Here we utilized a NNE-specific genetic labeling system to assess NNE dynamics during murine NTC and identified different NNE cell behaviors as the cranial region undergoes NTC. These results bring valuable new insight into regional differences in cellular behavior during NTC that may be driven by different molecular regulators and which may underlie the various positional disruptions of NTC observed in humans with neural tube defects.
Keywords: Neural tube closure, non-neural ectoderm, live embryo imaging, cellular projections, cellular dynamics
Introduction
Proper development of the embryonic neural tube is essential to set up the precursor of the central nervous system in vertebrates. The process of neural tube closure (NTC) is highly complex, and this is highlighted by the frequency of human neural tube defects (NTDs) that occur in approximately one in 1000 live births worldwide. Research to reveal the processes underlying NTC and to determine how disruptions in these processes result in NTDs has grown over the past decade, but major gaps remain in our understanding. Studies using amphibian, chick, fish and rodent model systems have combined to greatly increase our understanding of the ultrastructural, as well as the molecular and cellular basis, of NTC. While these studies have shown many similarities between NTC in different organisms, they have also found fundamental differences, including the relative size of the neural folds, how the neural folds come together and seal at the midline, and how the process initiates and proceeds along the rostral-caudal axis 1-3. These differences highlight the need to study NTC, especially at the cellular and tissue level, in a mammalian system that closely represents human NTC in order to better understand what underlies human pathology.
The neural tube initiates as a flat sheet of neuroepithelium, which then thickens to form a pseudostratified columnar neuroepithelium (NE) connected laterally to the squamous non-neural ectoderm (NNE). The NE bends at the midline to generate opposing neural folds which then bend towards each other and meet at the midline, at which time the NE and NNE separate from each other and seal with the corresponding tissue from the opposite fold to generate a solid neural tube (NT) that is overlaid with a layer of epithelium (NNE). In mammals, NTC initiates in the cervical spinal region (Closure I) and then proceeds in a bidirectional, zipper-like manner. Additional de novo closure points subsequently occur at the anterior-most point of the forebrain (Closure III) and at the forebrain-midbrain boundary (Closure II), and closure continues by zippering along the rostral-caudal axis from these points. In mice, the use of Closure II is strain dependant and there is debate as to whether Closure II occurs in human embryos, however the overall process of NTC is considered to be highly similar between mice and humans. Human neural tube defects (NTDs) can occur along the rostral-caudal axis and some may correspond to failures in NTC from these different closure points, although this cannot fully explain the spectrum of phenotypes and regional distinctions. The neural folds themselves appear structurally different along the rostral-caudal axis of the mammalian embryo, with regional differences in both fold size and hinge-point usage. Moreover, molecular information from mouse mutants points to regional differences in the underlying mechanisms of NTC. What is less well understood is whether rostral-caudal differences also occur at the level of cell form and function, and whether changes in regional cell behaviors can underlie NTDs.
Externally developing embryos have helped to reveal the dynamic cell and tissue movements associated with NTC. However, the mammalian embryo develops in utero and therefore similar dynamic studies have been more challenging; indeed much of our knowledge of mammalian NTC comes from analysis of fixed and sectioned embryos. Decades ago, scanning electron (SEM) and transmission electron microscopy (TEM) studies in mouse, rat and chick embryos undergoing NTC found that cells in both the NE and NNE extend cellular projections into the midline as the neural folds approach each other. These projections varied in appearance and were described as microvilli, ruffles, blebs and finger-like projections 3-6. Despite our long-standing knowledge of the existence of these cellular projections, to date there have been no functional studies to determine their role or importance in the NTC process. This may be due, in part, to the inability to preserve fine cellular structures in standard histology and immunohistochemistry preparations, and hence they have not been evaluated in experimental or genetic models. More recently, the development of ex vivo culture techniques has provided an opportunity to observe NTC in the living mouse embryo over time, which has been combined with time-lapse microscopy to study dynamic processes 7-9. For example, Pyrgaki et. al. used a live imaging approach to observe mouse NTC in the midbrain region and they identified many short bulbous and long thin cellular projections emanating from the NNE in optical cross sections. Additionally, they found that NNE cell membranes at the edge of the neural folds were fuzzy in appearance, suggesting that they are highly dynamic 7. Similar live imaging studies by Yamaguchi et. al. identified D-type (dancing) apoptotic cells in the midbrain-hindbrain region that were actively mobile and did not fragment 9. Thus, the ability to observe cellular projections through live imaging opens up a new opportunity to study how cellular dynamics are involved in NTC. Here we have expanded upon the previous work to describe the dynamic behavior of cellular projections specifically in the NNE at multiple time points and positions along the cranial neural folds. This work shows that there are regional differences in appearance and dynamic behaviour of NNE cellular projections and identifies novel behaviours that could not be observed in fixed sections, including membrane shuttling between NNE cells of the closing neural folds. Information gained from this live imaging platform sets the stage for future functional studies of cellular projections and their role in NTC.
Materials and Methods
Mice
For live embryo imaging, the following transgenic mouse strains were used: mTomato:mGFP (Jackson Laboratory #007576, 10) and Grhl3-Cre 11. Both strains were maintained on a C57/BL6 genetic background. Male breeder mice were used within one year of age and all female mice were mated between 6-16 weeks old. All mice were housed at University of Colorado Anschutz Medical Campus in accordance with IACUC approved protocol.
Live Embryo Imaging
Embryos were carefully dissected at various stages between 4 and 16 somites in age, mounted and imaged as in 8. Briefly, embryos were mounted in culture medium (three parts rat serum with one part culture media [DMEM:F12 without phenol red, 55 U/ml penicillin/streptomycin, 2.2 mM glutamax and 11 mM Hepes buffer]) and placed in a stage-top, temperature and gas controlled incubator (37°C, 5% O2, 5% CO2) on the motorized stage of an LSM 510 Meta confocal microscope (Zeiss). This system allows for time-lapse imaging of up to 5 embryos at once. For long term imaging (up to 12 hours), 10-20 z-planes were imaged at 20μm intervals, and acquired once every 5-8 minutes using a Plan-Achromat 10X/0.45 objective at 1X zoom. For high-speed imaging, embryos were imaged at one image every 15 seconds for 15 minutes total on a Zeiss 5Live line scanning confocal microscope system with images acquired using the same objective at 2.0X zoom. A total of 6 to 9 z-planes were imaged per experiment at an interval of 2-3 μm between z-planes. Multiple experiments were performed per time point and highly reproducible phenotypes are reported. Total numbers of embryos analyzed relative to each figure are listed in Table 1.
Table 1.
Numbers of embryos imaged and analyzed for each experiment. The cellular behaviors described were consistently observed in these biological and technical replicates.
Quantitative Analysis
Original imaging files were analyzed using Imaris software. Individual cellular projections were measured in 3D for length from the edge of the NNE to the farthest distal tip and width at half the distance along the length. Serial measurements were taken over the time period that the projection was visible and the average length and width were determined. To quantify the high-speed dynamics of cellular projections, movies taken over 15 minute time intervals were analyzed. Individual projections were followed as they extended away from the cell body (extensions) and retracted back towards the cell body (retractions), and the total number of these changes in directional movement were counted within the entire time course of the movie.
Results
NNE Dynamic Behaviours at Closure Points
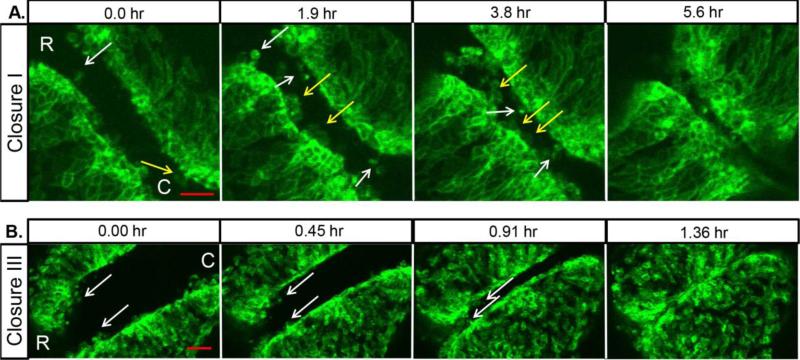
Previously our lab developed a robust live imaging system to visualize murine NTC throughout the rostral-caudal axis over long periods of time 8. Here we specifically followed the dynamics of the NNE during NTC by mating mTomato/mGFP fluorescent reporter mice (mTmG) 10 with mice expressing cre-recombinase under control of the Grhl3 promoter (Grhl3-cre) 11. This results in embryos that express a membrane-bound green fluorescent protein (mGFP) in NNE cells at the edges of the neural folds while the rest of the cells of the embryo express a membrane-bound red fluorescent protein (mTomato). This genetic cross was used throughout this study and the GFP signal was digitally isolated post-imaging in all figures to focus on behaviors of the NNE. In the mouse, NTC initiates at the hindbrain/cervical spine boundary in 4-5 somite stage embryos at a point referred to as closure I. Previous studies using SEM and TEM showed that the closely apposed neural folds just prior to closure I exhibit large membrane ruffles from the NNE at the tips of the folds which extend into the open space of the midline 5,6. Additionally, smaller finger-like projections were seen and these appeared to interdigitate with projections from the opposing fold, possibly aiding in fusion of the opposing tissues 6. To confirm and extend our knowledge of these projections and their dynamics over time, embryos were dissected at 4 somites in age and oriented to observe the initiation of closure I. Immediately upon the start of imaging, two types of cellular projections were observed extending from the NNE towards the gap at the midline (Fig. 1A, movie 1): wide membrane ruffles (Fig. 1A, yellow arrows) and more narrow bulbous projections (Fig. 1A, white arrows). Membrane ruffles emanated from multiple neighboring cells, and while they extended toward the midline individually, they often appear to make contact with each other. Both types of projections showed dynamic behaviour over time and additional projections of both types appeared as the folds moved closer together. These projections made the first points of contact across the gap of the opposing folds and then the individual projections became less distinct as the folds adhered to each other.
Fig. 1. NNE cells extend dynamic cellular projections at closure points.
(A) Imaging of the NNE in the hindbrain/cervical region of an initially 4 somite mouse embryo shows long narrow cellular projections (white arrows) and wide membrane ruffles (yellow arrows) that extend from NNE cells as the neural folds fuse at closure I. (B) Imaging of NNE at closure III starting at 13 somites shows a few cellular projections (white arrows) at the anterior-most end of the neural folds. Scale bars = 50μm. R=rostral, C=caudal
We then compared the cell behaviors of closure I with that of closure III, the de novo closure point at the anterior-most forebrain, using embryos that were ~13 somites at the start of imaging. In contrast to the multitude of projections seen during closure I, relatively few projections are apparent during closure III at the anterior prosencephalon (Fig. 1B, movie 2). As the folds in this anterior region approach each other, a few small narrow projections are seen at the most rostral end of the folds (white arrows), but the majority of the NNE caudal to this point appears smooth and uninterrupted as the folds meet at the midline. This suggests a model in which cellular projections help promote closure I, but that other mechanisms may drive initiation of closure III, such as large movements of the opposing neural folds towards one another.
NNE Cell Behaviours during Zipping
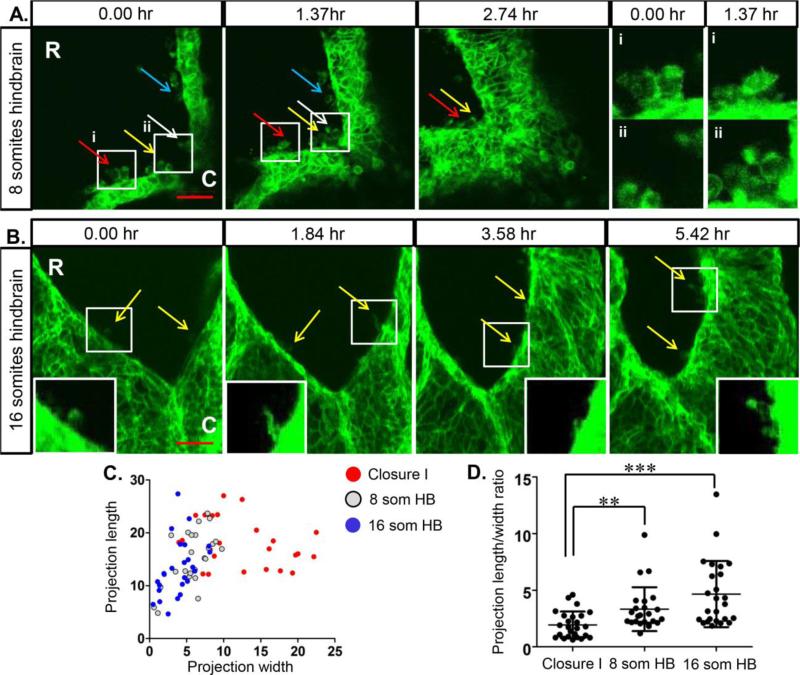
Following the completion of each closure point, the cranial neural tube continues to seal in a zipper-like fashion until final closure of the anterior neuropore (ANP) in the midbrain region. Imaging of the zippering in the hindbrain region in a 7 somite embryo shows multiple long cellular projections that are round in shape as well as some that appear ruffled (Fig. 2A, Movie 3), although all projections are more narrow than the ruffles seen during closure I. As zipping proceeds rostrally, these projections make contact across the folds at the midline and are sealed into the fusing folds (colored arrows follow individual projections over time). As closure proceeds to the ANP, it was previously shown that the NNE folds over the edge of the NE and that many cellular projections lined the edges of the ANP, even at a distance from the zip fork 5,10. When this region of a 16 somite embryo is imaged, many cellular projections can be seen that are thinner than those in other regions examined (Fig. 2B, movie 4) and they appear more filopodial in nature. These projections are also seen along the folds farther from the zip fork, although they seem sparser than what was observed through SEM. Interestingly, the sealed midline behind the zip fork shows a narrow line of higher GFP expression. As GFP is restricted to the plasma membrane, this suggests a region of tight association between opposing cells where the NNE cells are establishing new junctional complexes and/or it could be indicative of a force being generated along the midline that pulls cells in tight approximation.
Fig. 2. NNE cells extend dynamic cellular projections during neural tube zipping.
(A) Imaging of the NNE in the hindbrain of an 8 somite embryo shows multiple round bulbous and ruffled cellular projections during zipping of the neural folds. Coloured arrows follow individual projections over time. White boxes indicate regions of higher magnification of projections in (i) and (ii) showing changes in individual projections over time. (B) Multiple filopodial-like projections are seen (yellow arrows) extending from the NNE along the neural folds of the closing anterior neuropore in a 16 somite embryo. White boxes indicate regions of higher magnification of projections in insets. Scale bars = 50μm. R=rostral, C=caudal. (C) Individual cellular projections observed in time-lapse imaging were measured and plotted as length versus width by region. While filopodial-like projections seen in the two regions of hindbrain zipping have overlapping dimensions, the ruffled projections seen in the region of Closure I are much wider, indicative of lamellipodia. (D) Measurements from individual projections were calculated to determine the length to width ratio. The projections present during Closure I have a significantly smaller ratio than those present in the zipping hindbrain regions. ** p < 0.005, ***p < 0.001
We next assessed the dimensions of these regionally-distinct cell protrusions using Imaris image analysis software. Individual projections were measured over time and the average length and width were plotted (Fig. 2C). In the region of closure I, a large number of projections, correlating with the wide membrane ruffles, have a distinct shape being wider than they are long, indicative of lamellipodia A few other individual projections at closure I are much narrower and more similar to filopodia (Fig. 2C). In the two regions of hindbrain zipping, the cellular projections are similar in size and shape, and overall the projections are long and narrow, indicative of filopodia (Fig. 2C). We further calculated this data as a ratio of length to width (Fig. 2D) and this shows that the projections in the region of closure I are significantly different than those in both regions of the hindbrain. While these projections have yet to be molecularly characterized, this data suggests that the advancing edges of the NNE extend many lamellipodia during the initiation of closure I, but then the NNE changes its behavior to filopodia-like protrusions as the neural tube zippers anteriorly.
High Speed Dynamics of NNE Cells
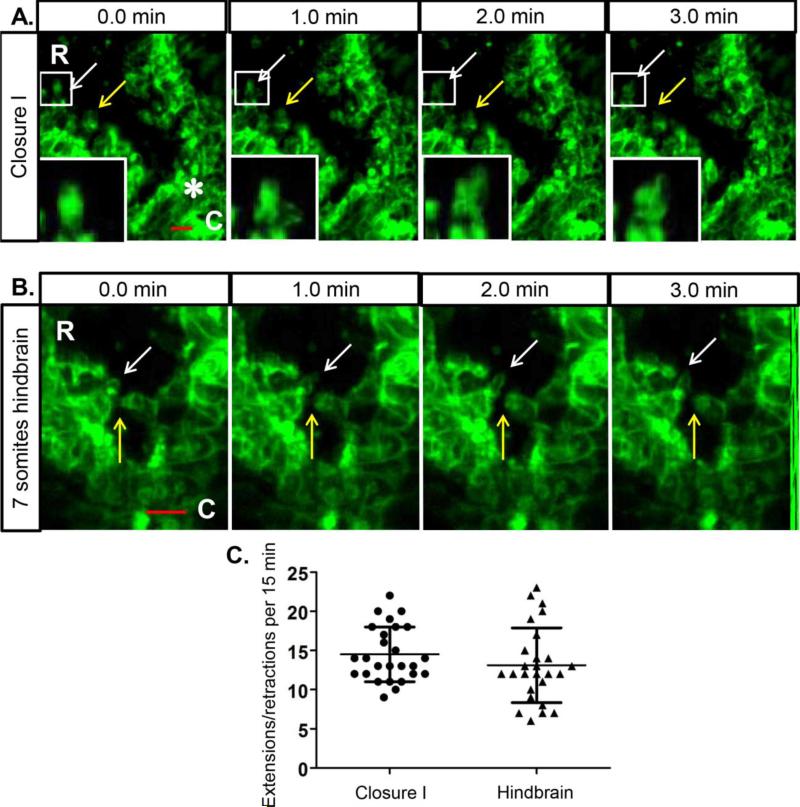
The experiments above gave a broad view of cellular projections at different postions along the length of the cranial neural folds. However, this relatively slow speed of imaging (images taken once every 5-8 minutes) at low magnification does not address how the cell projections behave as they closely approach and then come into contact with projections from opposing cells during neural fold convergence and zipping. To look more closely at individual projections and their high-speed membrane dynamics just at the time of fusion, we turned to a line scanning confocal platform whereby images were captured at higher magnification in 15 second increments. Figure 3A (movie 5) shows the region of closure I immediately rostral of the first contact point of the opposing neural folds (white asterisk). At this higher magnification, it is apparent that the apical edges of the NNE are rough in appearance, as many cellular projections extend toward the midline. The leading edges of these projections are highly dynamic and extend and retract in different directions over a short time frame of a few minutes (arrows follow individual projections over time, high magnification insets are the boxed region highlighted by the white arrow).
Fig. 3. NNE cells exhibit highly dynamic membrane projections.
(A) High speed imaging of NNE in the region just anterior to the initial contact of closure I (white asterisk) in a 6 somite embryo. Arrows indicate changing membrane shape in cellular projections over a 3 minute time course. White boxes indicate regions of further magnification of cellular projections in insets. (B) High speed imaging of the NNE in the zipping hindbrain in a 7 somite embryo show projections from opposite folds touching and then retracting (yellow arrows) and membrane dynamics (white arrows) over 3 minutes of time. Scale bars = 20μm. R=rostral, C=caudal. (C) Individual cellular projections were followed over a 15 minute time course of imaging and the number of extension and retraction movements relative to the cell body were calculated. Projections in both regions show similar dynamics with an average of 14 and 13 directional changes per 15 minutes respectively.
This highly dynamic behaviour is also evident in the zipping hindbrain rostral to closure I (Fig 3B, movie 6). In this region, NNE cells from opposing neural folds can be seen touching and then retracting (yellow arrow) while projections slightly rostral are extending and retracting into the open space (white arrow), again over a short time course of three minutes. Our previous data showed that the dimensions of cellular projections are significantly different in these two regions of closure I and the zipping hindbrain. To determine whether the projections in these two areas display different dynamic behaviors, we followed individual projections in each region over the 15 minute time course of high speed imaging and counted the number of extension (away from the cell body) and retraction (towards the cell body) movements (Fig. 3C). This data shows that there is no significant difference between the dynamic movements of cellular projections at closure I and the zipping hindbrain, with an average of 14 and 13 directional changes respectively.
It has been proposed previously that cellular projections are important for adhesion of the opposing neural folds 4,5. The present study provides a platform to begin to experimentally investigate this as well as to study at a molecular level the identity of these various cellular projections which have the appearance of filopodia and lamellipodia. In the future this robust imaging platform can be used along with conditional gene knockout in the NNE for detailed investigation into the nature of various cellular projections and to assess their requirement for neural tube fusion.
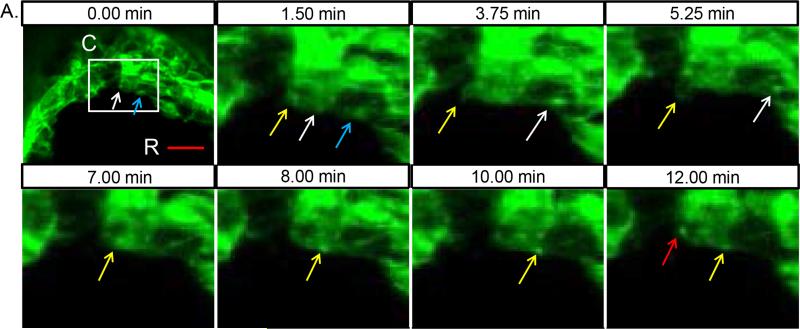
One phenomenon seen both here and previously by SEM is that cellular projections become prominent regionally just ahead of the zipping fork. Our new data also shows that these projections are highly dynamic. How do cells sense their positional identity relative to the time and place where closure will soon occur? One possibility is that cells transmit information to their neighbours ahead of the zip fork. In support of this idea, we observed an interesting phenomenon in which bright foci of GFP-labelled membrane moved from one cell to another. As shown in movie 7 over the course of 12 minutes (still images in Fig. 4), first one packet of membrane is seen moving from the cell undergoing fusion in the zip fork into the next cell rostral (Fig. 4, blue and white arrows), followed by another bleb of membrane which is internalized and shuttled to the neighbouring cell (yellow arrows). In all, over this 12 minute time-frame, three separate GFP foci move into the neighboring cell across what appears to be a single stable filopodial connection between these two cells. Sanders et al. previously showed that mesenchymal cells in the developing chick limb bud transmit Sonic hedgehog (Shh) ligand along specialized filopodia to signal to nearby cells 12. This suggests a more general mechanism whereby cells use filopodia to transmit information, including the potential for directly transferring membrane-bound proteins to other cells. Our observation offers an intriguing possibility whereby cells that have made contact with the opposing fold pass positional information on to neighbouring cells, readying them to make contact in turn.
Fig. 4. Membrane shuttling during zipping of the neural folds.
(A) High speed imaging of NNE during hindbrain zipping in a 9 somite embryo. White box indicates region of additional magnification in the remaining images taken over a 12 minute time course. A bright foci of membrane (highlighted by white arrow) moves from the cell at the midline to the neighbouring cell rostral along a filopodial connection. At the start of the movie, a membrane focus is already traversing along the filopodial extension between the two cells (blue arrow). Between 3.75 min and 5.25 min, a membrane bleb (highlighted by yellow arrow) is pulled back into the midline cell followed by the formation of a second focus of membrane that then moves into the next cell. At 12 min, an additional membrane focus is initiated (red arrow). Scale bar = 20μm. R=rostral, C=caudal
Conclusions
While much is currently known about the molecular mechanisms underlying development of the neural tube, little is known about what drives fusion of the opposing neural folds at the midline. Several previous electron microscopy studies of both chick and rodent embryos showed the presence of multiple cellular projections during the process of neural tube closure 4-6. These processes were seen emanating from both the NE and NNE cells and were hypothesized to help the folds adhere to each other at the midline during neural tube fusion. Recently, live imaging of mouse embryos undergoing NTC revealed cellular projections with the appearance of filopodia and lamellipodia 7, 8. Here, we have focused our attention on the NNE through genetic labelling and expanded on earlier studies to show that the NNE extends several phenotypically distinct cellular projections showing differential regional distribution along the developing cranial neural folds. Additionally, we used this method to determine the size and shape of the projections in these different regions and find that these projections display similar highly dynamic behaviour as shown by high magnification and high speed imaging. Additionally, we describe a new phenomenon of membrane “shuttling” that could be used by NNE cells to transmit information ahead of neural fold zipping. This study provides a platform that can be used to molecularly and functionally characterize these various cellular projections and determine their role in closure of the mammalian neural tube. Intriguingly, differential cellular behaviours along the rostral-caudal axis could underlie some of the different regional distributions seen in human neural tube defects.
Supplementary Material
Movie 1. Live imaging of NNE cellular projections during closure I.
mTmG:Grhl3-cre embryos were mounted for imaging of closure I starting at the 4 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 4 z-plane images were taken at 20 μm intervals continuously over a 7 hour period and combined in a maximum intensity projection. As the neural folds approach the midline, NNE cells extend long bulbous cellular projections and wide membrane ruffles. Scale bar = 50μm.
Movie 2. Live imaging of NNE cellular projections during closure III.
mTmG:Grhl3-cre embryos were mounted for imaging of closure III starting at the 13 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 6 z-plane images were taken at 20 μm intervals continuously over a 2 hour period and combined in a maximum intensity projection. As the neural folds approach the midline, a few long narrow cellular projections are extended from NNE cells at the anterior-most end of the neural folds, while the folds more caudal appear relatively smooth. Scale bar = 50μm.
Movie 3. Live imaging of NNE cellular projections during hindbrain zipping.
mTmG:Grhl3-cre embryos were mounted for imaging of hindbrain zipping starting at the 8 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 5 z-plane images were taken at 6 μm intervals continuously over a 4 hour period and combined in a maximum intensity projection. As the neural folds zipper closed, NNE cells extend multiple round bulbous and ruffled cellular projections that make contact with each other in the zipper fork. Scale bar = 50μm.
Movie 4. Live imaging of NNE cellular projections during zipping in the anterior neuropore. mTmG:Grhl3-cre embryos were mounted for imaging of closure of the anterior neuropore starting at the 16 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 14 z-plane images were taken at 20 μm intervals continuously over a 6 hour period and combined in a maximum intensity projection. Multiple filopodial-like projections are seen extending from the NNE along the neural folds. Scale bar = 50μm.
Movie 5. Live high-speed imaging of NNE cellular projections during closure I. mTmG:Grhl3-cre embryos were mounted for imaging of closure I at the 6 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 6 z-plane images were taken at 3 μm intervals every 15 seconds for 15 minutes and combined in a maximum intensity projection. Individual cellular projections show highly dynamic membrane movements and shape changes as they approach one another across the open midline. Scale bar = 20μm.
Movie 6. Live high-speed imaging of NNE cellular projections during hindbrain zipping. mTmG:Grhl3-cre embryos were mounted for imaging of closure I at the 7 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 9 z-plane images were taken at 2 μm intervals every 15 seconds for 15 minutes and combined in a maximum intensity projection. Cellular projections in the zip fork touch and retract, and show highly dynamic membrane movements. Scale bar = 20μm.
Movie 7. Live high-speed imaging of membrane shuttling during zipping of the neural folds. mTmG:Grhl3-cre embryos were mounted for imaging of hindbrain zipping at the 9 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 8 z-plane images were taken at 3 μm intervals every 15 seconds for 15 minutes and combined in a maximum intensity projection. Several bright foci of membrane are seen moving from the NNE cell undergoing fusion to the next cell rostral across an apparently stable filopodial connection. Scale bar = 20μm.
Highlights.
Live imaging shows dynamic cellular projections during mouse neural tube closure
The non-neural ectoderm extends regionally different cellular projections
Cellular projections have filopodia- and lamellipodia-like appearance
Membrane “shuttling” may transmit positional cues to neighboring cells
Acknowledgements
LN was an investigator of the Howard Hughes Medical Institute and this work was supported by NIH #1F31CA180438-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greene ND, Copp AJ. Development of the vertebrate central nervous system: formation of the neural tube. Prenat. Diagn. 2009;29:303–11. doi: 10.1002/pd.2206. doi:10.1002/pd.2206. [DOI] [PubMed] [Google Scholar]
- 2.Lawson A, England MA. Neural fold fusion in the cranial region of the chick embryo. Developmental Dynamics. 1998;212:473–81. doi: 10.1002/(SICI)1097-0177(199808)212:4<473::AID-AJA1>3.0.CO;2-E. doi:10.1002/(SICI)1097-0177(199808)212:43.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 3.Morriss-Kay GM. Growth and development of pattern in the cranial neural epithelium of rat embryos during neurulation. Journal of Embryology and Experimental Morphology. 1981;65(Suppl):225–41. [PubMed] [Google Scholar]
- 4.Bancroft M, Bellairs R. Differentiation of the neural plate and neural tube in the young chick embryo. A study by scanning and transmission electron microscopy. Anat. Embryol. 1975;147(3):309–35. doi: 10.1007/BF00315078. [DOI] [PubMed] [Google Scholar]
- 5.Waterman RE. Topographical changes along the neural fold associated with neurulation in the hamster and mouse. The American journal of anatomy. 1976;146(2):151–71. doi: 10.1002/aja.1001460204. doi:10.1002/aja.1001460204. [DOI] [PubMed] [Google Scholar]
- 6.Geelen JA, Langman J. Ultrastructural observations on closure of the neural tube in the mouse. Anat. Embryol. 1979;156:73–88. doi: 10.1007/BF00315716. [DOI] [PubMed] [Google Scholar]
- 7.Pyrgaki C, Trainor P, Hadjantonakis A, Niswander L. Dynamic imaging of mammalian neural tube closure. Dev. Biol. 2010;344(2):941–947. doi: 10.1016/j.ydbio.2010.06.010. doi:10.1016/j.ydbio.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massarwa R, Niswander L. In toto live imaging of mouse morphogenesis and new insights into neural tube closure. Development. 2013;140(1):226–36. doi: 10.1242/dev.085001. doi:10.1242/dev.085001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi Y, Shinotsuka N, Nonomura K, et al. Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. Journal of Cell Biology. 2011;195:1047–60. doi: 10.1083/jcb.201104057. doi:10.1083/jcb.201104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis (New York, N.Y. : 2000) 2007;45(9):593–605. doi: 10.1002/dvg.20335. doi:10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 11.Camerer E, Barker A, Duong DN, et al. Local protease signaling contributes to neural tube closure in the mouse embryo. Dev. Cell. 2010;18:25–38. doi: 10.1016/j.devcel.2009.11.014. doi:10.1016/j.devcel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497(7451):628–32. doi: 10.1038/nature12157. doi:10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Live imaging of NNE cellular projections during closure I.
mTmG:Grhl3-cre embryos were mounted for imaging of closure I starting at the 4 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 4 z-plane images were taken at 20 μm intervals continuously over a 7 hour period and combined in a maximum intensity projection. As the neural folds approach the midline, NNE cells extend long bulbous cellular projections and wide membrane ruffles. Scale bar = 50μm.
Movie 2. Live imaging of NNE cellular projections during closure III.
mTmG:Grhl3-cre embryos were mounted for imaging of closure III starting at the 13 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 6 z-plane images were taken at 20 μm intervals continuously over a 2 hour period and combined in a maximum intensity projection. As the neural folds approach the midline, a few long narrow cellular projections are extended from NNE cells at the anterior-most end of the neural folds, while the folds more caudal appear relatively smooth. Scale bar = 50μm.
Movie 3. Live imaging of NNE cellular projections during hindbrain zipping.
mTmG:Grhl3-cre embryos were mounted for imaging of hindbrain zipping starting at the 8 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 5 z-plane images were taken at 6 μm intervals continuously over a 4 hour period and combined in a maximum intensity projection. As the neural folds zipper closed, NNE cells extend multiple round bulbous and ruffled cellular projections that make contact with each other in the zipper fork. Scale bar = 50μm.
Movie 4. Live imaging of NNE cellular projections during zipping in the anterior neuropore. mTmG:Grhl3-cre embryos were mounted for imaging of closure of the anterior neuropore starting at the 16 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 14 z-plane images were taken at 20 μm intervals continuously over a 6 hour period and combined in a maximum intensity projection. Multiple filopodial-like projections are seen extending from the NNE along the neural folds. Scale bar = 50μm.
Movie 5. Live high-speed imaging of NNE cellular projections during closure I. mTmG:Grhl3-cre embryos were mounted for imaging of closure I at the 6 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 6 z-plane images were taken at 3 μm intervals every 15 seconds for 15 minutes and combined in a maximum intensity projection. Individual cellular projections show highly dynamic membrane movements and shape changes as they approach one another across the open midline. Scale bar = 20μm.
Movie 6. Live high-speed imaging of NNE cellular projections during hindbrain zipping. mTmG:Grhl3-cre embryos were mounted for imaging of closure I at the 7 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 9 z-plane images were taken at 2 μm intervals every 15 seconds for 15 minutes and combined in a maximum intensity projection. Cellular projections in the zip fork touch and retract, and show highly dynamic membrane movements. Scale bar = 20μm.
Movie 7. Live high-speed imaging of membrane shuttling during zipping of the neural folds. mTmG:Grhl3-cre embryos were mounted for imaging of hindbrain zipping at the 9 somite stage. Grhl3-expressing NNE cells are highlighted with membrane-bound green fluorescent protein (GFP). 8 z-plane images were taken at 3 μm intervals every 15 seconds for 15 minutes and combined in a maximum intensity projection. Several bright foci of membrane are seen moving from the NNE cell undergoing fusion to the next cell rostral across an apparently stable filopodial connection. Scale bar = 20μm.