Abstract
The WWTR1 (protein is known as TAZ) -CAMTA1 (WC) fusion gene defines epithelioid hemangioendothelioma, a malignant vascular cancer. TAZ (transcriptional coactivator with PDZ binding motif) is a transcriptional coactivator and end effector of the Hippo tumor suppressor pathway. It is inhibited by phosphorylation by the Hippo kinases LATS1 and LATS2. Such phosphorylation causes cytoplasmic localization, 14-3-3 protein binding and the phorphorylation of a terminal phosphodegron promotes ubiquitin-dependent degradation (the phosphorylation of the different motifs has several effects). CAMTA1 is a putative tumor suppressive transcription factor. Here we demonstrate that TAZ-CAMTA1 (TC) fusion results in its nuclear localization and constitutive activation. Consequently, cells expressing TC display a TAZ-like transcriptional program that causes resistance to anoikis and oncogenic transformation. Our findings elucidate the mechanistic basis of TC oncogenic properties, highlight that TC is an important model to understand how the Hippo pathway can be inhibited in cancer, and provide approaches for targeting this chimeric protein.
INTRODUCTION
The WWTR1 (protein known as TAZ)-CAMTA1 (WC) gene fusion encoded by a t(1;3)(p36;q25) translocation is present in > 90% of epithelioid hemangioendotheliomas (EHEs), a vascular cancer.1,2 Subsequent studies have revealed a less frequent YAP1-TFE3 (YT) fusion gene, which is present in < 10% of EHEs.3 EHE has a simple karyotype; other than the recurrent t(1;3) translocation, no consistent cytogenetic alterations have been described in this tumor,4 suggesting that the WC gene fusion likely constitutes the initiating tumorigenic event.
TC fuses the N-terminus of WW domain-containing transcription regulator 1 (WWTR1 protein known as TAZ (transcriptional coactivator with PDZ binding motif)) to the C terminus of Calmodulin binding transcription activator 1 (CAMTA1).1,2 TAZ and its human paralogue YAP have emerged as potential oncogenes in cancer.5 TAZ and YAP are expressed strongly in several cancers including breast cancer, colon cancer, liver cancer, lung cancer, and thyroid cancer.5,6 Expression of human YAP1 S127A in satellite skeletal muscle cells in a mouse model is sufficient to cause embryonal rhabdomyosarcoma.7 Thus expression of an activated form of YAP1 by itself is sufficient to drive cancer formation in the correct cell lineage.
The Hippo pathway is named because loss of Hippo (HPO) function results in tissue overgrowth in Drosophila melanogaster, causing ‘hippopotamus-like’ folds. Subsequent studies have also revealed that the hippo tumor suppressor is an important negative regulator of tissue growth in mammals.8,9 The Hippo pathway is a serine/threonine kinase cascade composed of the core kinases mammalian STE20-like protein kinases 1 and 2 (MST1/2, the homologs of D. melanogaster HPO),10–13 and the large tumor suppressor 1 and 2 (LATS1/2) (13,14).14,15 TAZ and YAP are both transcriptional coactivators and are the end effectors of the Hippo pathway.8,9 When TAZ and YAP are phosphorylated by LATS1/2, they translocate from the nucleus to the cytoplasm, where they undergo ubiquitin-mediated degradation, thus inactivating their transcriptional activity.8,9 During cellular states such as confluence or when cells are detached, Hippo is activated, resulting in phosphorylation of TAZ and YAP and their retention in the cytoplasm, where they are subsequently degraded.9,16
CAMTA1 is a transcription factor that has been implicated as a candidate tumor suppressor in neural cancers.17–20 The CAMTA1 gene is found in essentially all multicellular organisms. It is evolutionarily conserved down to Arabidopsis where it has been shown to have a role in drought tolerance.21 In humans, its function is not completely understood, although a role in memory has been suggested, given the high mRNA levels of CAMTA1 in memory-related regions of the brain.22 In addition, genetic alterations of CAMTA1 have been identified in neuropsychological diseases.23,24
Here we sought to define the mechanistic basis of how TC functions to promote cancer. We anticipate that understanding the mechanistic basis of TC function will provide insights into how TAZ and the Hippo pathway may be involved not only in the genesis of EHE, but also in other cancers, offering insights into approaches that target the TAZ and TC chimeric proteins therapeutically.
RESULTS
TAZ-CAMTA1 (TC) exhibits oncogenic activity in vitro
The TC fusion protein contains a 189 AA amino-terminal portion of TAZ including the TEA domain containing DNA (TEAD) binding domain, the 14-3-3 protein binding motif, and most of the WW domain fused in frame to a 1405 AA carboxy terminal portion of CAMTA1 containing the transactivation domain, TIG domain, ankyrin repeats, and IQ domains (Figure 1a). Because TC is the defining molecular alteration in the vast majority of EHEs, we hypothesized that it would have oncogenic properties.
Figure 1.
TC structure and oncogenic effects in vitro (a) Structure of full-length TAZ, CAMTA1, and the TC fusion protein. TAZ is 400 amino acids in length (44Kd) and contains a TEAD binding domain, a 14-3-3 protein binding motif, a WW domain, a transactivation domain including a coiled coil domain and a PDZ protein binding motif.31 CAMTA1 is 1673 amino acids in length (184Kd) and contains a CG-1 domain thought to bind CG-rich DNA sequences, a transactivation domain (TAD), a transcription factor immunoglobulin (TIG) domain, a series of ankyrin repeats (ANK) and three IQ calmodulin-binding motifs (IQ refers to the first two amino acids of the motif, isoleucine (I) and glutamine (Q)).32 While there is some microheterogeneity between different WWTR1–CAMTA1 gene fusions found in different EHEs, the most inclusive TAZ-CAMTA1 fusion protein is 1594 AA’s in length and predicts a 173Kd protein.2 The TAZ-CAMTA1 fusion protein is composed of the TEAD binding domain, 14-3-3 protein binding motif, and WW domain donated from TAZ fused in frame to the TAD TIG domain, ankyrin repeats, IQ domain and nuclear localization signal (NLS). S51 is critical for binding to TEAD proteins. (b) Soft agar assay with NIH/3T3 cells expressing empty vector, the TC fusion protein, full-length TAZ, full-length CAMTA1, as well as the truncated portions of TAZ and CAMTA1 involved in the fusion protein. Quantitative results graphically represented on the right. (c) Proliferation assay of cells to grow in suspension. NIH/3T3 cells expressing TC, but not TAZ or EV were able to grow in suspension. Error bars (b and c) represent one s.d.; experiments were performed at least twice. EV= empty vector.
Because there are no EHE cell lines or other models, we opted to study the function of TC by expressing it in several nontransformed cell lines (see Supplementary Table S1). The resultant TC-expressing cell lines were assayed for hallmarks of cancer, including proliferation, migration/invasion and anchorage-independent growth.25 Forced expression of TC in NIH/3T3 (TC-NIH/3T3) cells resulted in colony formation in soft agar (Figure 1b), comparable to that caused by expression of the N-Ras G12V mutant (Supplementary Figure S1). To ascertain whether the colony formation in soft agar was caused by TC itself, rather than the truncated portions of TAZ or CAMTA1 that are contained in the TC fusion protein, we forced the expression of full length and truncated TAZ or CAMTA1 in NIH/3T3 cells. TC, but not full length or truncated TAZ or CAMTA1, induced colony formation. These observations demonstrate that components of both TAZ and CAMTA1 are required for oncogenic transformation (Figure 1b).
NIH/3T3 cells are unable to grow in suspension and thus fail to show resistance to anoikis, another hallmark of cancer. Given that YAP has been implicated in promoting resistance to anoikis,16 we sought to determine whether TC could also confer resistance to anoikis by facilitating growth in suspension. Forced expression of TC was able to drive the proliferation of NIH/3T3 cells cultured in suspension on poly-HEMA coated plates (Figure 1c), whereas cells containing empty vector did not, mirroring the results found in soft agar.
TC is a transcription factor and activates a predominantly TAZ transcriptional program
As TAZ and CAMTA1 are both transcription factors/coactivators, we hypothesized that the TC fusion protein would function as a neomorphic transcription factor. We sought to interrogate the expression program in human cells in addition to mouse NIH/3T3 cells because we reasoned that comparisons of the transcriptional output of TC across species would strengthen the significance of the transcriptional program. Although HEK293 cells are not useful to analyze the effects of TC on growth in soft agar and in suspension because they grow on the substrates in the absence of TC (that is, they are already transformed), they do drive a reporter plasmid that is relevant to TC-TEAD4 transcription (See Figure 3c). Thus, we reasoned that HEK293 cells would be useful in establishing the TC transcriptional output. We stably transfected empty vector, TAZ, CAMTA1 and TC into NIH/3T3 cells and HEK293 cells and sought to define the impact of the different constructs on the transcriptional programs induced by TC. We also asked if TC expression would recapitulate those caused by expression of wild-type TAZ or CAMTA1. RNA samples extracted from the transfected cell lines were subjected to RNA-sequencing to determine their transcriptomes. Differential gene expression analysis comparing cell lines transfected with TAZ, CAMTA1 and TC with empty vector-transfected cells revealed 668, 18 and 137 genes differentially expressed by twofold in HEK293 cells and 619, 81 and 444 differentially expressed by twofold in NIH/3T3 cells, respectively (Figure 2 and Supplementary Data File 1). Comparison of the different sets of differentially expressed genes revealed a striking correlation between the set of RNAs induced/repressed by TAZ and the set of RNAs induced/repressed by TC in both HEK293 and NIH/3T3 cells (Hypergeometric test P-values of 8.4 × 10−50 and 2.7 × 10−55, respectively). On the basis of a Jaccard similarity analysis, we observed that the transcriptional programs induced by TC were substantially more similar to those caused by TAZ (Jaccard indices 0.11 and 0.17) than by CAMTA1 (Jaccard indices of 0.01 and 0.03) in both HEK293 and NIH/3T3 cells (Figure 2a). A comparison of genes induced/repressed by TAZ and CAMTA1 yielded hypergeometric test P-values of 2.3 × 10−2 and 2.7 × 10−21 and Jaccard indices of 0.01 and 0.06, respectively. Taken together, these results demonstrate that forced expression of TC results in a TAZ-like transcriptomic program. This result is consistent with the fact that the portion of TAZ that is included in TC retains its TEAD4 binding domain, which specifies the promoters to which TAZ binds, while the putative DNA-binding domain of CAMTA1 is lost.
Figure 3.
TC binds DNA through interaction with Tead4. Substitution of serine 51 for alanine (S51A) in TC resulted in abrogation of colony formation in soft agar (a) and decreased proliferation (b) in suspension. (c) TC S51A showed significantly reduced transcriptional activity by a Tead4 binding site luciferase reporter assay in TC S51A-HEK293 cells. (d) Effective RNA interference silencing of Tead4 in TC-NIH/3T3 cells. TC (TC-expressing cells), shEV (short hairpin empty vector control), shScr (scrambled empty vector control), sh1 (short hairpin Tead4 knock-down 1), sh2 (short hairpin Tead4 knock-down construct 2). (e) Silencing of Tead4 resulted in abrogation of colony formation in soft agar with sh1 and sh2 in TC-NIH/3T3 cells. (f) Silencing of Tead4 inhibited the ability of TC-NIH/3T3 cells to grow in suspension. (g) TC-NIH/3T3 cells and TCS51A-NIH/3T3 cells were transfected with Myc-Tead4, immunoprecipitated with anti-Myc antibody, then probed with anti-Flag antibody, showing that TC co-IPs with TEAD4 whereas TCS51A does not. Error bars (B, C, F) represent one s.d.; all experiments performed at least twice.
Figure 2.
TC promotes a predominantly TAZ-like gene expression signature (a) Diagrammatic representation of the overlap of differentially expressed genes between TAZ-CAMTA1 (TC), TAZ and CAMTA1 in HEK293 and NIH/3T3 cells. (b) Quantitative RT-PCR expression of CTGF in HEK293 and NIH/3T3 derivative cell lines. Error bars (b) represent one s.d.; values are averages of three separate wells from a single experiment. Values are normalized to EV. EV=empty vector.
Connective tissue growth factor (CTGF) is a canonical TAZ-TEAD4 target that was induced in all cell lines expressing TAZ and TC.26 To validate our RNA-seq results, CTGF expression was determined by RT-PCR in HEK293 and NIH/3T3 cells harboring the various expression constructs, revealing greatly induced expression of CTGF by TAZ and TC but not CAMTA1 or empty vector (Figure 2b). Also of interest, functional annotation of the genes in the intersection of the Venn diagrams also revealed several genes including TYMP (HEK293 cells) and Anxa2, Igsf10, Mmp2, and Plcd3 (NIH-3T3 cells) that are involved in ‘vasculature development’ and/or ‘blood vessel development.
TC interacts with TEAD4 to drive its transcriptional program
TEAD4, a TEAD transcription factor, is known to interact with TAZ and is responsible for tethering TAZ to TEAD4 bound promoters.26 A serine to alanine substitution on residue 51 of TAZ has been shown to ablate the interaction of TAZ with TEAD4.26 Serine 51 of TAZ is in the same position as serine 51 of TC (see Figure 1a). As TC was shown to activate a TAZ-like transcriptional program, we hypothesized that interaction with TEAD4 was necessary to activate its transcriptional program.
We tested the ability of the TC S51A mutant to promote colony formation in soft agar and growth in suspension of NIH/3T3 cells. As predicted, the TC S51A mutation abrogated TC-mediated NIH/3T3 colony formation in soft agar and growth in suspension (Figures 3a and b). Using a reporter containing eight consecutive Tead1–4 binding sites (8XGTIIC plasmid; GTIIC is also known as MCAT and Hippo response element), TC S51A significantly reduced TC transcriptional activity (Figure 3c). Silencing Tead4 in TC NIH/3T3 cells using two different shRNA constructs (Figure 3d) also resulted in abrogation of colony formation in soft agar and growth/proliferation in suspension (Figures 3e and f). We further confirmed the interaction of TC with Tead4 by co-immunoprecipitation (co-IP). While TC was able to co-IP with Tead4, the S51A mutation disrupted this interaction, confirming that the loss-of-function phenotypes were due to the loss of interaction between TC S51A and Tead4 (Figure 3g).
TC is insensitive to regulation by the Hippo pathway
TAZ activity is negatively regulated by the Hippo pathway. As TC gave NIH/3T3 cells the ability to proliferate in detached conditions where the Hippo pathway is activated (that is, growth in suspension), we posited that fusion of TAZ to CAMTA1 could interfere with the ability of the Hippo pathway to regulate the TAZ portion of the fusion protein, resulting in its constitutive activation.
LATS kinase is a core Hippo kinase and the direct Hippo regulator of TAZ. It phosphorylates TAZ on several serine residues, the most important of which is serine 89, which is localized within a 14-3-3 protein binding motif HVRSHSSP.27 When TAZ is phosphorylated at serine 89, 14-3-3ε binds to TAZ and causes it to translocate out of the nucleus and into the cytoplasm where it subsequently undergoes ubiquitin-mediated degradation.27–29 Mutation of TAZ serine 89 to alanine functionally ablates the 14-3-3 protein binding site, and has been shown to result in constitutive activation of TAZ.27 We confirmed that TAZ S89A causes NIH/3T3 cells to grow as colonies in soft agar (Figure 4a).30 In contrast to TAZ, the TC S89A substitution did not result in an increase in colony formation, suggesting that TC is not regulated by the Hippo pathway (Figure 4b). We also predicted that TC would reside in the nucleus constitutively when TC NIH/3T3 cells were grown in suspension. Indeed, cellular fractionation experiments demonstrated that in contrast to TAZ, nuclear TC levels remained constant when cells were grown in suspension (Figure 4c).
Figure 4.
TC is constitutively active and not regulated by the Hippo pathway. (a) Expression of TAZ promotes minimal colony formation in NIH/3T3 cells in soft agar, whereas expression of TAZ S89A in NIH/3T3 cells shows a marked increase in the number of colonies formed in soft agar, showing that TAZ is negatively regulated by the Hippo pathway. Expression of TCS89A in NIH/3T3 cells does not result in an increase of colonies when compared with expression of TC in NIH/3T3 cells, indicating that TC is not regulated by Hippo. (b) In NIH/3T3 cells grown in suspension, TAZ translocates out of the nucleus into the cytoplasm, consistent with its regulation by the Hippo pathway. When detached, the nuclear to cytoplasmic equilibrium of TC does not change; further supporting that TC is not regulated by Hippo. (c) Immunofluorescence (IF) reveals TAZ to be located within the nucleus of NIH/3T3 cells when they are plated sparsely. When they are plated at confluent conditions, the Hippo pathway is activated and the TAZ signal is absent because TAZ translocates into the cytoplasm and is subsequently degraded. TC is located within the nucleus during both sparse and confluent conditions, indicating that it is no longer regulated by the Hippo pathway. Scale bar=20 microns. (d) Western blot shows that when TAZ-NIH/3T3 cells are plated under confluent conditions, addition of MG132 (MG) rescues TAZ levels to those found under sparse conditions with DMSO. This confirms that TAZ is degraded under confluent conditions and explains the lack of TAZ signal seen with IF under confluent conditions. (e) Co-immunoprecipitation experiments - Flag-TAZ and Flag-TC were pulled down with anti-Flag antibody and probed with 14-3-3ε antibody. Whereas flag-TAZ coimmunoprecipitates with 14-3-3ε, flag-TC does not, providing a putative mechanism for TC escape from Hippo pathway regulation. Evaluation of S89 phosphorylation during confluent conditions (f) and when grown in suspension (g). Immunoprecipitation with anti-Flag antibody is done first to eliminate non-specific bands with the phospho-S127 YAP antibody. Both TAZ and TC are phosphorylated on serine 89 (homologous to S127 in YAP) during confluent conditions and while grown in suspension. Serine 89 on TC is phosphorylated even though Hippo functionally is not regulating TC. Error bars (a, b, c) represent one s.d.; all experiments performed at least twice.
The Hippo pathway is also activated during cell crowding, presumably as a mechanism to control organ size.9 The localization of TC during sparse and confluent conditions was therefore evaluated. Immunofluorescence analysis revealed that TC is located within the nucleus during both sparse and confluent conditions (Figure 4d). In contrast, during confluent conditions TAZ translocates from the nucleus into the cytoplasm where it is subsequently degraded (Figure 4d). To confirm that the absence of cytoplasmic signal for TAZ in confluent conditions was due to TAZ degradation, MG132, a proteasome inhibitor, was added to cells plated at both sparse and confluent conditions, which rescued TAZ levels to those seen at sparse conditions (Figure 4e).
To investigate the basis for the Hippo pathway independence of TC, we sought to determine whether 14-3-3ε interacts with TC. As previously reported,27,31 upon TAZ IP under confluent conditions, 14-3-3ε co-immunoprecipitates with TAZ (Figure 4f). However, upon TC IP under confluent conditions, 14-3-3ε does not coimmunoprecipitate with TC (Figure 4f). To determine whether LATS1/2 phosphorylates TC, we employed an antibody against phospho-S127 YAP, also known to bind to phospho-S89 TAZ, the analogous site phosphorylated by LATS1/2 in TAZ. Western blot analysis using this antibody showed that TC was phosphorylated on serine 89 (Figure 4g; control blots Supplementary Figure S2C). Although TC is phosphorylated at serine 89, it no longer binds to 14-3-3ε inhibiting the Hippo pathway’s ability to target TC for cytoplasmic translocation and degradation.
CAMTA1 contributes a nuclear localization signal to TC
We next investigated whether CAMTA1 might also contribute additional functional domains to the fusion protein which would be important for its constitutive activation. Given that TAZ does not contain a nuclear localization signal (NLS) and that we have observed that constitutive nuclear localization of TC is important for its function, we hypothesized that CAMTA1 might be contributing a NLS to the fusion protein. A well-described NLS is present within the N-terminus of CAMTA1;32 however, this portion of the protein is absent from the TC fusion. We noted that CAMTA2,33 a highly homologous CAMTA family member, contains an additional NLS in its C-terminus. Hence, we wondered whether CAMTA1 might also contain a NLS in its C terminus. Taking a bioinformatics-based approach, we used the NLStradamus algorithm34 and identified a putative monopartite NLS, KKCGKRRQ, involving amino acids 1608–1615 of CAMTA1 (amino acids 1529–1536 of TC).
Cells expressing a mutant form of TC that deleted the NLS (TCΔNLS) showed a markedly reduced ability to form colonies in soft agar and to grow in suspension (Figures 5a and b) as well as a corresponding decrease in transcriptional activity in the Tead4 luciferase reporter assay (Figure 5c). Immunofluorescence analysis revealed that the TC protein localized to the nuclei of a few cells when the TCΔNLS mutant was stably expressed (Figure 5d) as compared to the expression of wild-type TC, which was localized to the nuclear compartment in 100% of the cells (Figure 5d). MG132 proteasome inhibition resulted in a significant increase in the cytoplasmic amounts of TCΔNLS (Figure 5e). These results suggest that the equilibrium of the TCΔNLS mutant is shifted so that it no longer resides predominantly in the nucleus, but is exported to the cytoplasm, where it is subsequently degraded. Finally, we fused the 64 terminal amino acids of CAMTA1, starting with the eight aminoacid NLS signal that was deleted in the TCΔNLS mutant to green fluorescent protein (NLS-GFP) and expressed this mutant in NIH-3T3 cells. When we examined the cellular localization of NLS-GFP, we found that fluorescence was localized exclusively to the nucleus whereas a GFP that was not fused to the NLS was expressed predominantly in the cytoplasm (Figure 5f). This provides direct proof that the amino terminal 64 amino acids of CAMTA1 contains a NLS.
Figure 5.
CAMTA1 contributes a NLS to TC. (a) NIH/3T3 cells expressing the TCΔNLS construct (lacking the putative NLS) demonstrate a markedly reduced ability to form colonies in soft agar. (b) TCΔNLS expressing cells display a reduced ability to proliferate in suspension compared with TC. (c) TCΔNLS-NIH/3T3 cells show a reduced ability to stimulate transcription in a Tead4 luciferase reporter assay. (d) TCΔNLS is rarely expressed in the nuclear compartment of NIH/3T3 cells. MG132 treatment results in TCΔNLS accumulation within the cytoplasm, indicating that without a NLS, the equilibrium of TCΔNLS is shifted to the cytoplasm where it is subsequently degraded. (d) In whole cell lysates, TCΔNLS expression is negligible. With MG132 treatment, TCΔNLS can be detected supporting the idea that it is retained in the cytoplasm and degraded. Nuclear and cytoplasmic fractionation of TCΔNLS treated with MG132 shows that TCΔNLS is localized predominantly within the cytoplasm of cells, consistent with the IF findings. (e) Immunofluoresence reveals that GFP is localized predominantly to the cytoplasm but also to the nucleus (top row) while GFP-NLS (bottom row) is localized exclusively to the nucleus. Error bars (a, b, c) represent one s.d.; all experiments performed at least twice.
DISCUSSION
In this paper, we describe the mechanism of action of the TC fusion oncoprotein. The TC oncoprotein activates a TAZ-like transcriptional program. Although CAMTA1 constitutes 90% of the fusion protein, surprisingly, it is the TAZ portion that specifies the TC transcriptional program. The TAZ portion of TC contains the TEAD binding domain of TAZ, and it is the TEAD family of transcription factors that tether TC to DNA, similar to DNA tethering of wild-type TAZ (Figure 3).
We show that the CAMTA1 portion of TC plays a pivotal role in the constitutive nuclear localization of WC, by donating a strong NLS (Figure 5). This is critical because the NLS of TAZ is likely located within the PDZ motif which is lost in the TC fusion. This is supported by previous work demonstrating that the PDZ motif of YAP2, which is a highly homologous TAZ orthologue, contains its NLS within the PDZ motif.35 In addition to donating a NLS to TC, a major consequence of CAMTA1 fusion to TAZ is that the TAZ portion of the fusion protein is no longer responsive to the Hippo pathway. The Hippo pathway, when activated in conditions such as cell confluence, phosphorylates TC on serine 89. Despite this, TC maintains its nuclear localization because14-3-3ε does not bind S89 phosphorylated TC (Figures 4f and g). 14-3-3 proteins are known to mediate cytoplasmic localization of various proteins.36 One possibility is that the CAMTA1 portion of the fusion protein physically disrupts the interaction between 14-3-3ε and TC. Alternatively, another property of CAMTA1, such as the NLS donated to the TC fusion protein, results in constitutive TC nuclear localization which denies access to 14-3-3ε. In this context, the absence of 14-3-3ε binding to TC is not the cause of the constitutive activation of the fusion protein, rather it is the effect. Future studies to distinguish between these two possibilities are warranted.
In karyotypically simple cancers such as EHE, where chimeric transcription factors are the initiating and main tumorigenic event driving the cancer, blocking these oncogenic transcription programs would likely yield a significant therapeutic effect. Previously, Liu-Chittenden et al.37 demonstrated that disruption of YAP–TEAD4 interaction interfered with YAP oncological properties in a mouse model. Along with our own findings, where we inhibited TC function by blocking the TC-TEAD 4 interaction genetically and by depleting TEAD4 (Figure 3), these results suggest that YAP-TEAD4, TAZ-TEAD 4 and TC-TEAD4 interaction represent a potential Achilles heel for targeted therapy in EHE and other cancers driven by activated YAP or TAZ.
Although rare sarcomas with simple cytogenetic alterations offer opportunities to investigate important roles of oncogenes in cancer initiation and progression,38 they also pose logistical problems. As is the case with EHE, relevant cell lines and mouse models have not been developed, thus limiting the ability to perform a mechanistic dissection of the oncogene in its proper cancer context. Thus, we are obligated to study TC in the context of NIH/3T3 cells and HEK293 cells. These are standard cell lines used to study the functional consequences of expressing various oncogenes, especially when other model systems are not available for a specific cancer. The approach employed also offers opportunities, as studying the impact of the TC fusion gene in systems outside of their normal cancer context allows one to examine the generalized properties of the oncogene. Development of EHE cell lines, which would allow testing of the potential therapeutic approaches to target TC described in this study, is desired and warranted.
In summary, we have defined the mechanistic basis of the oncogenic functions of the TC fusion protein, which is the defining feature of EHE. We have shown that the TC oncoprotein activates a TAZ-like transcriptional program, that the fusion of CAMTA1 to TAZ results in constitutive nuclear localization due to a previously unknown C-terminal NLS in CAMTA1 that is present in TC (Figure 5), and that the TC oncoprotein is not regulated by the Hippo pathway, rendering it constitutively active (Figures 4 and 6). Further studies are under way to elucidate the downstream consequences of the TC transcriptional program and how they activate oncogenic transformation.
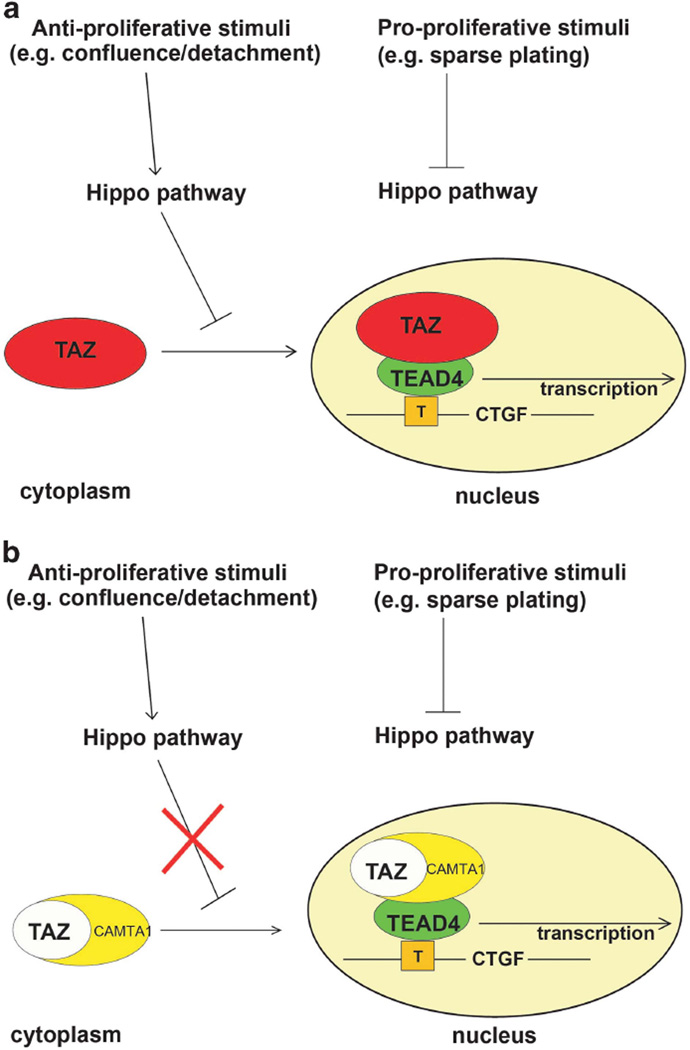
Figure 6.
Working model of TC function. (a) In cellular contexts which favor proliferation (for example, sparse plating or developing organs), the Hippo pathway is inhibited, leading to TAZ localization within the nucleus where it complexes with TEAD4 and activates its transcriptional program. Importantly, TAZ is negatively regulated by the Hippo pathway in cell contexts where the Hippo pathway is activated (confluence/detachment). (b) During cellular contexts where the Hippo pathway is inactivated (for example, sparse plating or in organs of normal size), TC functions in the same way as TAZ and is located in the nucleus, where it activates a TAZ-like transcriptional program, including the activation of canonical TAZ targets such as CTGF. In contrast to TAZ, WC is no longer regulated by the Hippo pathway, and, utilizing a NLS from the C-terminus of CAMTA1, is constitutively located within the nucleus even in conditions where the Hippo pathway is activated (confluence/detachment).
MATERIALS AND METHODS
Expression constructs
pDONR223WWTR1 ORF was obtained from ThermoScientific (Waltham, MA USA). pF1KSDA0833 (KIAA0833/CAMTA1 ORF) was obtained from Kazusa DNA Res. Inst. (Kisarazu, Chiba, Japan). WWTR1, CAMTA1 and WWTR1-CAMTA1 ORFs were fused to an N-terminus double-flag epitope tag and subsequently subcloned into the HindIII-XhoI site of pcDNA3Neo (Invitrogen-Life Technologies, Carlsbad, CA, USA).
The WWTR1-CAMTA1 ORF was constructed by fusing exons 1–3 of WWTR1 in frame to exons 9–23 of CAMTA1, based on the breakpoint of EHE5.2 The WWTR1-CAMTA1 ORF was constructed using overlapping primers and inserted into the HindIII-XhoI site of pcDNA3Neo. S51A and S89A mutants of WWTR1 and WWTR1-CAMTA1 were generated by site-directed mutagenesis. The nuclear localization mutant was generated by PCR-based mutagenesis. A 4.6-kb fragment of TC 5′ to the NLS sequence and a 0.2 kb fragment of TC 3’ to the NLS sequence was PCR amplified using overlapping primers which deleted the NLS sequence and hybridized to portions of the other PCR fragment. The 3’ fragment was then fused to the 5’ fragment in frame using PCR amplification to generate the TC ΔNLS mutant which deletes the NLS sequence from the TC protein. Expression of all constructs was verified (Supplementary Figure S2A and S2B). The GFP-NLS ORF was constructed by fusing the terminal 64 amino acids of CAMTA1 including the NLS to the N-terminus of enhanced green fluorescent protein, separated by a six amino-acid linker. The GFP-NLS ORF and GFP ORF were obtained as Gblocks from Integrated DNA Technologies (Coralville, IA, USA) and inserted into the BamH1-Sal1 site of pBABENeo by using overlapping primers. WWTR1, CAMTA1 and WWTR1-CAMTA1 ORFs were subcloned into the BamHI-SalI site of pBabeNeo.39 Cloning primers and site directed mutagenesis primers are listed in Supplementary Table S2. pRK5-Myc-TEAD4 was kindly provided by Dr Kun-Liang Guan.40
Antibodies and other materials
Anti-14-3-3ε polyclonal antibody (catalog #9635) and anti-phospho-YAP (S127; catalog #4911; rabbit polyclonal) were obtained from Cell Signaling (Danvers, MA, USA). Anti-TEAD4 (5H3; catalog #H00007004-M01) monoclonal antibody was obtained from Abnova (Taipei, Taiwan). Anti-FLAG monoclonal antibody (M2; catalog #F1804), β-actin (AC-15; catalog #A5441) and α-tubulin antibody (DM1A; catalog #T9026) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Anti-Histone H3 (FL-136; catlalogue #SC-10809) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-GFP monocolonal (DSHB-GFP-12A6) was obtained from Developmental Studies Hybridoma Bank (Iowa city, Iowa USA). Alexa Fluor 568- and 488-conjugated secondary antibodies (catalog #A-11004 and #A-10680, respectively) were obtained from Invitrogen-Life Technologies. Horseradish peroxidase-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology.
Cell culture, transfection, and retroviral infection
All cell lines were obtained directly from the American Type Tissue Collection (ATCC, Manassas, VA, USA) and tested every three months for mycoplasma contamination. NIH/3T3 cells were cultured in DMEM containing 10% Bovine Serum (Invitrogen-Life Technologies) and 50 µg/ml penicillin/streptomycin. HEK293 cells were cultured in DMEM containing 10% Fetal Bovine Serum and 50 µg/ml penicillin/streptomycin. All cells were cultured at 37° C and 5% CO2. Transfection with pcDNA3Neo was performed with Lipofectamine Plus Reagent (Invitrogen-Life Technologies) according to the manufacturer’s instructions. Pooled stable transfectants were generated by selecting for 2 weeks with G418 600 µg/ml for NIH/3T3 cells and 400 µg/ml for HEK293 cells. Retroviral transfection with pBabeNeo was performed by transfecting 293 PhoenixA retroviral packaging cells and supernatant was collected at 48 and 72 h after transfection, filtered with a 0.45-µm filter and supplemented with 8 µg/ml polybrene. Supernatant from 48 and 72 h were applied to cells for 8 h (serial infection). Pooled stable lines were generated by selecting in G418.
RNA sequencing
NIH/3T3 cells and HEK293 cells were stably transfused in duplicate (biological replicates) with empty vector (pBabeNeo), WWTR1, CAMTA1, or WWTR1-CAMTA1 and total RNA was extracted using Trizol Reagent (Invitrogen-Life Technologies). RNA samples were column purified using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) followed by DNase treatment (Invitrogen-Life Technologies). RNA quality was assessed using the Agilent Bio-analyzer (Agilent Technologies, Santa Clara, CA, USA). RNA-sequencing libraries were generated using Illumina TruSeq Stranded mRNA kit (Illumina, San Diego, CA, USA) and quantitated using the Agilent Bioanalyzer. Library pools were sequenced using the Illumina HiSeq2500 (Illumina). RNA sequencing data have been deposited in the Sequence Read Archive (SRA) under accessions SRP055005 and SRP055004. Reads from HEK293 and NIH3T3 cells were aligned to GRCh37 and GRCm38 using Bowtie 2.41 Aligned reads were summarized to the gene level using the Bioconductor package GenomicRanges.42 Differential expression was determined using the Bioconductor package DESeq.43 Genes consistently differentially expressed in all of TC, TAZ and CAMTA1 when compared with the empty vector were considered background effect and removed from further analysis. Comparison between lists of differentially expressed genes was performed using hypergeometric test and similarity was measured by the Jaccard index. Gene expression results were confirmed by quantitative RT-PCR (see below). Functional annotation of the genes in the intersection of the Venn diagrams was performed for the Gene Ontology terms using the ‘Functional Annotation Table’ tool of DAVID.44
TEAD4 RNA interference-mediated silencing
The following pLKO.1-puro constructs were obtained from Sigma-Aldrich. Empty vector construct (SHC001), scrambled negative control (SHC002), and Tead4 knock-down constructs TRCN0000054773 (TD1), TRCN00 00054774 (TD2), TRCN0000054775 (TD3), TRCN0000054776 (TD4), TRCN0000054777 (TD5). Constructs were transfected into HEK 293T cells along with pCMV8.12 and pVSVG packaging plasmids and virus was isolated for infection as previously described.42 Pooled stable lines were generated by selection in puromycin.
Quantitative RT–PCR
Total RNA was column purified from biological triplicates followed by DNase treatment. cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen-Life Technologies) starting with 1 µg total RNA and 250 ng of random primers (Promega, Madison, WI USA). PCR amplification was performed in technical duplicates on the LightCycler 480 SW 1.5 qPCR machine (Roche Diagnostics Corporation, Indianapolis, IN, USA) with monocolor hydrolysis probes from the Universal Probe Library System (Roche Diagnostics Corporation) and LightCycler 480 qPCR mixture (Roche Diagnostics Corporation). Relative quantitation was done using the delta-delta method and GAPDH as the reference control. The primers and probes used were as follows: Ctgf forward primer: 5′-tgacctggaggaaaacatt aaga-3′, Ctgf reverse primer: 5′-agccctgtatgtcttcacactg-3′, Probe: #71. CTGF forward primer: 5′-cctgcaggctagagaagcag-3′, CTGF reverse primer: 5′-tgga gattttgggagtacgg-3′, Probe: #85. qPCR cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s, 72 °C for 1 s, followed by 40 °C for 10 s.
Soft agar colony formation assay was performed according to previously published procedures.45 Each experimental condition was performed in technical triplicate and each experiment was performed at least twice.
Suspension culture
Tissue culture plates were coated with Poly 2-hydroxyethyl methacrylate (poly-HEMA) (Sigma-Aldrich) at 20 mg/ml and then UV sterilized overnight. Cells were plated and then collected/measured at time points required for the particular experiment.
Proliferation assay of suspension cultures
Cells (5 × 103 cells per 100 µl per well) were seeded into 10 individual wells for each time point in poly-HEMA-coated 96-well plates. Proliferation was assessed as previously described and measured as fold increase in absorbance over Day 0.42 Each experiment was performed at least twice.
Western blot
Western blotting was performed as previously described.42 Membranes were probed with anti-FLAG monoclonal antibody (1:1000), anti-14-3-3ε polyclonal antibody (1:1000), anti-phospho-S127-YAP (1:1000), anti-TEAD4 monoclonal antibody 5H3 (1:500), β-actin (1:5000), alpha-tubulin antibody (1:1000) and histone H3 (1:200). Each experiment was repeated at least twice.
Immunoprecipitation was performed as previously described.45 Each experiment was repeated at least twice.
Immunofluorescence staining
NIH/3T3 cells were fixed with 4% paraformaldehyde in 1XPBS for 15 min. After washing, cells were permeabilized and blocked with 0.3% Triton X-100 and 3% fetal bovine serum for 30 min. Cells were incubated with anti-Flag antibody diluted (1:1000) in 3% fetal bovine serum at 4° C overnight in a humidity chamber. Primary antibody was removed and cells were washed and then incubated with Alexa Fluor 568-conjugated secondary antibody (Invitrogen-Life Technologies) for 45 min to 1 h at room temperature. Slides were then washed and mounted with Vectashield (Vector Labs, Burlingame, CA, USA) with DAPI. Cells were visualized using the Leica DMR upright fluorescent microscope (Leica Microsystems, GmbH, Wetzlar, Germany), Retiga EXi Cooled CCD camera (QImaging, Surrey, BC, Canada), and ImagePro Plus software (Media Cybernetics, Rockville, MD, USA). Each experiment was repeated at least twice.
Luciferase reporter assay
HEK293 cells (1 × 105) stably containing empty vector, WC, or WC mutants were transfected with the pGL3b-8xGTIIC firefly luciferase reporter plasmid (containing 8 TEAD1-4 binding sites; GTIIC also known as MCAT and Hippo response element)46 and the renilla luciferase reporter plasmid (pRL-TK Renilla, (Promega, Madison, WI, USA) in a well from a 6-well plate. Fortyeight hours after transfection, the cells were collected and lysed, and extracts were assayed in technical triplicate for firefly and renilla luciferase activity using the Dual Luciferase Reporter Assay (Promega) and photometer reader (Wallac VICTOR 1420 Multilabel Counter:Perkin Elmer, Waltham, MA, USA). Each experiment was repeated at least twice.
Statistics
For soft agar colony formation assays, statistical significance was evaluated with an unpaired two-tailed t-test. For proliferation assays, statistical significance was evaluated using fold increase in proliferation at the terminal time point with an unpaired two-tailed t test (95% confidence intervals, P < 0.05). The mean was used to represent the average of technical replicates. Error bars were used to define one s.d.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U54CA168512 to BR. This work was also supported by grants from the Liddy Shriver Sarcoma Initiative, the Center for Research and Analysis of Vascular Tumors (CRAVAT), and the Friends of EHE research to BR.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, et al. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- 3.Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendlick MR, Nelson M, Pickering D, Johansson SL, Seemayer TA, Neff JR, et al. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am J Surg Pathol. 2001;25:684–687. doi: 10.1097/00000478-200105000-00019. [DOI] [PubMed] [Google Scholar]
- 5.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 6.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremblay AM, Missiaglia E, Galli GG, Hettmer S, Urcia R, Carrara M, et al. The Hippo transducer YAP1transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell. 2014;26:273–287. doi: 10.1016/j.ccr.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 8.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 11.Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 12.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Huang J, Dong J, Pan D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 14.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–1128. [PubMed] [Google Scholar]
- 18.Henrich KO, Bauer T, Schulte J, Ehemann V, Deubzer H, Gogolin S, et al. CAMTA1, a 1p36 tumor suppressor candidate, inhibits growth and activates differentiation programs in neuroblastoma cells. Cancer Res. 2011;71:3142–3151. doi: 10.1158/0008-5472.CAN-10-3014. [DOI] [PubMed] [Google Scholar]
- 19.Henrich KO, Schwab M, Westermann F. 1p36 tumor suppression--a matter of dosage? Cancer Res. 2012;72:6079–6088. doi: 10.1158/0008-5472.CAN-12-2230. [DOI] [PubMed] [Google Scholar]
- 20.Schraivogel D, Weinmann L, Beier D, Tabatabai G, Eichner A, Zhu JY, et al. CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 2011;30:4309–4322. doi: 10.1038/emboj.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey N, Ranjan A, Pant P, Tripathi RK, Ateek F, Pandey HP, et al. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics. 2013;14:216. doi: 10.1186/1471-2164-14-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huentelman MJ, Papassotiropoulos A, Craig DW, Hoerndli FJ, Pearson JV, Huynh KD, et al. Calmodulin-binding transcription activator 1 (CAMTA1) alleles predispose human episodic memory performance. Hum Mol Genet. 2007;16:1469–1477. doi: 10.1093/hmg/ddm097. [DOI] [PubMed] [Google Scholar]
- 23.Miller LA, Gunstad J, Spitznagel MB, McCaffery J, McGeary J, Poppas A, et al. CAMTA1 T polymorphism is associated with neuropsychological test performance in older adults with cardiovascular disease. Psychogeriatrics. 2011;11:135–140. doi: 10.1111/j.1479-8301.2011.00357.x. [DOI] [PubMed] [Google Scholar]
- 24.Thevenon J, Lopez E, Keren B, Heron D, Mignot C, Altuzarra C, et al. Intragenic CAMTA1 rearrangements cause non-progressive congenital ataxia with or without intellectual disability. J Med Genet. 2012;49:400–408. doi: 10.1136/jmedgenet-2012-100856. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Lv X, Liu C, Zha Z, Zhang H, Jiang Y, et al. The N-terminal phosphodegron targets TAZ/WWTR1 protein for SCFbeta-TrCP-dependent degradation in response to phosphatidylinositol 3-kinase inhibition. J Biol Chem. 2012;287:26245–26253. doi: 10.1074/jbc.M112.382036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkler A, Ashery-Padan R, Fromm H. CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 2007;581:3893–3898. doi: 10.1016/j.febslet.2007.07.051. [DOI] [PubMed] [Google Scholar]
- 33.Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent and regulate YAP2 nuclear localization and signalling. Biochem J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 36.Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 37.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang DA, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 45.Ma S, Rubin BP. Apoptosis-associated tyrosine kinase 1 inhibits growth and migration and promotes apoptosis in melanoma. Lab Invest. 2014;94:430–438. doi: 10.1038/labinvest.2014.13. [DOI] [PubMed] [Google Scholar]
- 46.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.