Abstract
We propose a similarity measure based on the multivariate hypergeometric distribution for the pairwise comparison of images and data vectors. The formulation and performance of the proposed measure are compared with other similarity measures using synthetic data. A method of piecewise approximation is also implemented to facilitate application of the proposed measure to large samples. Example applications of the proposed similarity measure are presented using mass spectrometry imaging data and gene expression microarray data. Results from synthetic and biological data indicate that the proposed measure is capable of providing meaningful discrimination between samples, and that it can be a useful tool for identifying potentially related samples in large-scale biological data sets.
Index Terms: Similarity measures, contingency tables, multivariate statistics, biology and genetics, chemistry
Introduction
Similarity measures are an important tool in the analysis of a wide range of biomedical data, with applications such as the comparison of peptide sequences [1] and gene expression data [2], as well as in text mining [3] and in image analysis [4], [5]. An important application of similarity measures is in the detection of new and potentially significant patterns in large-scale biological data sets [2], [5], [6]. For example, if a particular gene is known to be associated with a disease, other genes potentially related to the disease may be detected by identifying highly similar patterns of expression. In this respect, similarity measures can be used to provide a shortlist of targets for further research.
Different similarity measures exhibit considerable variation in properties and performance [7], [8]. For example, many common measures do not have a probabilistic framework, although this is a useful property in terms of the interpretation of assigned similarity scores [9]. In this paper, we propose a similarity measure with a probabilistic interpretation, utilizing the multivariate hypergeometric distribution and the Fisher-Freeman-Halton test. Previously, we developed a similarity measure utilizing the hypergeometric distribution and Fisher’s exact test [10]; this measure was restricted to two-class data, i.e., the comparison of binary images and data vectors. However, many types of biological data are not inherently binary in nature, and the process of binarization can discard useful information. Here, we extend our earlier result to present a general similarity measure that accommodates the comparison of nonbinary, “multiclass” data.
After defining the proposed multivariate hypergeometric similarity measure, we describe several tests using synthetic and biological data to investigate its performance. First, its patterns of sample ranking are compared with those of cosine similarity, Pearson correlation and mutual information, three similarity measures which are used in the analysis of many types of biomedical data [2], [3], [4], [5], [11]. Next, we test and implement a method of approximation that facilitates the application of the proposed similarity measure to large samples.
We then consider example applications on biological data sets: first, on mass spectrometry imaging (MSI) data, and second, on gene expression microarray data. MSI is a technique in which mass spectra are acquired over the surface of a sample, such as a tissue slice, to generate a 3D (x,y : spatial, z : spectral) data set as shown in Fig. 1. MSI has numerous biomedical applications, including the study of cancer and neurodegenerative diseases, and pharmaceutical-related research [12], [13]. MSI generates large-scale data sets; the spectral dimension may contain thousands of m/z(mass-to-charge ratio) values. Depending on the data acquisition modality, each m/z value can be interpreted as a molecule or molecular fragment [13]. The data set can be interpreted as a collection of images, each describing the presence and abundance of a single m/z value at every (x, y) point over the sample surface. Spatial comparison of these images can be useful for identifying patterns among m/z values. For example, a single tissue sample may contain healthy, diseased, and marginal regions. Certain m/z values may be spatially localized in particular regions [12]. It may be informative to identify other m/z values expressed predominantly within a region of interest [14], or with similar spatial distributions as an m/z value of interest.
Fig. 1.
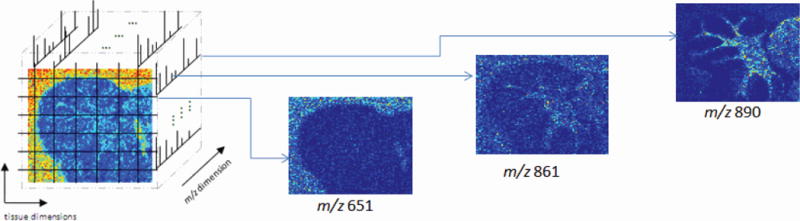
Description of 3D data structure of MSI data sets, and three example m/z images.
In this paper, the performance of the proposed similarity measure on biological data is assessed by applying it to an MSI data set to identify m/z images with similar spatial characteristics, and by applying it to a gene expression microarray data set to identify genes with similar expression patterns. Results from synthetic and biological data indicate that the proposed multivariate hypergeometric similarity measure is capable of providing meaningful discrimination among samples, and can be a useful tool for identifying potentially related samples in large-scale biological data sets.
Methods
2.1 Definition of Similarity Measure
Consider an image containing N pixels (or a data vector containing N samples), with all intensities quantized into n bins, where N and n are positive integers. When comparing two such images, there are n2 possible types of overlap between spatially corresponding pixels. These overlaps can be represented as an n×n contingency table, as shown in Fig. 2. Each class kij, for indices i=1,…,n and j=1,…,n, represents the number of spatially corresponding pixels which are in bin i in the first (“reference”) image and in bin j in the second (“query”) image. The terminology is used in the sense that a given image would be selected as a “reference” and other images in a data set would be compared, or “queried” against it to find images similar to the reference. The margins of the contingency table are fixed for a given pair of images: for each row i, Σnj=1kij=ri, the number of pixels in bin i in the reference image, and similarly for each column j, Σni=1kij=qj, the number of pixels in bin j in the query image. By definition, Σijkij=N. The probability of observing a particular distribution of overlaps kij, i.e., the probability of observing a given contingency table, can be represented as the product of probability mass functions of the multivariate hypergeometric distribution. Considering only the first column of an n×ncontingency table with row marginal totals ri, the column sum is q1=k11+k21+⋯kn1. Each component ki1 is “drawn” from its row sum ri. Since each draw is independent, the probability of observing a particular distribution of pixels is given as
Fig. 2.
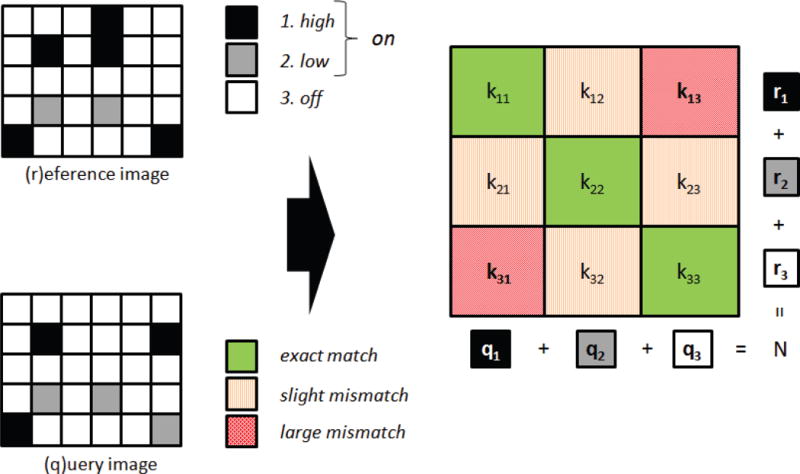
An image pair (reference and query images) with pixel intensities binned into three levels is represented as a 3×3 contingency table with fixed marginal totals.
This quantity is a probability mass function of the multivariate hypergeometric distribution. The probability of observing the second column is described similarly, but accounts for the pixels already assigned in the previous column:
The same pattern is followed through the (n−1)th column. Because the row and column sums are fixed, the configuration of the nth column is determined by the preceding columns. Since the configuration of the available pixels in each column (aside from the nth column) is independent of the other columns, the probability p of the complete n×n contingency table is given by the product of the column probabilities. This quantity, shown in (1), is known as the probability for k -variate contingency tables [15], [16]. Here, q=[q1,q2,…,qn], r=[r1,r2,…,rn], and k=[k11,k12,…,knn]:
| (1) |
In our previous work focusing on binary data, we defined a similarity measure based on the hypergeometric distribution; the probability mass function of this distribution gives the probability of a 2×2 contingency table [10]. The similarity measure was defined as the difference between the lower and upper tails of the hypergeometric distribution defined by the marginal totals r and q. The values of r and q are a function of the particular reference image and query image being compared. The tails were defined with respect to the observed overlap, which was defined as the number of spatially corresponding pixels which are “on” in both images, i.e., k11 in this terminology. To extend this approach from the two classes in binary data to n classes, we utilize the probability mass function of the n×n contingency table.
The statistical significance of a contingency table is evaluated by performing Fisher’s exact test (in the binary case) or the Fisher-Freeman-Halton test (in the general case)[15], [17]. In both cases, the isomarginal family of tables (i.e., those tables having the same fixed margins r and q as the original table representing the reference and query image pair) is first generated, and the probability of each table is calculated. In the binary case, the hypergeometric distribution describes the isomarginal family. For each table in the isomarginal family, the value of a chosen statistic S(k) is compared to that of the original table. With respect to S(k), tables in the isomarginal family may be more extreme than the original table in two directions. The set of tables which are “more extremely large” have a larger than or equal value of the statistic, while the set of tables which are “more extremely small” have a smaller than or equal value of the statistic. The significance of a table in a particular direction is found by summing the probabilities of all tables within the respective set.
In the binary case, the choice of the statistic S(k) is straightforward because due to the fixed margins, there is only one degree of freedom. S(k)=k11 completely defines the table, and is reasonable because more similar images will have greater numbers of overlapping pixels. In the general case, however, there are n2−2n+1 degrees of freedom, and for n>2, the choice of a statistic is not obvious. Here, we choose a vector of statistics—the set of diagonal elements of the n×n table—as S(k), as shown in (2). These diagonal elements represent the exact matches— the spatially corresponding pixels in the reference and query images which are in the same class. While S(k) may be defined in many alternative ways, we propose (2) as a reasonable choice for multiclass data because images which are more similar will have a greater number of each of the n types of exact matches:
| (2) |
For each table in the isomarginal family, we perform an index-wise comparison of each diagonal element to the corresponding diagonal element in the original table. In other words, we compare each element in S(k) with the corresponding element in S0, which is the instance of S(k) observed for the original table. If each diagonal element in the table is greater than or equal to its corresponding element in S0, the table is assigned to set G, the set of “more extremely large” tables with respect to all elements of S(k). If each diagonal element is less than or equal to its corresponding element in S0, the table is assigned to set L, the set of “more extremely small” tables. Equation (3) defines the proposed multivariate hypergeometric similarity measure h:
| (3) |
2.2 Similarity Measure Comparison
The sample rankings obtained from the proposed measure are compared with those from cosine similarity, Pearson correlation, and mutual information. Cosine similarity and Pearson correlation are defined for vectors V1 and V2 in (4) and (5), respectively. Mutual information is defined in (6), where xi and yj are the elements of V1 and V2, respectively:
| (4) |
| (5) |
| (6) |
2.3. Synthetic Data
First, the performance of the multivariate hypergeometric similarity measure is evaluated on synthetic data. While the proposed similarity measure is defined for any n≥2 classes, these synthetic experiments are performed using only three classes to clearly illustrate the method. Two synthetic data sets are used for this comparison. The first consists of the three-class isomarginal family defined by marginal totals (r1,r2,r3,q1,q2,q3)=(5,5,5,5,5,5) and N=15. The second consists of all three-class tables with N=5.
2.4 Piecewise Approximation
Testing the significance of n×n contingency tables obtained from biomedical data, such as MSI data, poses a challenge due to data size. As the numbers of pixels in the images, and hence the marginal totals, increase, generating the isomarginal family of tables to perform the Fisher-Freeman-Halton test becomes demanding. The number of possible tables increases factorially as the numbers of rows, columns, or total pixels increase [18], [19]. As an analytical example, the number of three-class contingency tables where all rows and columns sum to r is given by (r+22)+3(r+34) [20], [21]. When faced with a very large number of tables to enumerate in the isomarginal family, approximate solutions can be found through Monte Carlo testing [17]. However, in practice, this may demand a very large numbers of permutations to achieve satisfactory separation of similarity rankings.
Here, we propose a piecewise method of approximation, in which the two images or data vectors to be compared are divided into a number of smaller subsections. The motivating idea is that similar samples will also have similar corresponding subsections. For each pair of reference and query subsections, an n×n contingency table is constructed and the multivariate hypergeometric similarity measure is calculated. The overall similarity of the image pair is computed as a function of the similarities of all subsections. Fig. 3 illustrates this process.
Fig. 3.
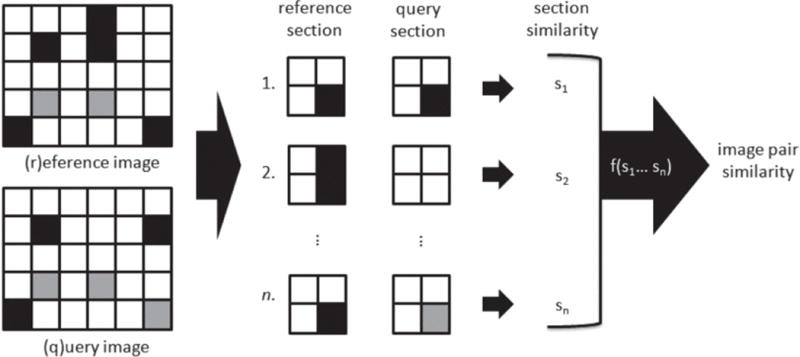
Overview of piecewise approximation process: subsection (1) corresponds to the top-left4×4 blocks of the reference and query images; (2) to the 4×4 blocks to the immediate right of (1); and (n) to the bottomright 4×4 blocks. The similarity is calculated for each spatially corresponding reference and query section, and the overall similarity of the reference and query is calculated as a function of the subsection scores.
Piecewise approximation requires choices in how images or data vectors are separated into subsections (e.g., different subsection sizes) and how the similarity scores for the subsections are combined to obtain an overall similarity score for the image pair (e.g., different functions). Alternative choices are examined here through experiments on synthetic and biomedical data. First, the previously described synthetic data set (for N=15) is used to examine whether there is a pattern between subsection size and the extent of difference observed between the piecewise approximation rankings and the exact rankings. In this test, the rankings for each sample obtained by using subsections of sizes 3, 4, and 5 pixels are compared with the ranking calculated using the whole sample. To avoid the comparison of single-pixel sections, if the sample is not evenly divisible at a particular increment size, the remainder pixels are added to the previous subsection to create one subsection larger than the others. In the same experiment, the effect of permuting the reference and query samples which correspond to a single n×n table is considered. While a given pair of samples yields a single n×n table, mapping a given table back to the sample space yields nonunique indexing of spatially corresponding pixels. This type of indexing difference would not affect the similarity score of a given reference and query pair if the whole sample is utilized. However, when piecewise approximation is employed, different subsections may contain different proportions of the pixels for each type of overlap kij. To examine how this may affect results, the “randperm” function in MATLAB was used to generate a permutation of the sample indices, which was applied to both the reference and query samples before they were divided into subsections. This was repeated 10, 000 times. The purpose of this step is to confirm that overall sample rankings in the synthetic data set are not an artifact of arbitrary methods of generating synthetic samples from tables and subsectioning samples. For each subsection size, the sample ranking results shown are the mean across all permutations. Next, biomedical data was used to empirically compare alternative functions for aggregating the subsection similarity scores into an overall score for the image pair.
2.5 Experimental Data
The experimental MSI data used in this study is from a mouse model of Tay-Sachs/Sandhoff disease [22]. The image corresponding to m/z 889.6 (located at index 783 within the data set) was selected as the reference image due to its distinctive spatial pattern. The MSI data set has a spectral dimension of 4, 438 m/z values, each corresponding to an m/z image, and all of these images were tested as query images against the reference image of m/z 889.6. The three-class case was used for this experiment: pixel intensities in the data set were binned into “high” (≥thresholdx), “low” (0< and <x), and “zero” (=0) classes. The threshold x was arbitrarily selected as the 50th percentile of the mean spectrum of the data set. The piecewise approximation approach was used with a primary subsection size of 4×4, chosen after testing several sizes in an effort to balance section size and computational time. The proposed similarity measure score was calculated for each subsection. Several functions for combining subsection scores into an aggregate image pair score were then compared by qualitatively evaluating the images which were ranked by each as highly similar to the reference. Finally, the top m/z images selected by the proposed similarity measure using three classes were compared to the three-class results for cosine similarity, Pearson correlation and mutual information.
The gene expression data used in this study consists of 230 breast cancer samples. The raw Affymetrix *.CEL files were downloaded from the Gene Expression Omnibus (GEO) website (accession: GSE20194). The Affymetrix Expression console software was used to compute log2 normalized gene expression values. Clinical information available on GEO was used to label each sample within the data set as estrogen receptor (ER) positive or negative, and the 141 ER+ samples were used for further analysis. The three-class case was also used for this experiment: gene-specific, percentile-based threshold pairs (x, y) were used to bin each gene expression value into the “high” (>y), “medium” (x< and ≤y) or “low” (≤x) class. Results from several alternative threshold pairs were compared. Probe “205225_at,” for the estrogen receptor gene (ESR1), was used as the reference sample.
Results
This section describes four sets of results. First, the performance of the proposed multivariate hypergeometric similarity measure is compared with the other similarity measures using synthetic data. Second, the effects of subsection size and combination functions on the piecewise approximation method are investigated using synthetic and experimental MSI data. Third, the performance of the proposed similarity measure on experimental MSI data is evaluated. Fourth, its performance is evaluated on experimental gene expression data.
In the first set of results, the rankings of samples in synthetic data sets by the four similarity measures are compared in Figs. 4 and 5. Each sample (horizontal bar) represents a single table, with the green, yellow, and red segments representing the number of exact matches (k11+k22+k33), slight mismatches (k12+k21+k23+k32), and large mismatches (k13+k31), respectively. In Fig. 4, there are 231 tables represented; these tables comprise the isomarginal family defined by marginal totals (r1,r2,r3,q1, q2,q3)=(5,5,5,5,5,5). All four similarity measures agree in that the highest score is assigned to the table with the largest number of exact matches. None of the similarity measures are monotonic with respect to the number of exact matches, but rankings from the proposed similarity measure are much closer to this trend than rankings from cosine similarity and Pearson correlation. Cosine similarity and Pearson correlation more closely sort by the number of large mismatches. For a single isomarginal family, the magnitudes and means of the two vectors are constant. The rankings of cosine similarity and Pearson correlation therefore depend on the value of the dot product, and the minimum dot product is observed when the number of large mismatches is maximized. The proposed similarity measure does not provide such distinction between slight and large mismatches, but it does provide a probabilistic interpretation which cosine similarity and Pearson correlation do not: the samples associated with extreme scores are the most “surprising” patterns of overlap observed. Mutual information assigns higher scores to cases where most pixels are concentrated in a few classes, but does not differentiate among the classes. For example, the tables with [k11,k22,k33]=[5,5,5] (i.e., all exact matches) and [k31,k22,k13]=[5,5,5] (i.e., many large mismatches) both receive equally high scores; as a result, the mutual information results do not show any trend with respect to exact matches, slight mismatches or large mismatches. In contrast, [k11,k22,k33]=[5,5,5] is ranked highly by the proposed similarity measure, while [k31,k22,k13]=[5,5,5] receives a much lower score.
Fig. 4.
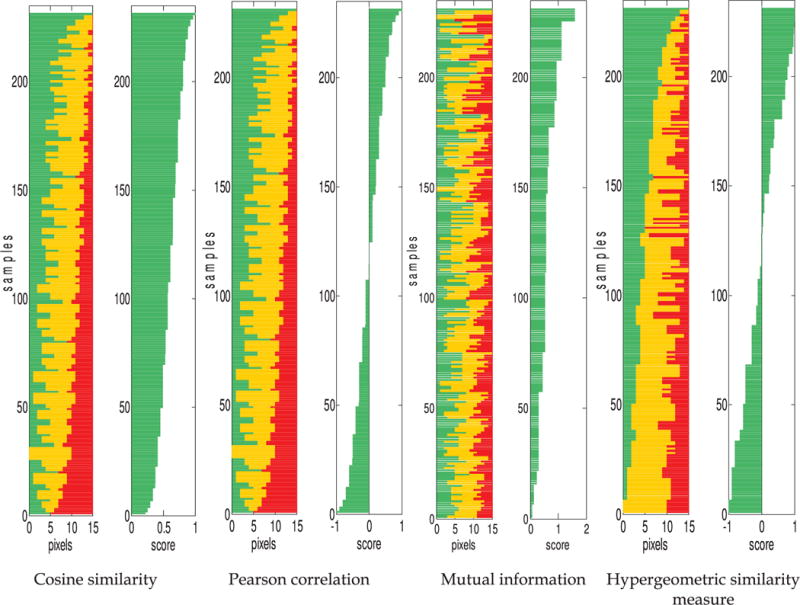
Comparison of sample rankings by the four similarity measures for the synthetic data set comprising the isomarginal family given by (r1,r2,r3,q1,q2,q3)=(5,5,5,5,5,5). Each sample (horizontal bar) represents a certain number of exact matches, slight mismatches, and large mismatches (corresponding to [green, yellow, and red], or [medium, light, and dark] in grayscale). The length of each color segment corresponds to the number of that type of match in the sample. For each similarity measure, the similarity score corresponding to each sample is shown on the right.
Fig. 5.
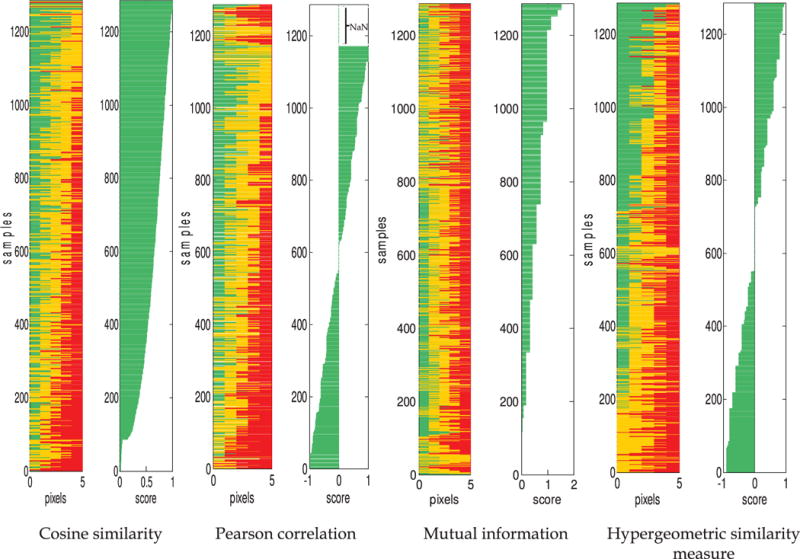
Comparison of sample rankings by the four similarity measures for the synthetic data set containing all tables for N=5. Each sample (horizontal bar) contains a certain number of exact matches, slight mismatches, and large mismatches (corresponding to [green, yellow, and red], or [medium, light, and dark] in grayscale). The length of each color segment corresponds to the number of that type of match in the sample. For each similarity measure, the similarity score for each sample is shown on the right.
Fig. 5 considers the rankings of the 1, 287 tables generated by considering every possible combination of marginal totals such that (r1+r2+r3=5) and (q1+q2+ q3=5). Again, all four similarity measures agree in that the highest score is assigned to the table with the largest number of exact matches, but the proposed similarity measure more consistently assigns lower scores to tables with fewer exact matches. The rankings in this set of all tables for N=5 illustrate additional probabilistic aspects of the proposed similarity measure. For example, the proposed measure can distinguish between instances of overlap with different distribution magnitudes. It assigns identical scores to the set of tables with [k11,k22,k33 ] as [3,1,1],[1,3,1], and [1,1,3], and a different identical score to the other possible set of tables describing only exact overlap, with [k11,k22,k33 ] as [2,2,1],[2,1,2], and [1,2,2]. Pearson correlation and cosine similarity do not distinguish between these two sets of tables. Mutual information distinguishes the two sets of tables, but again does not distinguish between cases of exact matches and many large mismatches; for example, the cases where [k11,k22,k33]=[3,1,1] and [k31,k22,k13]=[3,1,1] are assigned the same score, and[k11,k22,k33]=[2,2,1] and [k31,k22,k13]=[2,1,2] are assigned the same score. For a second example, in the proposed measure, all tables which have marginal totals such that only one n×n table is possible are mapped to a score of zero. If only one set of overlaps kij can be observed for a particular pair of images or data vectors, then the overlap which is observed can be considered inherently “unsurprising.” In contrast, this set of tables is undefined for Pearson correlation (i.e., these tables are assigned the value NaN, as shown at the top of the Pearson correlation plot in Fig. 5). Cosine similarity does not group these tables together or otherwise distinguish them.
In the second set of results, the effects of subsection size on the piecewise approximation result are described in Fig. 6. The 231 samples in the synthetic data set shown in Fig. 4 are plotted in order of increasing exact score. The piecewise approximation scores for each sample, across increments of sizes 3, 4, and 5, are compared. For all three subsection sizes, the mean score from 10, 000 permutations of the reference and query vectors is shown. Overall, the piecewise approximation scores follow the trend of the exact score, but there are notable deviations. In such cases, samples are ranked higher or lower as an artifact of the piecewise sectioning process. Interestingly, these cases tend to correspond across all of the subsection sizes; if a sample is scored much higher or lower than its adjacent samples by the piecewise method, the same jump or dip in score is observed across all three subsection sizes. However, the magnitudes of the piecewise scores indicate that, as expected, larger sections give scores closer to the exact result.
Fig. 6.
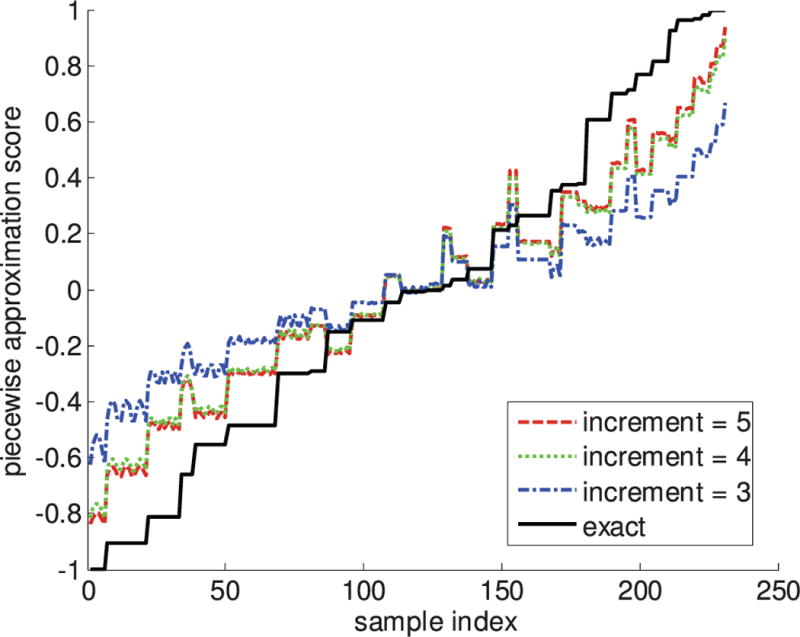
The mean rankings of synthetic samples using piecewise approximation at different subsection sizes (size 3: blue dash-dot line; size 4: green dotted line; and size 5: red dashed line) compared to rankings from using the whole sample (black, solid line).
Next, different statistics for combining the similarity scores of sections into a single overall score for the sample pair are compared empirically, using MSI data with a subsection size of 4×4 pixels for piecewise approximation. Fig. 7 shows the image pair similarity scores for each of the 4, 438 m/z values, computed as the mean, median, mode, variance, skewness, or kurtosis of all of their subsection scores. The x-axis of these plots, showing indices 1 through 4, 438, represents the query m/z images; each is associated with a single score (dot) on the y -axis. This score is obtained by evaluating the specified function (e.g., the mean) over the set of subsection scores obtained for that query image when it was compared to the reference image. To interpret these results, it is necessary to consider that the reference m/z image corresponds to index 783. Since the most similar image in the data set to the reference image should be the reference image itself, a well-performing function should assign the most extreme score to this index. This result is observed for the mean, median, variance, and kurtosis. During previous study of this data set, 47 of the 4, 438 images were observed to be qualitatively very similar to the reference m/z image, and those images were observed to be associated with indices relatively close to the reference index [10]. In contrast, lower indices were associated with noisy images (an artifact of MALDI MSI data acquisition), and higher indices with sparse images. A well-performing function would, therefore, exhibit a peak centered at the reference index of 783. The mean and kurtosis both show this feature by assigning extreme (higher and lower than most others, respectively) scores to indices close to the reference index.
Fig. 7.
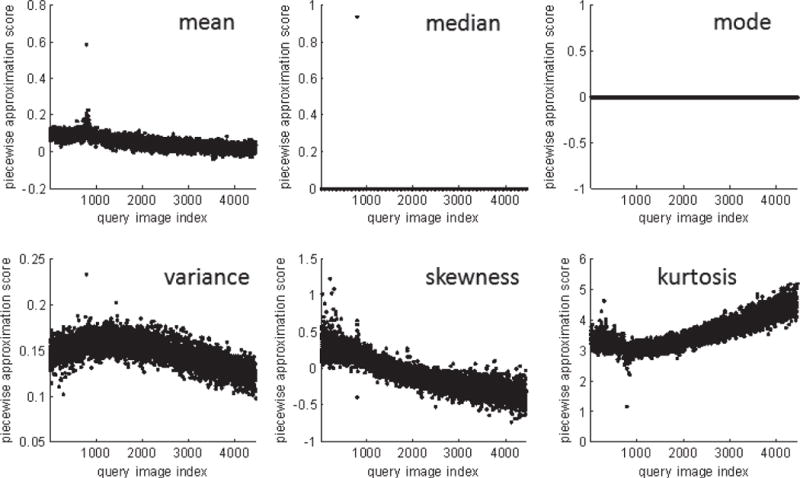
Empirical comparison of alternative functions for combining subsection similarity scores into an overall similarity score through piecewise approximation. Each dot on the scatter plot represents one query image (m/z value); 4, 438 are in the data set. The value (image pair score) of the dot represents the similarity score assigned to the query image based on the specified function of its subsection scores. For example, in the “mean” plot, the image pair score of each query image is the average similarity score of its subsections.
In the third set of results, these findings are applied to experimental MSI data. The 12m/z images selected by each measure as most similar to the reference image are shown in Fig. 8. All measures agree that the reference itself is the most similar (selection 1:m/z 889.6). Notably, the proposed similarity measure gives results which are qualitatively very similar to the reference m/z image. The Pearson correlation and mutual information results for three classes also closely resemble the reference m/z image, while the cosine similarity results for three classes include several noisy images without a clearly discernible pattern. Interestingly, the top 12 results selected by the proposed similarity measure using the mean and kurtosis as combination functions are not identical. Moreover, neither set of results overlaps completely with the results from Pearson correlation, cosine similarity, and mutual information. For example, the proposed similarity measure, using the mean as the combination function, selects m/z908.9, which none of the others select. Similarly, m/z 894.5, another unique selection, is picked by the proposed similarity measure when using the kurtosis as the combination function. Examining the top n results is common when applying a similarity measure to a data set, and these observations indicate that applying the proposed multivariate hypergeometric similarity measure can yield relevant and potentially useful results.
Fig. 8.
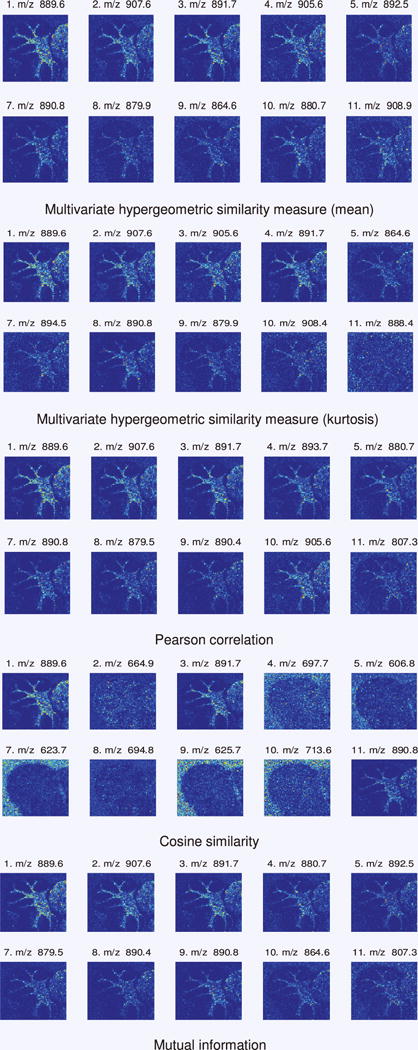
The 12 most similar m/z images, as ranked by the four similarity measures. The multivariate hypergeometric similarity measure results are shown with the mean and kurtosis as combination functions.
In the fourth set of results, Tables 1, 2, and 3 show the top 20 gene rankings for each of the similarity measures for a single ESR1 reference probe across three alternative percentile-based thresholds. First, in all cases, the reference probe “205225_at” is selected as the most similar, as it should be. Second, the multivariate hypergeometric similarity measure (using the mean as a combination function) identifies some of the other ESR1 probes as highly similar to the reference, but highlights the other probes to a lesser extent than the other three measures. Third, this set of results shows that the multivariate hypergeometric similarity measure is successful in identifying genes which are known to be associated with breast cancer, such as BCL2 and FOXA1 in Table 3. Fourth, notably, the proposed similarity measure highlights several genes—WWP1 [23] and NME5 [24] in Table 1, PGRMC2 [25] and HPN [26] in Table 2—which have recently been shown to be relevant in breast cancer, but which are not included in the top rankings of the other three similarity measures.
TABLE 1.
Top 20 Rankings by Similarity Measures, Using Percentiles [0, 25] as Thresholds
| Multivariate hypergeometric similarity measure | Pearson correlation | Cosine similarity | Mutual intormation | |||||
|---|---|---|---|---|---|---|---|---|
| Rank | Probe | Gene | Probe | Gene | Probe | Gene | Probe | Gene |
| 1 | 205225_at | ESR1 | 205225_at | ESR1 | 205225_at | ESR1 | 205225_at | ESR1 |
| 2 | 215552_s_at | ESR1 | 215552_s_at | ESR1 | 215552_s_at | ESR1 | 215552_s_at | ESR1 |
| 3 | 212960_at | TBC1D9 | 205253_at | PBX1 | 2052S3_at | PBX1 | 211235_s_at | ESR1 |
| 4 | 212637_s_at | WWP1 | 209603_at | GATA3 | 209603_at | GATA3 | 205253_at | PBX1 |
| 5 | 219717_at | DCAF16 | 218186_at | RAB25 | 218186_at | RAB25 | 209603_at | GATA3 |
| 6 | 217190_x_at | ESR1 | 211235_s_at | ESR1 | 211235_s_at | ESR1 | 218186_at | RAB25 |
| 7 | 218394_at | ROGDI | 204508_s_at | CA12 | 200711_s_at | SKP1 | 212446_s_at | LASS6 |
| 8 | 203928_x_at | MAPT | 200711_s_at | SKP1 | 203963_at | CA12 | 203928_x_at | MAPT |
| 9 | 204242_s_at | ACOX3 | 203963_at | CA12 | 204508_s_at | CA12 | 200711_s_at | SKP1 |
| 10 | 209064_x_at | PAIP1 | 215867_x_at | CA12 | 215867_x_at | CA12 | 203963_at | CA12 |
| 11 | 212638_s_at | WWP1 | 217190_x_at | ESR1 | 217190_x_at | ESR1 | 204508_s_at | CA12 |
| 12 | 204045_at | TCEAL1 | 212446_s_at | LASS6 | 212446_s_at | LASS6 | 215867_x_at | CA12 |
| 13 | 203359_s_at | MYCBP | 203928_x_at | MAPT | 203928_x_at | MAPT | 217190_x_at | ESR1 |
| 14 | 202596_at | ENSA | 209759_s_at | DC1 | 204798_at | MYB | 209064_x_at | PAIP1 |
| 15 | 218524_at | E4F1 | 212956_at | TBC1D9 | 209602_s_at | GATA3 | 209730_at | SEMA3F |
| 16 | 211233_x_at | ESR1 | 218086_at | NPDC1 | 209604_s_at | GATA3 | 211778_s_at | OVOL2 |
| 17 | 221934_s_at | DALRD3 | 218195_at | C6orf211 | 209681_at | SLC19A2 | 208865_at | CSNK1A1 |
| 18 | 201353_s_at | BAZ2A | 204798_at | MYB | 209759_s_at | DCI | 211234_x_at | ESR1 |
| 19 | 206197_at | NME5 | 210735_s_at | CA12 | 210735_s_at | CA12 | 202596_at | ENSA |
| 20 | 200719_at | SKP1 | 212099_at | RHOB | 212099_at | RHOB | 208614_s_at | FLNB |
TABLE 2.
Top 20 Rankings by Similarity Measures, Using Percentiles [25, 75] as Thresholds
| Multivariate hypergeometric similarity measure | Pearson correlation | Cosine similarity | Mutual information | |||||
|---|---|---|---|---|---|---|---|---|
| Rank | Probe | Gene | Probe | Gene | Probe | Gene | Probe | Gene |
| 1 | 205225_at | ESR1 | 205225_at | ESR1 | 205225_at | ESR1 | 205225_at | ESR1 |
| 2 | 211234_x_at | ESR1 | 215552_s_at | ESR1 | 215552_s_at | ESR1 | 215552_s_at | ESR1 |
| 3 | 203359_s_at | MYCBP | 211235_s_at | ESR1 | 211235_s_at | ESR1 | 211235_s_at | ESR1 |
| 4 | 215552_s_at | ESR1 | 211234_x_at | ESR1 | 211234_x_at | ESR1 | 211234_x_at | ESR1 |
| 5 | 218259_at | MKL2 | 211233_x_at | ESR1 | 211233_x_at | ESR1 | 211233_x_at | ESR1 |
| 6 | 212960_at | TBC1D9 | 204798_at | MYB | 204798_at | MYB | 204798_at | MYB |
| 7 | 204798_at | MYB | 204667_at | FOXA1 | 204667_at | FOXA1 | 217190_x_at | ESR1 |
| 8 | 209739_s_at | PNPLA4 | 217190_x_at | ESR1 | 217190_x_at | ESR1 | 204667_at | FOXA1 |
| 9 | 201701_s_at | PGRMC2 | 218437_s_at | LZTFL1 | 218437_s_at | LZTFL1 | 209603_at | GATA3 |
| 10 | 205597_at | SLC44A4 | 215014_at | KCND3 | 215014_at | KCND3 | 205253_at | PBX1 |
| 11 | 206869_at | CHAD | 209681_at | SLC19A2 | 209681_at | SLC19A2 | 211596_s_at | LRIG1 |
| 12 | 203928_x_at | MAPT | 208613_s_at | FLNB | 208613_s_at | FLNB | 203072_at | MYO1E |
| 13 | 205140_at | FPGT | 209603_at | GATA3 | 203359_s_at | MYCBP | 201426_s_at | VIM |
| 14 | 212956_at | TBC1D9 | 203359_s_at | MYCBP | 208716_s_at | TMCO1 | 203359_s_at | MYCBP |
| 15 | 213648_at | EXOSC7 | 208716_s_at | TMCO1 | 209603_at | GATA3 | 218259_at | MKL2 |
| 16 | 204242_s_at | ACOX3 | 218259_at | MKL2 | 218259_at | MKL2 | 209681_at | SLC19A2 |
| 17 | 204934_s_at | HPN | 205158_at | RNASE4 | 205158_at | RNASE4 | 218437_s_at | LZTFL1 |
| 18 | 211233_x_at | ESR1 | 209696_at | FBP1 | 202454_s_at | ERBB3 | 208613_s_at | FLNB |
| 19 | 200776_s_at | BZW1 | 202454_s_at | ERBB3 | 209696_at | FBP1 | 215014_at | KCND3 |
| 20 | 203420_at | FAM8A1 | 217770_at | PIGT | 217770_at | PIGT | 204508_s_at | CA12 |
TABLE 3.
Top 20 Rankings by Similarity Measures, Using Percentiles [50, 100] as Thresholds
| Multivariate hypergeometric similarity measure | Pearson correlation | Cosine similarity | Mutual information | |||||
|---|---|---|---|---|---|---|---|---|
| Rank | Probe | Gene | Probe | Gene | Probe | Gene | Probe | Gene |
| 1 | 205225_at | ESR1 | 205225_at | ESR1 | 205225_at | ESR1 | 205225_at | ESR1 |
| 2 | 215552_s_at | ESR1 | 215552_s_at | ESR1 | 215552_s_at | ESR1 | 215552_s_at | ESR1 |
| 3 | 211235_s_at | ESR1 | 211235_s_at | ESR1 | 211235_s_at | ESR1 | 211235_s_at | ESR1 |
| 4 | 200711_s_at | SKP1 | 204667_at | FOXA1 | 204667_at | FOXA1 | 204667_at | FOXA1 |
| 5 | 215014_at | KCND3 | 211234_x_at | ESR1 | 211234_x_at | ESR1 | 211234_x_at | ESR1 |
| 6 | 211234_x_at | ESR1 | 218437_s_at | LZTFL1 | 218094_s_at | DBNDD2 SYS1/SYS1-DBNDD2 | 218094_s_at | DBNDD2/SYS1/SYS1-DBNDD2 |
| 7 | 204667_at | FOXA1 | 218094_s_at | DBNDD2/SYS1/SYS1-DBNDD2 | 218437_s_at | LZTFL1 | 218437_s_at | LZTFL1 |
| 8 | 21052_s_at | TIC39A | 200711_s_at | SKP1 | 200711_s_at | SKP1 | 206571_s_at | MAP4K4 |
| 9 | 218035_s_at | RBM47 | 211233_x_at | ESR1 | 202088_at | SLC39A6 | 200711_s_at | SKP1 |
| 10 | 203928_x_at | MAPT | 202088_at | SLC39A6 | 202089_s_at | SLC39A6 | 202088_at | SLC39A6 |
| 11 | 41660_at | CELSR1 | 202089_s_at | SLC39A6 | 211233_x_at | ESR1 | 202089_s_at | SLC39A6 |
| 12 | 217190_x_at | ESR1 | 204798_at | MYB | 201169_s_at | BHLHE40 | 211233_x_at | ESR1 |
| 13 | 208810_at | DNAJB6/TMEM135 | 208336_s_at | TECR | 204798_at | MYB | 219155_at | PITPNC1 |
| 14 | 208336_s_at | TECR | 210652_s_at | TIC39A | 205186_at | DNALI1 | 219506_at | C1orf54 |
| 15 | 201845_s_at | RYBP | 217860_at | NDUFA10 | 208336_s_at | TECR | 219806_s_at | C11orf75 |
| 16 | 204798_at | MYB | 205186_at | DNALI1 | 209604_s_at | GATA3 | 201169_s_at | BHLHE40 |
| 17 | 218094_s_at | DBNDD2/SYS1/SYS1-DBNDD2 | 209604_s_at | GATA3 | 210652_s_at | TTC39A | 204798_at | MYB |
| 18 | 201826_s_at | SCCPDH | 222125_s_at | P4HTM | 217860_at | NDUFA10 | 205186_at | DNALI1 |
| 19 | 206401_s_at | MAPT | 201169_s_at | BHLHE40 | 222125_s_at | P4HTM | 208336_s_at | TECR |
| 20 | 203685_at | BCL2 | 218026_at | CCDC56 | 200946_x_at | GLUD1 | 209604_s_at | GATA3 |
These results also emphasize the value of integrating multiple forms of analysis to leverage complementary findings. For example, recent investigations have examined the role of MAPT [27] and GATA3 [28] in breast cancer; while MAPT appears in the top rankings of all four similarity measures in Table 1, GATA3 appears in the rankings of the other three similarity measures, but not in those of the multivariate hypergeometric similarity measure. Additionally, the benefits of examining a single data set across alternative thresholds can be clearly observed through the notably different gene lists for each measure in Tables 1, 2, and 3. In addition, parallel assessments with different probes for the same gene are also important. For example, the top 20 rankings by the multivariate hypergeometric similarity measure for ESR1 probe “211235_s_at” in the same data set (rankings not shown) highlight several other genes—IGF1R [29], [30],GMPR2 [31], FGFR3 [32]—which have recently been shown to be relevant in breast cancer. Again, these observations indicate that applying the proposed multivariate hypergeometric similarity measure can yield relevant and potentially useful results.
Discussion
In this paper, we propose a multivariate hypergeometric similarity measure for the pairwise comparison of images and data vectors featuring any positive integer number of intensity levels. This is an extension of our previous work on a hypergeometric similarity measure, which was restricted to binary data. Using synthetic data sets, we compared the proposed measure to Pearson correlation, cosine similarity and mutual information in terms of sample rankings, and identified several favorable properties of the proposed measure. Next, we developed a method of piecewise approximation to facilitate the application of this approach to large data sets. Piecewise approximation was tested at several different subsection sizes on synthetic data, and was observed to follow the trend of the exact score. Then, biological MSI data was used to empirically assess functions for combining subsection similarity scores found through piecewise approximation. The proposed similarity measure was demonstrated to be effective in identifying qualitatively similar images in MSI data and breast cancer-related genes in microarray data. For both data types, it made relevant selections which were not identified by other similarity measures in their top selections.
While the current results of this study are encouraging, they also highlight several avenues for further research on the proposed similarity measure. For instance, this approach is defined for any positive integer number of classes, but the results in this study have considered only three classes. Three classes were chosen both for simplicity in examining similarity measure properties and to highlight the difference between the binary case and the multiclass case. Future work will assess the effect of increasing the number of classes. However, as previously noted, the generation of the isomarginal family becomes increasingly demanding as the number of classes increases [18], [19],[20], [21]. Additionally, alternative definitions of the statistic S (k) will be explored. Here, we chose S (k) as the set of diagonal elements of the contingency table. However, it may be desirable to include sub- and superdiagonal terms when larger numbers of classes are considered. The selection of the appropriate number of classes—and of appropriate thresholds for separating classes—is another issue of interest; in this study, the thresholds between classes for the MSI and gene expression data sets were arbitrarily selected. From the perspective of practical biomedical applications, choices of thresholds for a particular data set may be based on examination of descriptive data statistics, or by applying selected tests as a preliminary step [33]. The selection of functions for aggregating subsection scores obtained from piecewise approximation is another area for further study. Six statistics were tested in this study, and many additional functions could be tested. Interestingly, the set of top selections for MSI data using the mean and kurtosis were not identical, indicating that it may be useful to consider which combination functions may be complimentary.
A notable constraint is that the molecular identities of the m/z values are not known for the MSI data set investigated here; future testing on labeled (MS/MS) data will enable biological interpretation of similarity measure performances for MSI data, similar to what was done for the microarray data. In addition, only one MSI and one microarray data set have been investigated here; implementation on multiple data sets will provide another key measure of performance. Finally, this paper focuses on the application to MSI and gene expression data, but the proposed similarity measure could also be applied to other types of biological data. Future work will also examine the performance of the proposed multivariate hypergeometric similarity measure for detecting patterns in other types of large-scale biological data sets, with which other similarity measures like mutual information, cosine similarity, and Pearson correlation are currently used.
Acknowledgments
This research was supported by grants from the Parker H. Petit Institute for Bioengineering and Bioscience, Johnson & Johnson, the Bio Imaging Mass Spectrometry Initiative at Georgia Tech, the US National Institutes of Health (Bioengineering Research Partnership R01CA108468, Center for Cancer Nanotechnology Excellence U54CA119338), the Georgia Cancer Coalition (Distinguished Cancer Scholar Award to MDW), Microsoft Research, and the US National Science Foundation (GRFP to CK). The authors would like to thank Drs. M. Cameron Sullards and Yanfeng Chen for sharing the MSI data set used in this study and Dr. John H. Phan for assistance with the gene expression data used in this study.
Contributor Information
Chanchala D. Kaddi, Email: gtg538v@mail.gatech.edu, Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA 30332.
R. Mitchell Parry, Email: rmp@cs.appstate.edu, Department of Computer Science, Appalachian State University, Boone, NC 28608.
May D. Wang, Email: maywang@bme.gatech.edu, Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA 30332.
References
- 1.Sadygov RG, Yates JR. A Hypergeometric Probability Model for Protein Identification and Validation Using Tandem Mass Spectral Data and Protein Sequence Databases. Analytical Chemistry. 2003;75:3792–3798. doi: 10.1021/ac034157w. [DOI] [PubMed] [Google Scholar]
- 2.Shih IM, Nakayama K, Wu G, Nakayama N, Zhang JH, Wang TL. Amplification of the Ch19P13.2 NACC1 Locus in Ovarian High-Grade Serous Carcinoma. Modern Pathology. 2011;24:638–645. doi: 10.1038/modpathol.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyack KW, Newman D, Duhon RJ, Klavans R, Patek M, Biberstine JR, Schijvenaars B, Skupin A, Ma NAL, Borner K. Clustering More than Two Million Biomedical Publications: Comparing the Accuracies of Nine Text-Based Similarity Approaches. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0018029. article e18029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Megalooikonomou V, Barnathan M, Kontos D, Bakic PR, Maidment ADA. A Representation and Classification Scheme for Tree-Like Structures in Medical Images: Analyzing the Branching Pattern of Ductal Trees in X-Ray Galactograms. IEEE Trans Medical Imaging. 2009 Apr;28(4):487–493. doi: 10.1109/TMI.2008.929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell TM, Shinkareva SV, Carlson A, Chang KM, Malave VL, Mason RA, Just MA. Predicting Human Brain Activity Associated with the Meanings of Nouns. Science. 2008;320:1191–1195. doi: 10.1126/science.1152876. [DOI] [PubMed] [Google Scholar]
- 6.Perlman L, Gottlieb A, Atias N, Ruppin E, Sharan R. Combining Drug and Gene Similarity Measures for Drug-Target Elucidation. J Computational Biology. 2011;18:133–145. doi: 10.1089/cmb.2010.0213. [DOI] [PubMed] [Google Scholar]
- 7.Yona G, Dirks W, Rahman S, Lin DM. Effective Similarity Measures for Expression Profiles. Bioinformatics. 2006;22:1616–1622. doi: 10.1093/bioinformatics/btl127. [DOI] [PubMed] [Google Scholar]
- 8.Cha S-H. Comprehensive Survey on Distance/Similarity Measures between Probability Density Functions. Int’l J Math Models and Methods in Applied Sciences. 2007;1:300–307. [Google Scholar]
- 9.Li XB, Dubes RC. A Probabilistic Measure of Similarity for Binary Data in Pattern-Recognition. Pattern Recognition. 1989;22:397–409. [Google Scholar]
- 10.Kaddi C, Parry RM, Wang MD. Hypergeometric Similarity Measure for Spatial Analysis in Tissue Imaging Mass Spectrometry. Proc IEEE Int’l Conf Bioinformatics and Biomedicine (BIBM ’11), pp. 2011:604–607. doi: 10.1109/BIBM.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The Mutual Information: Detecting and Evaluating Dependencies between Variables. Bioinformatics. 2002 Oct;18:S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Chen Y, Momin A, Shaner R, Wang E, Bowen NJ, Matyunina LV, Walker LD, McDonald JF, Sullards MC, Merrill AH., Jr Elevation of Sulfatides in Ovarian Cancer: An Integrated Transcriptomic and Lipidomic Analysis Including Tissue-Imaging Mass Spectrometry. Molecular Cancer. 2010;9 doi: 10.1186/1476-4598-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonnell LA, Heeren RMA. Imaging Mass Spectrometry. Mass Spectrometry Revs. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 14.Van de Plas R, Pelckmans K, De Moor B, Waelkens E. Spatial Querying of Imaging Mass Spectrometry Data: A Nonnegative Least Squares Approach. Proc Neural Information Processing Systems Workshop Machine Learning in Computational Biology. 2007 [Google Scholar]
- 15.Freeman GH, Halton JH. Note on an Exact Treatment of Contingency, Goodness of Fit and Other Problems of Significance. Biometrika. 1951;38:141–149. [PubMed] [Google Scholar]
- 16.Sprent P, Smeeton NC. Applied Nonparametric Statistical Methods. Chapman & Hall/CRC; 2001. [Google Scholar]
- 17.Verbeek A, Kroonenberg PM. A Survey of Algorithms for Exact Distributions of Test Statistics in R C Contingency Tables with Fixed Margins. Computational Statistics and Data Analysis. 1985;3:159–185. [Google Scholar]
- 18.Greselin F. Counting and Enumerating Frequency Tables with Given Margins. Statistica and Applicazioni. 2003;1:87–104. [Google Scholar]
- 19.Gail M, Mantel N. Counting the Number of r c Contingency Tables with Fixed Margins. J Am Statistical Assoc. 1977;72:859–862. [Google Scholar]
- 20.Nath GB, Iyer PVK. Note on the Combinatorial Formula for nHr. J Australian Math Soc. 1972;14:264–268. [Google Scholar]
- 21.Anand H, Dumir VC, Gupta H. A Combinatorial Distribution Problem. Duke Math J. 1966;33:757–769. [Google Scholar]
- 22.Chen YF, Allegood J, Liu Y, Wang E, Cachon-Gonzalez B, Cox TM, Merrill AH, Sullards MC. Imaging MALDI Mass Spectrometry Using an Oscillating Capillary Nebulizer Matrix Coating System and Its Application to Analysis of Lipids in Brain from a Mouse Model of Tay-Sachs/Sandhoff Disease. Analytical Chemistry. 2008;80:2780–2788. doi: 10.1021/ac702350g. [DOI] [PubMed] [Google Scholar]
- 23.Chen CS, Zhou ZM, Sheehan CE, Slodkowska E, Sheehan CB, Boguniewicz A, Ross JS. Overexpression of WWP1 is Associated with the Estrogen Receptor and Insulin-Like Growth Factor Receptor 1 in Breast Carcinoma. Int’l J Cancer. 2009 Jun;124:2829–2836. doi: 10.1002/ijc.24266. [DOI] [PubMed] [Google Scholar]
- 24.Parris TZ, Danielsson A, Nemes S, Kovacs A, Delle U, Fallenius G, Mollerstrom E, Karlsson P, Helou K. Clinical Implications of Gene Dosage and Gene Expression Patterns in Diploid Breast Carcinoma. Clinical Cancer Research. 2010 Aug;16:3860–3874. doi: 10.1158/1078-0432.CCR-10-0889. [DOI] [PubMed] [Google Scholar]
- 25.Causey MW, Huston LJ, Harold DM, Charaba CJ, Ippolito DL, Hoffer ZS, Brown TA, Stallings JD. Transcriptional Analysis of Novel Hormone Receptors PGRMC1 and PGRMC2 as Potential Biomarkers of Breast Adenocarcinoma Staging. J Surgical Research. 2011 Dec;171:615–622. doi: 10.1016/j.jss.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 26.Ferguson DA, Muenster MR, Zang Q, Spencer JA, Schageman JJ, Lian Y, Garner HR, Gaynor RB, Huff JW, Pertsemlidis A, Ashfaq R, Schorge J, Becerra C, Williams NS, Graff JM. Selective Identification of Secreted and Transmembrane Breast Cancer Markers Using Escherichia coli Ampicillin Secretion Trap. Cancer Research. 2005 Sep;65:8209–8217. doi: 10.1158/0008-5472.CAN-04-3726. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda H, Taira N, Hara F, Fujita T, Yamamoto H, Soh J, Toyooka S, Nogami T, Shien T, Doihara H, Miyoshi S. The Estrogen Receptor Influences Microtubule-Associated Protein Tau (MAPT) Expression and the Selective Estrogen Receptor Inhibitor Fulvestrant Downregulates MAPT and Increases the Sensitivity to Taxane in Breast Cancer Cells. Breast Cancer Research. 2010;12(3) doi: 10.1186/bcr2598. article R43. KADDI ET AL.: MULTIVARIATE HYPERGEOMETRIC SIMILARITY MEASURE 1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voduc D, Cheang M, Nielsen T. GATA-3 Expression in Breast Cancer Has a Strong Association with Estrogen Receptor but Lacks Independent Prognostic Value. Cancer Epidemiology Biomarkers and Prevention. 2008 Feb;17:365–373. doi: 10.1158/1055-9965.EPI-06-1090. [DOI] [PubMed] [Google Scholar]
- 29.Drury SC, Detre S, Leary A, Salter J, Reis J, Barbashina V, Marchio C, Lopez-Knowles E, Ghazoui Z, Habben K, Arbogast S, Johnston S, Dowsett M. Changes in Breast Cancer Biomarkers in the IGF1R/PI3K Pathway in Recurrent Breast Cancer after Tamoxifen Treatment. Endocrine-Related Cancer. 2011 Oct;18:565–577. doi: 10.1530/ERC-10-0046. [DOI] [PubMed] [Google Scholar]
- 30.Fagan D, Yee D. Crosstalk between IGF1R and Estrogen Receptor Signaling in Breast Cancer. J Mammary Gland Biology and Neoplasia. 2008 Dec;13:423–429. doi: 10.1007/s10911-008-9098-0. [DOI] [PubMed] [Google Scholar]
- 31.Baker BG, Ball GR, Rakha EA, Nolan CC, Caldas C, Ellis IO, Green AR. Lack of Expression of the Proteins GMPR2 and PPAR are Associated with the Basal Phenotype and Patient Outcome in Breast Cancer. Breast Cancer Research and Treatment. 2013;137:127–137. doi: 10.1007/s10549-012-2302-3. [DOI] [PubMed] [Google Scholar]
- 32.Tomlinson DC, Knowles MA, Speirs V. Mechanisms of FGFR3 Actions in Endocrine Resistant Breast Cancer. Int’l J Cancer. 2012 Jun;130:2857–2866. doi: 10.1002/ijc.26304. [DOI] [PubMed] [Google Scholar]
- 33.Irigoien I, Arenas C. INCA: New Statistic for Estimating the Number of Clusters and Identifying Atypical Units. Statistics in Medicine. 2008;27:2948–2973. doi: 10.1002/sim.3143. [DOI] [PubMed] [Google Scholar]


