Abstract
Diagnosis, clinical management and research of psychiatric disorders remains subjective - largely guided by historically developed categories which may not effectively capture underlying pathophysiological mechanisms of dysfunction. Here, we report a novel approach of identifying and validating distinct and biologically meaningful clinical phenotypes of bipolar disorders using both unsupervised and supervised machine learning techniques. First, neurocognitive data were analyzed using an unsupervised machine learning approach and two distinct clinical phenotypes identified namely; phenotype I and phenotype II. Second, diffusion weighted imaging scans were pre-processed using the tract-based spatial statistics (TBSS) method and ‘skeletonized’ white matter fractional anisotropy (FA) and mean diffusivity (MD) maps extracted. The ‘skeletonized’ white matter FA and MD maps were entered into the Elastic Net machine learning algorithm to distinguish individual subjects' phenotypic labels (e.g. phenotype I vs phenotype II). This calculation was performed to ascertain whether the identified clinical phenotypes were biologically distinct. Original neurocognitive measurements distinguished individual subjects' phenotypic labels with 94% accuracy (sensitivity = 92%, specificity = 97%). TBSS derived FA and MD measurements predicted individual subjects' phenotypic labels with 76% and 65 % accuracy respectively. In addition, individual subjects belonging to phenotypes I and II were distinguished from healthy controls with 57% and 92% accuracy respectively. Neurocognitive task variables identified as most relevant in distinguishing phenotypic labels included; affective go/no-go (AGN), Cambridge gambling task (CGT) coupled with inferior fronto-occipital fasciculus and callosal white matter pathways. These results suggest that there may exist two biologically distinct clinical phenotypes in bipolar disorders which can be identified from healthy controls with high accuracy and at an individual subject level. We suggest a strong clinical utility of the proposed approach in defining and validating biologically meaningful and less heterogenous clinical sub-phenotypes of major psychiatric disorders.
Keywords: bipolar disorder, neuroimaging, neuropsychology, machine learning, heterogeneity, research domain criteria (RDOC), big data
Introduction
Diagnosis and clinical management of psychiatric disorders largely relies on traditional symptom-based classification systems such as the diagnostic and statistical manual (DSM) and the international classification of disease (ICD). However, it has been suggested that these symptom-based classification systems do not necessarily align with pathophysiological mechanisms of dysfunction (Cuthbert and Insel, 2010; Frangou, 2013; Hickie et al., 2013; Insel et al., 2010; Insel and Cuthbert, 2015; Morris and Cuthbert, 2012). Furthermore, current classification systems consider psychiatric disorders as unique phenotypic entities – whilst there is evidence of symptom overlap and diagnostic heterogeneity within disorders (Wardenaar and de Jonge, 2013). The current impasse has undoubtedly slowed down development of objective diagnostic markers of disease and effective treatments. To address this issue the National Institute of Mental Health (NIMH) has recently proposed the Research Domain Criteria (RDoC) initiative which is aimed at “developing, for research purposes, new ways of classifying mental disorders based on dimensions of observable behavior and neurobiological measures” (National Institute of Mental Health, 2008). Notably, the RDoC framework offers a promising alternative to standard classification systems as it views psychiatric disorders from a multidimensional viewpoint and integrates molecular, behavioral, motivational and cognitive data (Insel, 2014). To date, few studies have adopted the RDoC classification system to describe the clinical status and neurocognitive functioning of patients with neuropsychiatric disorders.
In line with this goal, the two main objectives of this study were; first, to determine whether we can identify distinct clinical phenotypes based on a composite representation of neurocognitive data. Second, investigate whether neuroimaging measurements acquired from diffusion weighted scans are predictive of identified phenotypes at an individual subject level and with above chance level accuracy. Notably, in this study we refer to disease phenotypes as subgroups or sub-classifications of patient groups with unique patterns of neurocognitive functioning. As a first step, this approach was applied on a sample of patients with bipolar disorders and validated against matched healthy controls.
Bipolar disorder (BD) is a common psychiatric disorder characterized by mania or hypomania, mostly alternating with depression and ranked amongst top 20 causes of disability worldwide (Hirschfeld and Vornik, 2005; Murray et al., 2013; Rajkowska et al., 2001). BD is associated with a high socioeconomic burden including suicidality, substance abuse and comorbidity with other disorders (Galvez et al., 2014; Hirschfeld and Vornik, 2005; Müller-Oerlinghausen et al., 2002). Clinically, BD is categorized into two main subtypes; 1) bipolar I disorder (BD I) – characterized by one or more manic or mixed episodes, accompanied by one or multiple depressive episodes (Association, 2000; Müller-Oerlinghausen et al., 2002). 2) Bipolar II disorder (BD II) – characterized by one or more hypomanic episodes with recurrent depressive episodes (Association, 2000; Müller-Oerlinghausen et al., 2002). Noticeably, these clinical subtypes are based on observed and potentially heterogeneous ‘signs and symptoms’ as opposed to measureable neurobiological or behavioral characteristics. Consequently, the main motivation of this study was not to investigate neurobiological differences across subtypes of BD but to investigate distinct neurobiologically relevant phenotypes within the entire bipolar disorders spectrum. Markedly, identifying neurobiologically distinct phenotypes may lead to development of more effective, better-targeted treatments and most importantly personalized or individualized treatments. Indeed, this is also inline with the recently proposed P4 (predictive, preventive, personalized and participatory) medicine framework in investigating major medical conditions (Hood and Friend, 2011).
Multiple neurocognition studies have reported impaired cognition in BD patients as compared to healthy individuals. For example, a recent meta-analysis summarizing 42 studies observed significant impairments in BD patients across multiple domains such as attention, working memory, verbal/non-verbal memory, visuospatial function, psychomotor speed, language and executive function (Kurtz and Gerraty, 2009). Most recently, several studies have successfully identified unique neurocognitive subgroups using data-driven machine learning algorithms (Brodersen et al., 2014; Burdick et al., 2014; Fair et al., 2012; Heinrichs and Awad, 1993; LEE et al., 2014). Distinctively, a recent longitudinal study reported a data-driven approach able to identify neurocognition subgroups which proved to be useful in predicting longitudinal functional outcomes (Hermens et al., 2011). However, whilst these studies have undoubtedly offered significant insights on unique and objective phenotypes of psychiatric disorders - the association between these data-driven phenotypes and other neurobiological measurements (e.g. white matter tissue diffusivity) remains largely unexplored. Furthermore, a significant advance would be to demonstrate that neuroimaging measurements are highly predictive of identified disease phenotypes albeit at an individual subject level. Notably though, there is active research work in this area (Brodersen et al., 2014; Geisler et al., 2015; Karalunas et al., 2014).
Multiple studies have recently demonstrated the utility of machine learning or pattern recognition algorithms in making clinically relevant predictions at an individual subject level (Johnston et al., 2014; Lavagnino et al., 2015; Mwangi et al., 2012a; Mwangi et al., 2012b; Mwangi et al., 2014b; Rocha-Rego et al., 2014). Specifically, previous studies have attempted to discriminate DSM defined categories from matched healthy controls using neuroanatomical scans (Mwangi et al., 2012a; Oliveira et al., 2013; Schnack et al., 2014). However, whilst these algorithms have been received within the neuropsychiatric community with great optimism – a major criticism has been that these algorithms are ordinarily ‘trained’ to categorize patients based on symptom-based DSM defined categories – an assumption which may lead to circularity (Mwangi et al., 2014a; Savitz et al., 2013). In this study though, we somewhat circumvent this limitation by first; identifying distinct behavioral phenotypes using neurocognitive data and an unsupervised machine-learning technique. Second, using diffusion weighted neuroimaging scan data such as mean diffusivity (MD) and fractional anisotropy (FA) we ‘train’ a machine learning algorithm to ditinguish individual subjects' among clinical phenotypes as well as healthy controls. Importantly, to allow inter-subject comparisons diffusion weighted imaging FA and MD maps were pre-processed using the tract-based spatial statistics (TBSS) method (Smith et al., 2006). This entailed spatial normalization of subjects' FA and MD volumes into a standardized template coupled with a ‘tract skeletonization’ process to account for any residual misalignments and resulting FA and MD volumes used in the machine learning analyses and phenotype validation.
Materials and Methods
Subjects
This study was approved by the local Institutional review board (IRB) at The University of North Carolina at Chapel Hill. Study participants included 70 patients with DSM-IV diagnosis of BD as shown in Table 1. A diagnosis of BD in patients was established through administration of the structured clinical interview for the diagnostic and statistical manual of mental disorders axis I (SCID I) (First et al., 2012). Subjects were excluded if they met criteria for substance abuse or dependence in the last 6 months preceding their participation in the study. Additional exclusion criteria were positive pregnancy test, neurological disorders, head injury with loss of consciousness and family history of hereditary neurologic disorders. Participants' affective state was assessed with the Hamilton Depression Rating Scale (HDRS) - 21 items and the Young Mania Rating Scale (YMRS) (Young et al., 1978), and the Montgomery-Asberg Depression Scale (MADRS) (Davidson et al., 1986).
Table 1.
Demographic and clinical details.
| Mean (SD) | |
|---|---|
| Age (years) | 33.87 (13.02) |
| Female/total (N) | 41 (70) |
| Age of onset (years) | 16.98 (6.43) |
| Education (Number of years) | 14.42 (2.67) |
| YMRS | 5.64 (5.49) |
| HDRS | 12.59 (8.17) |
| MADRS | 17.30 (11.33) |
| Hollingshead SES score | 29.59 (17.22) |
| BD subtype | |
| DSM IV bipolar disorder I (N) | 47 |
| DSM IV bipolar disorder II (N) | 12 |
| DSM IV bipolar disorder NOS (N) | 11 |
| Comorbidities | |
| Panic disorder (N) | 8 |
| Social phobia (N) | 5 |
| PTSD (N) | 13 |
| OCD (N) | 3 |
| GAD (N) | 8 |
| Substance abuse (N) | 15 |
| Specific/simple phobia (N) | 2 |
| Agoraphobia (N) | 7 |
| Anxiety disorder (N) | 1 |
| Anorexia (N) | 1 |
| Currently or previously taken any psychotropic medication (N) | 61 |
| Race | |
| White (N) | 51 |
| Black (N) | 5 |
| Hispanic (N) | 2 |
| Unknown (N) | 12 |
BPD- bipolar disorder, SD- standard deviation, YMRS- young mania rating scale, HDRS- Hamilton depression rating scale, MADRS- Montgomery-Asberg Depression Scale, SES – social economic status, PTSD- Post-traumatic stress disorder, OCD- obsessive compulsive disorder, GAD- generalized anxiety disorder.
Neurocognitive data
Participants performed the computerized Cambridge Neurocognitive Test Automated Battery (CANTAB - http://www.cantab.com). This cognitive battery was chosen based on the established sensitivity to cognitive impairments in psychiatric disorders (Sweeney et al., 2000). Participants completed tests evaluating visuomotor speed (Choice Reaction Time – CRT, Motor Screening –MOT), selective and continuous visual attention (Match to Sample Visual Search – MTS, Rapid Visual Processing – RVP), working memory and planning (Intra/Extradimensional Set Shift –IED, Spatial Span task – SSP, Spatial Recognition Memory – SRM), and cognitive control (Affective Go/No-Go-AGN, Cambridge Gambling Task – CGT). These neurocognitive tasks are briefly described in Table 2 and a detailed description is also given elsewhere (Robbins et al., 1994).
Table 2.
Cognitive tasks and measurements.
| No. | CANTAB Task | Evaluation | Measurements |
|---|---|---|---|
| 1 | Affective Go/No-Go | Affective and Cognitive control | Reaction time, accuracy |
| 2 | Cambridge Gambling Task | Decision-making | Reaction time, accuracy, proportion bets across trials with more/equally/less likely outcome |
| 3 | Choice Reaction Time | Cognitive processing speed | Reaction time, accuracy |
| 4 | Intra/Extradimensional Set Shift | Working memory and planning | Accuracy |
| 5 | Motor Screening | Motor processing speed | Reaction time |
| 6 | Match to Sample Visual Search | Visuo-motor speed | Reaction time, accuracy |
| 7 | Rapid Visual Processing | Sustained attention | Reaction time, accuracy |
| 8 | Spatial Recognition Memory | Spatial memory | Reaction time, accuracy |
| 9 | Spatial Span task | Spatial working memory | Span length, number of attempts, reaction time |
Reaction time is in milliseconds.
These cognitive tasks assess domains of the NIMH RDoC framework such as arousal, cognitive control, declarative memory, social communication and valence system. Therefore, based on the preliminary description of the RDoC cognitive constructs provided elsewhere (http://www.nimh.nih.gov/research-priorities/rdoc/rdoc-constructs.shtml) we mapped the CANTAB tasks onto the RDoC construct in the following manner:
Arousal: includes measures of basic motor and cognitive processing speed such as the RTs in the CRT, MTS and MOT tasks.
Cognitive control: includes measures of response inhibition such as AGN RTs, number of errors to neutral conditions, deliberation time and delay aversion (CGT) and commission errors in the IED/RVP tasks.
Declarative memory: includes aspects of memory retrieval assessed by the SSP task.
Social communication: includes measures of affective processing such as AGN RTs and number of commission errors in response to affective stimuli.
Valence system: measures of reward, reinforcement, expectancy and likelihood to attain reward such as quality of decision making and risk adjustement (CGT)
A conceptual diagram of our analytical approach and mapping of the CANTAB tasks onto the RDoC framework are presented in in Figure 1 and Table 2.
Figure 1.
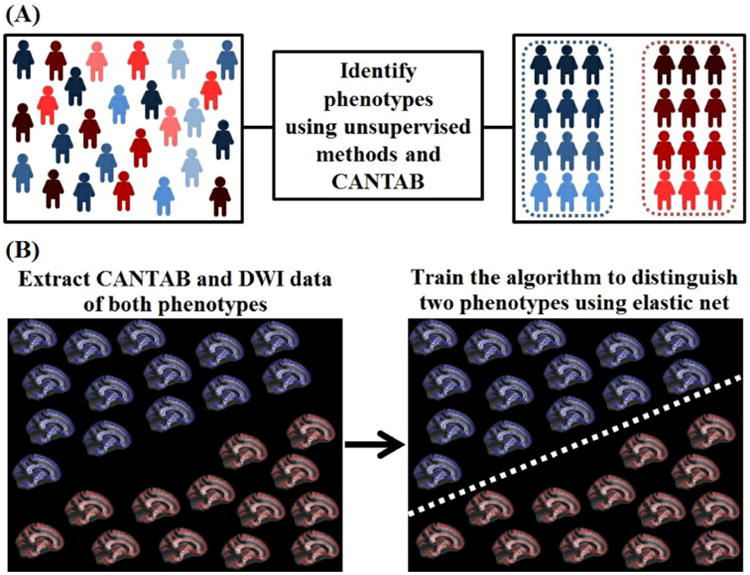
A flow diagram illustrating the ‘top-down’ conceptual approach used in this study. A) Phenotypes were identified using CANTAB neurocognitive scores through data dimensionality reduction and clustering using the principal component analysis and k-means clustering techniques. B) Phenotypes were validated using raw neurocognitive scores (without data dimensionality) and neuroimaging measurements (fractional anisotropy and mean diffusivity). Supervised machine learning techniques – LASSO and Elastic Net were ‘trained’ to predict individual subjects' phenotypic labels using neurocognitive scores or neuroimaging measurements. The supervised model was ‘trained’ and ‘tested’ using a leave-one-out cross-validation process.
Neuroimaging data acquisition and pre-processing
Diffusion weighted imaging (DWI) scans were acquired using a 3.0 T Siemens allegra scanner using a spin echo-planar imaging (EPI) protocol with the following acquisition parameters. Repetition time (TR) = 9200 ms, echo time = 79 ms, slice thickness = 2 mm, voxel size = 2 mm, image matrix = 104 × 128 with 21 diffusion encoding directions, b-value = 1000 s/mm2 and one non-diffusion weighted volume (b = 0). All DWI scans were inspected visually to rule out any gross artefacts (e.g. ghosting). Scans were pre-processed using the FMRIB software package (FSL) version 5.0.7 (Smith, 2002) through the following steps. 1) Head motion and ‘eddy currents’ correction using FSL's ‘eddy_correct’ routine. 2) Removal of non-brain tissue (e.g. skull) using the brain extraction tool (BET). 3) Calculation and diffusion tensor fitting at every voxel as detailed elsewhere (Johansen-Berg and Behrens, 2013). 4) Lastly, estimation of fractional anisotropy (FA) and mean diffusivity (MD) values was performed resulting into individual subjects' FA and MD image volumes. To allow inter-subject statistical comparisons, scans were pre-processed using the tract-based spatial statistics (TBSS) (Smith et al., 2006) routine as follows. FA volumes were spatially aligned into a standard template available with FSL (FMRIB58_FA) through a non-linear registration routine (FNIRT) (Smith et al., 2006). Spatially aligned FA volumes were averaged to create a group average FA volume which was input into a ‘tract skeletonization’ routine as implemented in FSL (Smith et al., 2006). The tract sekeleton represents white matter tracts as single lines going through the center of the tract – a process often used to account for any residual misalignmets due to the spatial normalization process (Johansen-Berg and Behrens, 2013). Lastly, individual subjects' spatially normalized FA images were ‘projected’ onto the mean FA skeleton and thresholded using mean FA = 0.2 to exclude voxels which are potentially within gray matter or cerebral spinal fluid (CSF) and therefore avoid partial voluming. The spatial normalization and a subsequent skeletonization calculation was also applied to MD volumes as described elsewhere (Smith et al., 2006). This process resulted in FA and MD variables or feature vector of dimension 1 × 113084 and subsequently used in the ensuing machine learning analyses.
Identification of data-driven phenotypes using neurocognitive data
90 cognitive scores of the CANTAB (see Supplementary Material, Table S1) were normalized by subtracting the mean and dividing with standard deviation (z-score) and entered into principal component analysis (PCA) (Jolliffe, 2005; Mwangi et al., 2014c) for data dimensionality reduction. Resulting principal components were entered into the k-means clustering algorithm (Hartigan and Wong, 1979) for clustering and phenotype identification. However, this approach required identification of two parameters. 1) Optimal number of principal components. 2) Ideal number of K-means clusters. These two data-driven parameters were identified using a ‘grid-search’ procedure which was optimized to maximize the average silhouette width value (Rousseeuw, 1987). Silhouette width value (SWV) is a statistical measure used to quantify similarity of a data point to other points within its own cluster as compared to data points in other clusters. SWV falls between -1 and 1, with a value of 1 signifying most optimal number of clusters as further described in the supplementary materials and elsewhere (Mwangi et al., 2014a). Lastly, resulting low-dimensional data from PCA were visualized using the t-distributed stochastic neighbor embedding (t-SNE) technique (Van der Maaten and Hinton, 2008). t-SNE is a data visualization technique used to embed high dimensional data (>3) into a low dimensional space (e.g. 2D) for visualization purposes (Mwangi et al., 2014a; Plis et al., 2014). The phenotype identification process is conceptually summarized in Figure 1. A summary of BD patients identified in two phenotypes (phenotype I and phenotype II) with matched healthy controls is shown in Table 3.
Table 3.
Demographic and clinical variables of Healthy controls, Phenotype I and Phenotype II patients. Notably, this table was generated ‘post-hoc’ – after the unsupervised machine learning step.
| Healthy controls mean (SD) | Phenotype I mean (SD) | Phenotype II mean (SD) | p-value | |
|---|---|---|---|---|
| Age (years) | 32.93 (13.38) | 33.47 (13.05) | 34.34 (13.17) | 0.9042 |
| Female/total (N) | 22 (38) | 24 (38) | 17 (32) | 0.6963 |
| Education (Number of years) | 16.53 (3.68) | 13.19 (3.09) | 15.71 (2.37) | < 0.001 |
| BD subtype | 0.9468 | |||
| DSM IV bipolar disorder I (N) | - | 26 | 21 | |
| DSM IV bipolar disorder II (N) | - | 6 | 6 | |
| DSM IV bipolar disorder NOS (N) | - | 6 | 5 | |
| Current mood status | - | 0.6746 | ||
| Euthymic (N) | - | 7 | 12 | |
| Depressed (N) | - | 20 | 12 | |
| Manic (N) | - | 2 | 1 | |
| Hypomanic (N) | - | 3 | 0 | |
| Mixed (N) | - | 2 | 3 | |
| Undertermined (N) | - | 4 | 4 | |
| Age of manic (years) | - | 15.82 (14.12) | 10.70 (9.09) | 0.1209 |
| Age of depression (years) | - | 18.27 (13.28) | 16.53 (11.11) | 0.6049 |
| Comorbidities (total) (N) | - | 21 | 20 | 0.4236 |
| Current lithium (N) | - | 1 | 4 | 0.1026 |
| Current severity | - | 2.27 (1.43) | 3.64 (2.06) | 0.0096 |
| HAMD (total) | - | 14.06 (7.33) | 11.93 (8.94) | 0.3031 |
| MADRAS (total) | - | 18.76 (10.38) | 15.59 (12.32) | 0.2707 |
| YMRS (total) | - | 6.74 (5.56) | 4.43 (4.60) | 0.0757 |
BD- bipolar disorder, SD- standard deviation, HAMD- Hamilton rating scale for depression, MADRS- Montgomery-Asberg Depression Scale, YMRS- young mania rating scale.
Phenotype validation using neurocognitive data
Neurocognitive scores (without PCA data reduction) were input into the least absolute shrinkage and selection operator (LASSO) algorithm (Tibshirani, 1996, 2011a) which was ‘trained’ to identify individual subjects' phenotypic labels and prediction accuracy (specificity and sensitivity) calculated. Specifically, we assumed a two group classification problem (e.g. phenotype I vs phenotype II) with predictor variables or featuers (neurocognitive data) represented as, xij where i = 1,2, … N represents number of subjects and j = 1,2, … P represents number of predictor variables or features. In addition, yi represents corresponding target labels (1- phenotype I, 2 - phenotype II). As a result, the LASSO algorithm was used to compute a set of coefficients (β̂) by minimizing the following objective function (Tibshirani, 1996; Tibshirani, 2011b).
The algorithm parameter (λ) which is used to encourage algorithm sparsity – translating into fewer coefficients with non-zero weights was selected using a 10-fold cross-validation approach. This parameter was selected using training data only to avoid circularity also known as double dipping (Kriegeskorte et al., 2009; Mwangi et al., 2014c). The LASSO objective function was optimized using the coordinate descent algorithm through a MATLAB (The Mathworks, Inc) toolbox provided by Friedman and colleagues (Friedman et al., 2010). Lastly, a generalized linear model was used to estimate the proability of an individual subject belonging to either phenotype I or phenotype II, given raw neurocognitive scores and the identified LASSO coefficients (β̂). In addition, the LASSO algorithm was also ‘trained’ to distinguish individual phenotype I or phenotype II patients from healthy controls. In each comparison (e.g. phenotype I vs healthy conrols) a leave-one-out cross-validation (LOOCV) approach was used to ‘train’ and ‘test’ the model whilst the penalty parameter (λ) was selected using a 10-fold cross-validation with training data only. LOOCV involves ‘training’ an algorithm with all subjects but one whilst the ‘left out’ subject is used for algorithm testing (Johnston et al., 2013). This procedure is repeated until all subjects are ‘left out’ for algorithm testing atleast once and prediction accuracy, sensitivity, specificity estimated. Clinical and demographic characteristics of BD patients included in both phenotype I and phenotype II with a matched healthy control group are summarized in Table 3.
Phenotype validation using tract-based spatial statistics diffusion weighted imaging data
To establish whether the identified behavioral phenotypes were neurobiologically distinct, we examined whether diffusion weighted FA and MD volumes pre-processed using tract-based spatial statistics (TBSS) can distinguish individual patients in phenotype I from those in phenotype II as well as healthy controls. Specifically, the TBSS pre-processed FA and MD volumes were entered into a two-group (e.g. phenotype I vs phenotype II) Elastic Net machine learning algorithm (Zou and Hastie, 2005)– which was ‘trained’ to predict individual subjects' phenotypic labels and resulting model prediction accuracy, specificity and sensitivity calculated. The Elastic Net algorithm follows a similar formulation as the LASSO albeit with an additional penalty term to encourage variable grouping and a more stable solution (Mwangi et al., 2014c; Ogutu et al., 2012; Zou and Hastie, 2005). The additional penalty term in the Elastic Net translates into the following objective function (Zou and Hastie, 2005)
Similar to above, xij stands for predictor variables (e.g. TBSS derived FA or MD) while yi represents corresponding target labels (e.g. 1 - phenotype I and 2 - phenotype II), where i = 1, 2, …N represents observations or subjects and j = 1, 2, …P represents number of predictor variables (features). The Elastic Net algorithm parameters (λ1 and λ2) were selected using a 10-fold grid-seach process using training data only and the algorithm's objective function solved through the coordinate descent algorithm using a MATLAB (The Mathworks, Inc) toolbox provided by Friedman and colleagues (Friedman et al., 2010). Importantly, the Elastic Net algorithm was used in the neuroimaging phenotype validation step as unlike the LASSO where number of relevant predictor variables (non-zero coefficients) may not exceed number of observations, the Elastic Net allows relevant variables to exceed number of observations (Zou and Hastie, 2005) and therefore more suitable in visualizing relevant group-level statistical maps. Similar to agove, a generalized linear model was used to estimate a proability of an individual subject belonging to phenotype I or phenotype II patieng groups given TBSS pre-processed FA and MD features. In addition, the Elastic Net algorithm was also ‘trained’ to distinguish individual phenotype I or phenotype II patients from healthy controls. In each comparison (e.g. phenotype I vs healthy conrols) a LOOCV approach was used to establish prediction accuracy whilst algorithm parameters were selected using a 10-fold cross-validation approach using training data only as shown in Figure 2. Importantly, a ‘consensus group-level map’ higlightinig FA or MD differences among groups (e.g. phenotype I vs phenotype II) was generated by identifying features identified by the algorithm as ‘relevant’ - by the virtue of having a non-zero model coefficients across all LOOCV iterations. A ‘consensus map’ approach has previously been used to summarize group-level differences in machine learning studies (Dosenbach et al., 2010; Mwangi et al., 2013). Lastly, prediction accuracy, sensitivity and specificity in identifying patients from both phenotypes using FA and MD features were summarized and reported. A similar calculation was performed between the healthy control and patient phenotypic groups.
Figure 2.
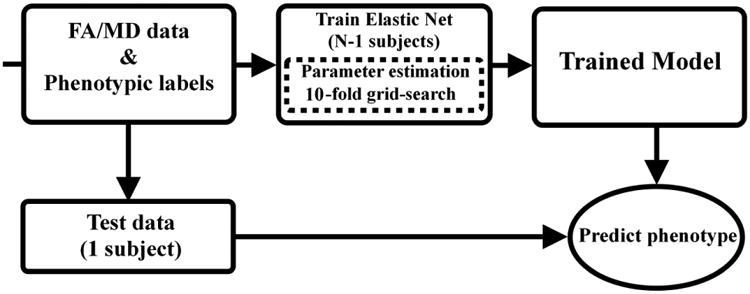
Flow diagram showing the leave-one-out cross-validation process used with the Elastic Net algorithm in predicting subjects' phenotypic lables. Algorithm regularization parameters were selected using a 10-fold grid-search process using training data only to avoid circularity or double dipping.
Results
Table 1 summarizes the sociodemographic and clinical details of BD patients used in the phenotype identification process. The unsupervised machine learning approach identified two phenotypes as shown in Figure 1. Detailed sociodemographic and clinical characteristics of both phenotypes are shown in Table 3. The optimal number of principal components in PCA was selected using a ‘grid-search’ procedure by maximizing the average silhouette width value as shown in Figure 3. The most optimal silhouette width value was 0.74. Raw CANTAB measurements (without PCA data reduction) predicted individual subjects' phenotypic labels with 94% accuracy, 97% specificity, 92% sensitivity, 0.945 area under receiver operating characteristic curve and chi-square (p < 0.005) as shown in Figure 4. As illustrated in Table 4 the CANTAB tasks identified by the LASSO algorithm as most relevant in distinguishing phenotype I and phenotype II subjects included the AGN (relevant variables included the mean RT to correct trials across non-shift blocks, in particular those starting with positive stimuli, and during shift blocks starting with negative stimuli, and the number of commission errors during non-shift blocks starting with neutral stimuli), the CGT (quality of decision making), the IED (accuracy on the extradimensional component) and CRT (general alertness and motor speed).
Figure 3.
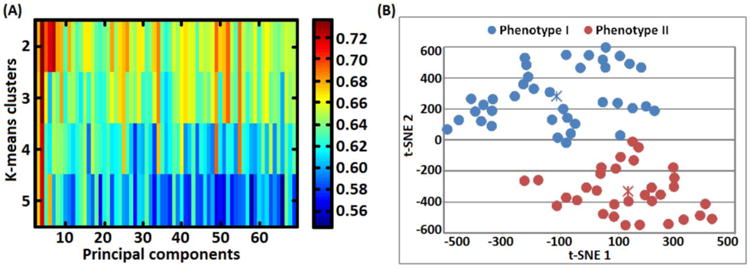
A) Grid-search plot illustrating a search for optimal number of principal components and k-means number of clusters that lead to a high silhouette width value. The grid-search process selected 4 components accounting for 43.09 % variance and 2 clusters with a silhouette width value of 0.735. B) 2-dimensional (2D) visualization of phenotype I and phenotype II patient groups. Low dimensional 2D points were generated using the t-distributed stochastic neighbor embedding (t-SNE) technique by mapping the most relevant principal components (leading to a high silhouette width value) from a high-dimensional (4D) into a lower visually plausible 2D space. Importantly, the two phenotypes did not overlap with DSM-IV related categories (BP I, II and BP-NOS). Notably, t-SNE units are arbitrary and they only depict subjects' similarities based on euclidean distances.
Figure 4.
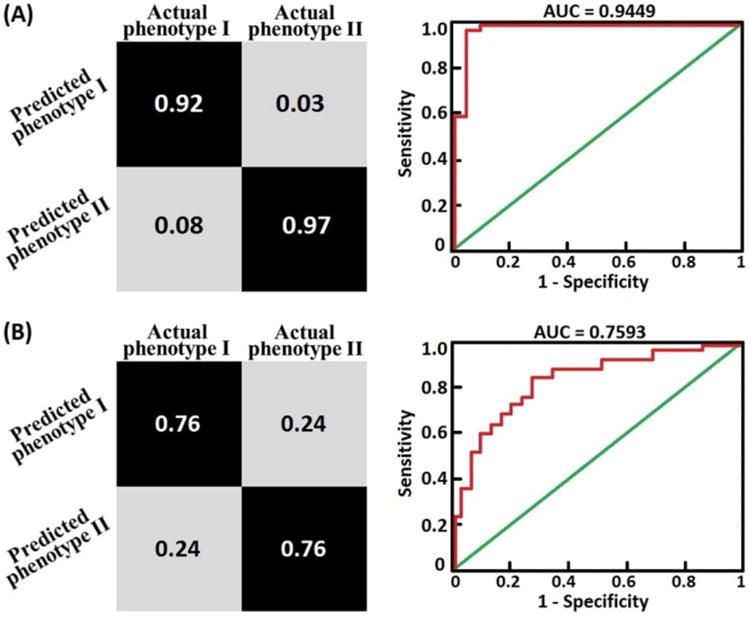
A) Confusion matrix and ROC curve generated from a LASSO algorithm which was trained using ‘raw’ neurocognitive tasks scores (see Table 2) to predict individual subjects phenotypic labels. The model predicted individual subjects phenotypic labels with 94% accuracy (sensitivity= 92%, specificity =97%) and area under ROC curve (AUC) = 0.9449. B) Confusion matrix and ROC curve generated from a Elastic Net algorithm which was trained using whole brain fractional anisotropy values to predict individual subjects phenotypic labels. The model predicted individual subjects phenotypic labels with 76% accuracy (sensitivity= 76%, specificity =76%) and area under ROC curve (AUC) = 0.7593.
Table 4.
Cognitive tasks scores most relevant in distinguishing phenotype I from phenotype II patients using LASSO.
| Cognitive task feature | Research domain criteria (RDOC) construct | Mean beta value | Beta % | Comparison |
|---|---|---|---|---|
| AGN mean RT nonshift blocks (ms) | Cognitive control | 1.5111 | 25.09% | PH2 > PH1 |
| AGN mean RT to nonshift blocks starting with positive stimuli) (ms) | Social communication | 0.7858 | 13.05% | PH2 > PH1 |
| AGN mean RT to shift blocks starting with negative stimuli (ms) | Social communication | 0.7283 | 12.09% | PH2 > PH1 |
| AGN N commissions errors in shift blocks starting with positive stimuli | Social communication | -0.6200 | 10.29% | PH1 > PH2 |
| AGN N commissions errors in nonshift blocks starting with neutral stimuli | Cognitive control | -0.5027 | 8.35% | PH1 > PH2 |
| CGT deliberation time descending blocks (ms) | Cognitive control | -0.3644 | 6.05% | PH1> PH2 |
| CRT minimum RT (ms) | Arousal | 0.3037 | 5.04% | PH2 > PH1 |
| CGT quality of decision making | Valence system | 0.2990 | 4.96% | PH2 > PH1 |
| IED N errors during Intradimensional component | Cognitive control | -0.2480 | 4.12% | PH1 > PH2 |
| CGT deliberation time (ms) | Cognitive control | -0.2428 | 4.03% | PH1 > PH2 |
PH1 – Phenotype I, PH2 – Phenotype II, AGN – Affective Go/No-go, CGT – Cambridge Gambling Task, CRT – Choice Reaction Time, IED – Intra/Extradimensional Set Shift, RT – Reaction time.
Table 5 summarizes prediction results in distinguishing individual patients in phenotype I from those in phenotype II using neurocognition and diffusion weighted measuremetns (FA and MD). Specifically, Elastic Net algorithm ‘trained’ using FA data distinguished both phenotypes with 75.9 % accuracy. In addition, individual phenotype II patients were distinguished from healthy controls with 92% accuracy as shown in Table 5. The most relevant anatomical regions in distinguishing phenotype I and phenotype II patients included frontal white matter tracts such as; inferior frontal-occipital fasciculus (IFOF) and the minor forceps of the corpus callosum. In particular, phenotype II showed reduced FA and increased MD values in a significant cluster within the IFOF as compared to phenotype I patients and healthy controls as shown in Figure 5.
Table 5.
Prediction and validation results of Healthy controls, Phenotype I and Phenotype II patients. Notably, this table was generated ‘post-hoc’ – after the unsupervised machine learning step.
| Feature domain | Group | Acc | Sens | Spec | PPV | NPV | P | AUC |
|---|---|---|---|---|---|---|---|---|
| Cognition | (PH1, PH2) | 0.9429 | 0.9211 | 0.9688 | 0.9722 | 0.9118 | < 0.0001 | 0.9449 |
| (HC, PH1) | 0.8553 | 0.8158 | 0.894 | 0.8857 | 0.8293 | < 0.0001 | 0.8553 | |
| (HC, PH2) | 0.7969 | 0.7500 | 0.8438 | 0.8276 | 0.7714 | < 0.0001 | 0.7969 | |
| FA | (PH1, PH2) | 0.7593 | 0.7600 | 0.7586 | 0.7308 | 0.7857 | 0.0001 | 0.7593 |
| (HC, PH1) | 0.5714 | 0.5517 | 0.5926 | 0.5926 | 0.5517 | 0.2802 | 0.5722 | |
| (HC, PH2) | 0.9231 | 0.8800 | 0.9630 | 0.9565 | 0.8966 | < 0.0001 | 0.9215 | |
| MD | (PH1, PH2) | 0.6481 | 0.6800 | 0.6207 | 0.6071 | 0.6923 | 0.0275 | 0.6503 |
| (HC, PH1) | 0.5714 | 0.5517 | 0.5926 | 0.5926 | 0.5517 | 0.2802 | 0.5722 | |
| (HC, PH2) | 0.8654 | 0.8800 | 0.8519 | 0.8462 | 0.8846 | < 0.0001 | 0.8659 |
FA- fractional anisotropy, MD- mean diffusivity, HC- healthy controls, PH1- Phenotype I, PH2- Phenotype II, Acc- Accuracy, Sens- Sensitivity, Spec- Specificity, PPV- Positive predictive values, NPV- Negative predictive values, AUC- Area under curve.
Figure 5.
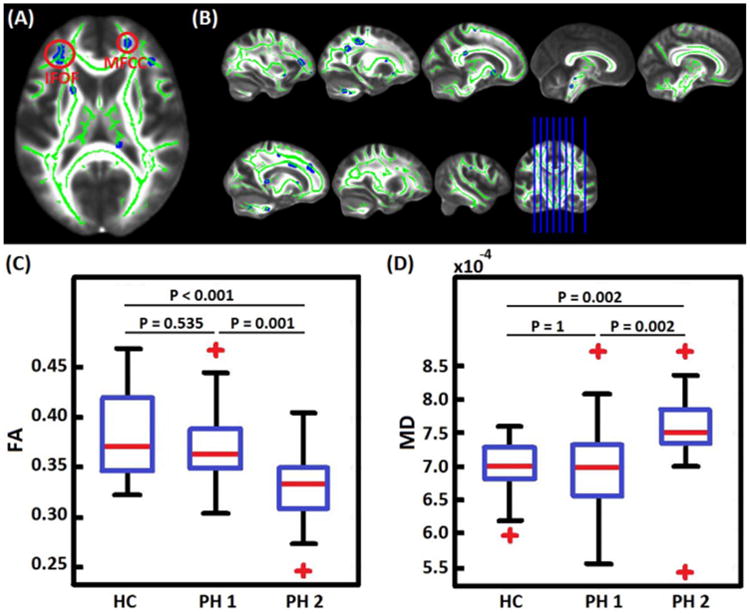
A) The most relevant white-matter tracts in distinguishing phenotype I from phenotype II patients at an individual subject level. IFOF – inferior fronto-occipital fasciculus, MFCC - minor forceps of the corpus callosum. B) A multisclice view of the most relevant clusters. C) A comparison of FA values within IFOF cluster in healthy controls (HC), phenotype I (PH1) and phenotype II (PH2). Phenotype II patients showed reduced FA values as compared to healthy controls and phenotype I patients. D) A comparison of MD values within IFOF cluster in healthy controls (HC), phenotype I (PH1) and phenotype II (PH2). Phenotype II patients showed increased MD values as compared to healthy controls and phenotype I patients. These analysis of Variance (ANOVA) statistical tests were performed using SPSS Version 20, IBM Inc and corrected for multiple comparisons using the Bonferroni method.
Discussion
We have presented a proof-of-concept study investigating the utility of an unsupervised data-driven machine learning approach to identify biologically distinct and less heterogenous phenotypes in bipolar disorders. Most importantly, our approach utilizes neurocognitive measurements to identify unique phenotypes which are validated using neuroanatomical markers. The most compelling finding of this study is the identification of two unique phenotypes (phenotype I, phenotype II) which did not overlap with DSM-IV derived categories (BD I, BD II and BD-NOS). Notably, phenotypes I and II display distinct cognitive profiles on the cognitive control, social communication and arousal RDoC constructs. Markedly, neuroimaging measurements of microstructural white matter diffusivity successfully identified individual subjects from both phenotypes with high accuracy. In addition, individualized prediction distinguished patients from both phenotypes from healthy controls – in particular phenotype II patients.
Most relevant neurocognitive and neuroanatomical measurements
The most relevant CANTAB measurements in separating the two phenotypes included measures of reaction time in the AGN task, response accuracy in the ID/ED, and quality of decision making during the CGT task. Importantly, we report two key observations and characteristics of both phenotypes from the CANTAB measurements. First, phenotype I patients exhibited reduced response accuracy during the AGN and IED tasks and slow decision making during the CGT task. Second, Phenotype II patients exhibited slower reaction time in the AGN and CRT tasks but their decision-making approach was more efficient than that of phenotype I patients. Therefore, it could be argued that Phenotype II patients completed the AGN and CGT tasks in a strategic manner, by trading speed for accuracy. The slow reaction times in both the AGN and CRT tasks may also be consistent with psychomotor slowing. Notably, in the AGN task individuals with phenotype I made a greater number of commission errors in response to positive and neutral stimuli but not to negative stimuli when compared to individuals with phenotype I. This pattern of responses is in line with concept of “negative affective bias”, a well-established feature of mood disorders including bipolar disoders (Bauer et al., 2015; Gotlib et al., 2005). Negative affective bias is defined by the inability to disengage from processing negative stimuli even after they disappear. This inability impairs the processing of subsequent stimuli and could explain the reduced accuracy in processing positive and neutral stimuli (Singhal et al., 2012), as observed in the current study. Previous studies found that verbal memory, psychomotor speed, executive functioning, and to a less extent visual memory and attention, are greatly impaired in BPD patients (Bora et al., 2009; Cavanagh et al., 2002; Glahn et al., 2007; Lopes and Fernandes, 2012; Martínez-Arán et al., 2004; Quraishi and Frangou, 2002; Sapin et al., 1987; Zubieta et al., 2001). In a recent study, patients with BD type I displayed a perseverative and risky behavior in the CGT (Linke et al., 2013). Notably, relatives of BD patients displayed a similar cognitive performance, which along with our present findings; indicate that the performance in the CGT task may be a potential marker of disease susceptibility. However, other studies examining executive functions in BD are more controversial as they suggest a possible effect of mood phase on cognitive functioning. For instance, Clark and colleagues found that euthymic BD encountered set-shifting difficulties during the IED task (Clark et al., 1999), while another study found that the performance of a bipolar sample in a depressed/mixed state was comparable to that of healthy volunteers (Sweeney et al., 2000). It is noteworthy that in the current study, phenotype I and phenotype II patients were comparable in regard to subjects' current level of mood symptoms. Therefore, differences in performance in the CANTAB tasks are unlikely to be related to the mood of the participants. Additionally, the reported executive function impairment (e.g. slow reaction times and errors in the AGN task) in both phenotypes is consistent with previous evidence of structural and functional abnormalities in anatomical regions responsible for executive control in pediatric and adult populations with BD (Brambilla et al., 2002; Hajek et al., 2005; Houenou et al., 2011; Lim et al., 2013; Sassi et al., 2004; Soares and Mann, 1997). Compared to individuals with phenotype I, those with phenotype II displayed slowed reaction times in the AGN and CRT tasks. Notably the quality of the decision making (e.g. gambling on stimuli with a more likely outcome) was higher in phenotype II than in phenotype I. Phenotype I is characterized by an overall higher number of commission errors in the AGN and ID/IED tasks, and slowed decision making in the CGT task.
In terms of imaging results, white matter diffusivity values within the frontal white matter tracts were most relevant in distinguishing individual patients from both phenotypes. As shown in Table 6, the most relevant clusters were within the inferior fronto-occipital fasciculus (IFOF) and the minor forceps of the corpus callosum. In particular, phenotype II patients showed reduced FA values and increased MD values within the IFOF as compared to phenotype I patients and healthy controls. The IFOF is a white matter pathway that connects the posterior or superior temporal regions with the dorsolateral prefrontal cortex (McCrea, 2008). Although the IFOF connects the frontal temporal and occipital lobes, it's main functions are still not very well understood although there is evidence that this white matter pathway connects the prefrontal cortex and auditory as well as visual association cortex and associated with functions such as semantic processing and attentional set-shifting ability (Kvickström et al., 2011). A recent meta-analysis reported reduced FA in the IFOF of bipolar disorder patients as compared to healthy controls (Vederine et al., 2011). However, in the current study, we observed reduced FA and increased MD in the IFOF of phenotype II patients as compared to healthy controls and not in phenotype I patients. Other white matter anatomical regions most relevant in distinguishing phenotype I from phenotype II patients included; isthmus of the corpus callosum, mid colossal pathways, anterior forceps of corpus callosum as well as the cingulum. In summary, phenotype II patients reported reduced FA and increased MD values within the IFOF white-matter tract and longer reaction time in the AGN cognitive task.
Table 6.
White matter tracts most relevant in distinguishing individual phenotype I patients from phenotype II patients using the Elastic Net algorithm.
| Rank | Anatomical region | MNI Coordinates (x, y, z) | Cluster size | *Average beta weight | Beta % |
|---|---|---|---|---|---|
| 1 | IFOF | -31, 35, 11 | 92 | -5.778 | 6.56% |
| 2 | Mid colossal pathways | -22, -30, 43 | 79 | 4.379 | 4.97% |
| 3 | Isthmus of the corpus callosum | -22, -52, 33 | 104 | 3.849 | 4.37% |
| 4 | MFCC | 21, 41, 19 | 108 | -2.749 | 3.12% |
| 5 | Cingulum | 17, 7, 34 | 62 | 0.846 | 0.96% |
IFOF – inferior fronto-occipital fasciculus, MFCC – minor forceps of the corpus callosum.
Beta weights were averaged within the cluster. Anatomical regions are listed based on absolute average beta weight within the cluster. Negative beta weights indicate FA reductions in phenotype II patients as compared to phenotype I
The current application of the RDoC approach is to be viewed as a first step in parsing out heterogeneity in complex psychiatric disorders. However, although the RDoC framework is increasingly being used in clinical research, to the best of our knowledge this is amongst the first studies that have used the RDoC cognitive constructs in combination with neuroimaging measures to identify and validate biologically distinct disease phenotypes. Consequently, given the lack of clear guidelines to “translate” classical neuropsychological measures into the RDoC framework, this approach is challenging as it relies on the subjective evaluation of the definition of each cognitive domains, but certainly constitutes a starting point to improve understanding of the RDoC dimensions.
Limitations
Potential limitations of this study should be noted. First, the study sample size was small and therefore this approach will need to be validated using a larger sample size. However, the generalization ability of the supervised machine learning algorithms using both neurocognitive data and diffusion weighted neuroimaging scan data were significant (chi-square p<0.05). This indicates that the sample size may have been adequate to identify biologically distinct phenotypes. Our sample did not include other DSM related categories (e.g. major depression or schizophrenia) and future work will focus on extending the sample in a ‘dimensional’ approach to include other psychiatric syndromes and spectrums (e.g. depression, psychosis) as also hypothesized elsewhere (Insel and Cuthbert, 2015; Mwangi et al., 2014a; Stephan et al., 2015). Markedly, future work will also focus on examining the utility of multi-class or multinomial classification methods in identifying subjects from all groups (e.g. phenotype I vs. phenotype II vs. healthy controls) as well as validating identified phenotypes using other biological markers such as genetics.
Conclusion
We highlight three notable observations from this proof-of-concept study. First, multivariate patterns of neurocognitive measurements are potentially useful in patient stratification and unearthing distinct or monolithic phenotypes in heterogeneous patient populations. Second, neuroimaging measurements are potential validators and predictors of phenotypic groups identified using behavioral data. Third, machine learning predictive classification techniques (e.g. LASSO, Elastic Net) give a strong indication that identified phenotypes are biologically relevant and may be useful clinically. Data-driven and biologically relevant disease phenotypes identified using this approach will give the psychiatric research community a unique opportunity to identify objective and biologically relevant disease categories. Markedly, if we are to determine objective and clinically meaningful phenotypic bio-signatures which can be identified through quick and hazard free biological tests – this will lead to improved patient care and better targeted therapeutic interventions.
Supplementary Material
Highlights.
An unsupervised machine learning method and neurocognitive data used to identify two phenotypes
LASSO distinguished two phenotypes using neurocognitive data with 94% accuracy.
Elastic Net validates differences of the two phenotypes using FA data with 76% accuracy.
Healthy controls are further used to validate differences between the two phenotypes.
Acknowledgments
Disclosures and Acknowledgments: Supported in part by NIMH grant R01 085667, the Dunn Foundation and the Pat Rutherford, Jr. Endowed Chair in Psychiatry to JCS.
Footnotes
Financial Disclosures: Jair C. Soares has participated in research funded by Forest, Merck, BMS, GSK and has been a speaker for Pfizer and Abbott. Marsal Sanches has received research grants from Janssen. All other authors report no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Association, A.P. Diagnostic And Statistical Manual Of Mental Disorders DSM-IV-TR Fourth Edition (Text Revision) Author: American Psychiatr; 2000. [Google Scholar]
- Bauer IE, Frazier TW, Zunta-Soares GB, Soares JC. Affective Processing in Pediatric Bipolar Disorder and Offspring of Bipolar Parents. 2015 doi: 10.1089/cap.2015.0076. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora E, Yucel M, Pantelis C. Cognitive endophenotypes of bipolar disorder: a meta-analysis of neuropsychological deficits in euthymic patients and their first-degree relatives. Journal of affective disorders. 2009;113:1–20. doi: 10.1016/j.jad.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Brambilla P, Nicoletti MA, Harenski K, Sassi RB, Mallinger AG, Frank E, Kupfer DJ, Keshavan MS, Soares JC. Anatomical MRI study of subgenual prefrontal cortex in bipolar and unipolar subjects. Neuropsychopharmacology. 2002;27:792–799. doi: 10.1016/S0893-133X(02)00352-4. [DOI] [PubMed] [Google Scholar]
- Brodersen KH, Deserno L, Schlagenhauf F, Lin Z, Penny WD, Buhmann JM, Stephan KE. Dissecting psychiatric spectrum disorders by generative embedding. NeuroImage: Clinical. 2014;4:98–111. doi: 10.1016/j.nicl.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick K, Russo M, Frangou S, Mahon K, Braga R, Shanahan M, Malhotra A. Empirical evidence for discrete neurocognitive subgroups in bipolar disorder: clinical implications. Psychological Medicine. 2014:1–14. doi: 10.1017/S0033291714000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Van Beck M, Muir W, Blackwood D. Case—control study of neurocognitive function in euthymic patients with bipolar disorder: an association with mania. The British Journal of Psychiatry. 2002;180:320–326. doi: 10.1192/bjp.180.4.320. [DOI] [PubMed] [Google Scholar]
- Clark L, Goodwin G, Iversen S. Frontal lobe function in the euthymic phase of bipolar disorder. Soc Neurosci Abstr 1999 [Google Scholar]
- Cuthbert BN, Insel TR. Toward new approaches to psychotic disorders: the NIMH Research Domain Criteria project. Schizophrenia bulletin. 2010;36:1061–1062. doi: 10.1093/schbul/sbq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson J, Turnbull CD, Strickland R, Miller R, Graves K. The Montgomery-Åsberg Depression Scale: reliability and validity. Acta Psychiatrica Scandinavica. 1986;73:544–548. doi: 10.1111/j.1600-0447.1986.tb02723.x. [DOI] [PubMed] [Google Scholar]
- Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Bathula D, Nikolas MA, Nigg JT. Distinct neuropsychological subgroups in typically developing youth inform heterogeneity in children with ADHD. Proceedings of the National Academy of Sciences. 2012;109:6769–6774. doi: 10.1073/pnas.1115365109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV® Axis I Disorders (SCID-I), Clinician Version, Administration Booklet. American Psychiatric Pub; 2012. [Google Scholar]
- Frangou S. Snipping at the Endophenotypic Space. American Journal of Psychiatry. 2013;170:1223–1225. doi: 10.1176/appi.ajp.2013.13081116. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of statistical software. 2010;33:1. [PMC free article] [PubMed] [Google Scholar]
- Galvez JF, Bauer IE, Sanches M, Wu HE, Hamilton JE, Mwangi B, Kapczinski FP, Zuntasoares G, Soares JC. Shared clinical Associations between obesity and impulsivity in rapid cycling bipolar disorder: A systematic review. Journal of Affective Disorders. 2014 doi: 10.1016/j.jad.2014.05.054. [DOI] [PubMed] [Google Scholar]
- Geisler D, Walton E, Naylor M, Roessner V, Lim KO, Schulz SC, Gollub RL, Calhoun VD, Sponheim SR, Ehrlich S. Brain structure and function correlates of cognitive subtypes in schizophrenia. Psychiatry Research: Neuroimaging. 2015 doi: 10.1016/j.pscychresns.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Bearden CE, Barguil M, Barrett J, Reichenberg A, Bowden CL, Soares JC, Velligan DI. The neurocognitive signature of psychotic bipolar disorder. Biological psychiatry. 2007;62:910–916. doi: 10.1016/j.biopsych.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Gotlib IH, Traill SK, Montoya RL, Joormann J, Chang K. Attention and memory biases in the offspring of parents with bipolar disorder: indications from a pilot study. Journal of Child Psychology and Psychiatry. 2005;46:84–93. doi: 10.1111/j.1469-7610.2004.00333.x. [DOI] [PubMed] [Google Scholar]
- Hajek T, Carrey N, Alda M. Neuroanatomical abnormalities as risk factors for bipolar disorder. Bipolar Disord. 2005;7:393–403. doi: 10.1111/j.1399-5618.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. Applied statistics. 1979:100–108. [Google Scholar]
- Heinrichs RW, Awad AG. Neurocognitive subtypes of chronic schizophrenia. Schizophrenia research. 1993;9:49–58. doi: 10.1016/0920-9964(93)90009-8. [DOI] [PubMed] [Google Scholar]
- Hermens DF, Redoblado Hodge MA, Naismith SL, Kaur M, Scott E, Hickie IB. Neuropsychological clustering highlights cognitive differences in young people presenting with depressive symptoms. Journal of the International Neuropsychological Society. 2011;17:267–276. doi: 10.1017/S1355617710001566. [DOI] [PubMed] [Google Scholar]
- Hickie IB, Scott J, Hermens DF, Scott EM, Naismith SL, Guastella AJ, Glozier N, McGorry PD. Clinical classification in mental health at the cross-roads: which direction next? BMC medicine. 2013;11:125. doi: 10.1186/1741-7015-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld R, Vornik LA. Bipolar disorder--costs and comorbidity. The American journal of managed care. 2005;11:S85–90. [PubMed] [Google Scholar]
- Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nature Reviews Clinical Oncology. 2011;8:184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- Houenou J, Frommberger J, Carde S, Glasbrenner M, Diener C, Leboyer M, Wessa M. Neuroimaging-based markers of bipolar disorder: evidence from two meta-analyses. J Affect Disord. 2011;132:344–355. doi: 10.1016/j.jad.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. American Journal of Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Insel TR. The NIMH research domain criteria (RDoC) project: precision medicine for psychiatry. American Journal of Psychiatry. 2014;171:395–397. doi: 10.1176/appi.ajp.2014.14020138. [DOI] [PubMed] [Google Scholar]
- Insel TR, Cuthbert BN. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE. Diffusion MRI: from quantitative measurement to in vivo neuroanatomy. Academic Press; 2013. [Google Scholar]
- Johnston B, Mwangi B, Matthews K, Coghill D, Steele J. Predictive classification of individual magnetic resonance imaging scans from children and adolescents. European child & adolescent psychiatry. 2013;22:733–744. doi: 10.1007/s00787-012-0319-0. [DOI] [PubMed] [Google Scholar]
- Johnston BA, Mwangi B, Matthews K, Coghill D, Konrad K, Steele JD. Brainstem abnormalities in attention deficit hyperactivity disorder support high accuracy individual diagnostic classification. Human brain mapping. 2014 doi: 10.1002/hbm.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe I. Principal component analysis. Wiley Online Library; 2005. [Google Scholar]
- Karalunas SL, Fair D, Musser ED, Aykes K, Iyer SP, Nigg JT. Subtyping Attention-Deficit/Hyperactivity Disorder Using Temperament Dimensions: Toward Biologically Based Nosologic Criteria. JAMA psychiatry. 2014 doi: 10.1001/jamapsychiatry.2014.763. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI. Circular analysis in systems neuroscience: the dangers of double dipping. Nature neuroscience. 2009;12:535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz MM, Gerraty RT. A meta-analytic investigation of neurocognitive deficits in bipolar illness: profile and effects of clinical state. Neuropsychology. 2009;23:551. doi: 10.1037/a0016277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvickström P, Eriksson B, van Westen D, Lätt J, Elfgren C, Nilsson C. Selective frontal neurodegeneration of the inferior fronto-occipital fasciculus in progressive supranuclear palsy (PSP) demonstrated by diffusion tensor tractography. BMC neurology. 2011;11:13. doi: 10.1186/1471-2377-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavagnino L, Amianto F, Mwangi B, D'Agata F, Spalatro A, Zunta-Soares G, Abbate Daga G, Mortara P, Fassino S, Soares J. Identifying neuroanatomical signatures of anorexia nervosa: a multivariate machine learning approach. Psychological medicine. 2015:1–8. doi: 10.1017/S0033291715000768. [DOI] [PubMed] [Google Scholar]
- Lee R, Hickie I, Hermens D. Letter to the Editor: Neuropsychological subgroups are evident in both mood and psychosis spectrum disorders. Psychological Medicine. 2014;44:2015–2015. doi: 10.1017/S0033291714001019. [DOI] [PubMed] [Google Scholar]
- Lim CS, Baldessarini RJ, Vieta E, Yucel M, Bora E, Sim K. Longitudinal neuroimaging and neuropsychological changes in bipolar disorder patients: Review of the evidence. Neuroscience & Biobehavioral Reviews. 2013;37:418–435. doi: 10.1016/j.neubiorev.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Linke J, King AV, Poupon C, Hennerici MG, Gass A, Wessa M. Impaired anatomical connectivity and related executive functions: differentiating vulnerability and disease marker in bipolar disorder. Biological psychiatry. 2013;74:908–916. doi: 10.1016/j.biopsych.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Lopes R, Fernandes L. Bipolar disorder: clinical perspectives and implications with cognitive dysfunction and dementia. Depression research and treatment. 2012;2012 doi: 10.1155/2012/275957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Arán A, Vieta E, Colom F, Torrent C, Sánchez-Moreno J, Reinares M, Benabarre A, Goikolea J, Brugue E, Daban C. Cognitive impairment in euthymic bipolar patients: implications for clinical and functional outcome. Bipolar disorders. 2004;6:224–232. doi: 10.1111/j.1399-5618.2004.00111.x. [DOI] [PubMed] [Google Scholar]
- McCrea SM. Bipolar disorder and neurophysiologic mechanisms. Neuropsychiatric disease and treatment. 2008;4:1129. doi: 10.2147/ndt.s4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Cuthbert BN. Research Domain Criteria: cognitive systems, neural circuits, and dimensions of behavior. Dialogues Clin Neurosci. 2012;14:29–37. doi: 10.31887/DCNS.2012.14.1/smorris. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Oerlinghausen B, Berghöfer A, Bauer M. Bipolar disorder. The Lancet. 2002;359:241–247. doi: 10.1016/S0140-6736(02)07450-0. [DOI] [PubMed] [Google Scholar]
- Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. The Lancet. 2013;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- Mwangi B, Ebmeier KP, Matthews K, Steele JD. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain. 2012a;135:1508–1521. doi: 10.1093/brain/aws084. [DOI] [PubMed] [Google Scholar]
- Mwangi B, Hasan KM, Soares JC. Prediction of individual subject's age across the human lifespan using diffusion tensor imaging: a machine learning approach. NeuroImage. 2013;75:58–67. doi: 10.1016/j.neuroimage.2013.02.055. [DOI] [PubMed] [Google Scholar]
- Mwangi B, Matthews K, Steele JD. Prediction of illness severity in patients with major depression using structural MR brain scans. Journal of Magnetic Resonance Imaging. 2012b;35:64–71. doi: 10.1002/jmri.22806. [DOI] [PubMed] [Google Scholar]
- Mwangi B, Soares JC, Hasan KM. Visualization and unsupervised predictive clustering of high-dimensional multimodal neuroimaging data. Journal of neuroscience methods. 2014a doi: 10.1016/j.jneumeth.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Mwangi B, Spiker D, Zunta-Soares GB, Soares JC. Prediction of pediatric bipolar disorder using neuroanatomical signatures of the amygdala. Bipolar disorders. 2014b doi: 10.1111/bdi.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi B, Tian TS, Soares JC. A review of feature reduction techniques in neuroimaging. Neuroinformatics. 2014c;12:229–244. doi: 10.1007/s12021-013-9204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Mental Health. Health, U.S.D.O.H.H.S.-N.I.o, editor. National Instutes of Mental Health Strategic Plan. 2008 http://www.nimh.nih.gov/about/strategic-planning-reports/nimh-strategic-plan-2008.pdf.
- Ogutu JO, Schulz-Streeck T, Piepho HP. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions. BMC proceedings BioMed Central Ltd. 2012:S10. doi: 10.1186/1753-6561-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L, Ladouceur CD, Phillips ML, Brammer M, Mourao-Miranda J. What does brain response to neutral faces tell us about major depression? Evidence from machine learning and fMRI. PloS one. 2013;8:e60121. doi: 10.1371/journal.pone.0060121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plis SM, Hjelm DR, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, Johnson HJ, Paulsen JS, Turner JA, Calhoun VD. Deep learning for neuroimaging: a validation study. Frontiers in neuroscience. 2014;8 doi: 10.3389/fnins.2014.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quraishi S, Frangou S. Neuropsychology of bipolar disorder: a review. Journal of affective disorders. 2002;72:209–226. doi: 10.1016/s0165-0327(02)00091-5. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Halaris A, Selemon LD. Reductions in neuronal and glial density characterize the dorsolateral prefrontal cortex in bipolar disorder. Biological psychiatry. 2001;49:741–752. doi: 10.1016/s0006-3223(01)01080-0. [DOI] [PubMed] [Google Scholar]
- Robbins T, James M, Owen A, Sahakian B, McInnes L, Rabbitt P. Cambridge Neuropsychological Test Automated Battery (CANTAB): a factor analytic study of a large sample of normal elderly volunteers. Dementia and Geriatric Cognitive Disorders. 1994;5:266–281. doi: 10.1159/000106735. [DOI] [PubMed] [Google Scholar]
- Rocha-Rego V, Jogia J, Marquand A, Mourao-Miranda J, Simmons A, Frangou S. Examination of the predictive value of structural magnetic resonance scans in bipolar disorder: a pattern classification approach. Psychological Medicine. 2014;44:519–532. doi: 10.1017/S0033291713001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of computational and applied mathematics. 1987;20:53–65. [Google Scholar]
- Sapin LR, Berrettini WH, Nurnberger JI, Jr, Rothblat LA. Mediational factors underlying cognitive changes and laterality in affective illness. Biological psychiatry. 1987;22:979–986. doi: 10.1016/0006-3223(87)90007-2. [DOI] [PubMed] [Google Scholar]
- Sassi RB, Brambilla P, Hatch JP, Nicoletti MA, Mallinger AG, Frank E, Kupfer DJ, Keshavan MS, Soares JC. Reduced left anterior cingulate volumes in untreated bipolar patients. Biological psychiatry. 2004;56:467–475. doi: 10.1016/j.biopsych.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Savitz J, Rauch SL, Drevets W. Clinical application of brain imaging for the diagnosis of mood disorders: the current state of play. Molecular psychiatry. 2013;18:528–539. doi: 10.1038/mp.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, Nieuwenhuis M, van Haren NE, Abramovic L, Scheewe TW, Brouwer RM, Hulshoff Pol HE, Kahn RS. Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. NeuroImage. 2014;84:299–306. doi: 10.1016/j.neuroimage.2013.08.053. [DOI] [PubMed] [Google Scholar]
- Singhal A, Shafer AT, Russell M, Gibson B, Wang L, Vohra S, Dolcos F. Electrophysiological correlates of fearful and sad distraction on target processing in adolescents with attention deficit-hyperactivity symptoms and affective disorders. Frontiers in integrative neuroscience. 2012;6 doi: 10.3389/fnint.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Soares JC, Mann JJ. The anatomy of mood disorders—review of structural neuroimaging studies. Biological Psychiatry. 1997;41:86–106. doi: 10.1016/s0006-3223(96)00006-6. [DOI] [PubMed] [Google Scholar]
- Stephan KE, Iglesias S, Heinzle J, Diaconescu AO. Translational perspectives for computational neuroimaging. Neuron. 2015;87:716–732. doi: 10.1016/j.neuron.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Sweeney JA, Kmiec JA, Kupfer DJ. Neuropsychologic impairments in bipolar and unipolar mood disorders on the CANTAB neurocognitive battery. Biological psychiatry. 2000;48:674–684. doi: 10.1016/s0006-3223(00)00910-0. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996:267–288. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2011a;73:273–282. [Google Scholar]
- Tibshirani RJ. The solution path of the generalized lasso. Stanford University; 2011b. [Google Scholar]
- Van der Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:85. [Google Scholar]
- Vederine FE, Wessa M, Leboyer M, Houenou J. A meta-analysis of whole-brain diffusion tensor imaging studies in bipolar disorder. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2011;35:1820–1826. doi: 10.1016/j.pnpbp.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Wardenaar KJ, de Jonge P. Diagnostic heterogeneity in psychiatry: towards an empirical solution. BMC medicine. 2013;11:201. doi: 10.1186/1741-7015-11-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, Biggs J, Ziegler V, Meyer D. A rating scale for mania: reliability, validity and sensitivity. The British Journal of Psychiatry. 1978;133:429–435. doi: 10.1192/bjp.133.5.429. [DOI] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. [Google Scholar]
- Zubieta JK, Huguelet P, O'Neil RL, Giordani BJ. Cognitive function in euthymic bipolar I disorder. Psychiatry research. 2001;102:9–20. doi: 10.1016/s0165-1781(01)00242-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


