Abstract
Polycystic ovary syndrome (PCOS) is a common endocrinopathy characterized by hyperandrogenism and metabolic disorders. The excess androgens may be of both ovarian and adrenal origin. PCOS has a strong genetic component, and genome-wide association studies have identified several candidate genes, notably DENND1A, which encodes connecdenn 1, involved in trafficking of endosomes. DENND1A encodes two principal variants, V1 (1009 amino acids) and V2 (559 amino acids). The androgen-producing ovarian theca cells of PCOS women over-express V2. Knockdown of V2 in these cells reduces androgen production, and overexpression of V2 in normal theca cells confers upon them a PCOS phenotype of increased androgen synthesis. We report that human adrenal NCI-H295A cells express V1 and V2 mRNA and that the V2 isoform is produced by exonization of sequences in intron 20, which generates a unique exon 20A, encoding the C-terminus of V2. As in human theca cells from normal women, forced expression of V2 in NCI-H295A cells resulted in increased abundance of CYP17A1 and CYP11A1 mRNAs. We also found genetic variation in the intronic region 330 bp upstream from exon 20A, which could have the potential to drive the selective expression of V2. There was no clear association with these variants with PCOS when we analyzed genomc DNA from normal women and women with PCOS. Using minigene expression vectors in NCI-H295A cells, this variable region did not consistently favor splicing of the V2 transcript. These findings suggest increased V2 expression in PCOS theca cells is not the result of genomic sequence variation in intron 20.
Keywords: adrenal, alternate splicing, androgens, NCI-H295A cells, ovary, RNA splicing
1. Introduction
The polycystic ovary syndrome (PCOS) affects about 6% of women of reproductive age across multiple ethnic groups, and is by far the most common disorder involving overproduction of sex steroids (1–3). PCOS is associated with hyperandrogenemia, hyperandrogenism, anovulation, infertility, and multiple small subcortical ovarian follicular “cysts” (2,3). PCOS women have a higher prevalence of impaired glucose tolerance, type 2 diabetes and metabolic syndrome, irrespective of their body mass index (2,4). Many definitions and clinical criteria have been proposed for PCOS, but hyperandrogenism and/or hyperandrogenemia are the key feature of all definitions (5–7). Androgen synthesis in normal and PCOS tissues is initiated by the conversion of cholesterol to pregnenolone by mitochondrial P450scc (CYP11A1) (8), and thence to the C19 androgen precursor dehydroepiandrosterone (DHEA) by P450c17 (CYP17A1), the microsomal cytochrome P450 enzyme that catalyzes both 17α-hydroxylase and 17,20 lyase activity (9). Despite its name, both the adrenal and ovary independently contribute to the hyperandrogenism of PCOS. Ovarian hyperandrogenemia persists when the adrenals of PCOS women are suppressed (10,11), and PCOS theca cells in vitro secrete more androgen than theca cells from regularly ovulating women (12–16). Three lines of evidence show that PCOS women also overproduce C19 (19-carbon) androgen precursors in the adrenal (17). First, dehydroepiandrosterone sulfate (DHEAS), an almost uniquely adrenal steroid (9), is elevated in at least 25% of PCOS patients (18) and remains high when ovarian C19 steroids are suppressed by agonists of gonadotropin releasing hormone (GnRH) (19); second, siblings of many PCOS women have elevated circulating DHEAS (20,21); third, PCOS is frequently preceded by premature, exaggerated adrenarche, in which the pre-adolescent adrenal overproduces C19 steroids (22–27).
PCOS is a heterogeneous disorder with strong evidence for a genetic component, either autosomal dominant or oligogenic/polygenic (20,28). Incomplete penetrance, epigenetic modification, and environmental contributions have hindered attempts to clarify the underlying mode of inheritance. Numerous candidate gene association studies have been conducted, but few have yielded statistically significant associations that have been replicated. Genome wide association studies (GWAS) with large populations of Han Chinese individuals identified eleven PCOS candidate loci; these loci (and their encoded products) are: DENND1A (a GTPase), INSR (insulin receptor), YAP1 (yes-associated protein 1), C9orf3 (chromosome 9 open reading frame 3), RAB5B (a RAS oncogene), HMGA2 (High Mobility Group AT-Hook 2), TOX3 (TOX high mobility group box family member 3), SUMO1P1/ZNF217 (SUMO1 pseudogene 1/zinc finger protein 217), THADA (thyroid adenoma associated), FSHR (follicle stimulating hormone receptor) and LHCGR (luteinizing hormone/choriogonadotropin receptor) (29,30). Three studies in European populations confirmed the association of DENND1A with PCOS (31–33), strongly supporting DENND1A as a PCOS candidate gene.
The 18 human genes in the DENND family encode proteins that function as Rab-specific GTPases involved in membrane trafficking. The DENND1A gene consists of 22 exons and encodes the protein DENND1A (connecdenn 1), which has a clathrin-binding domain thought to facilitate endocytosis (34). DENND1A yields two principal transcripts: variant 1 (V1), encodes a 1009 AA protein with C-terminal proline-rich domain; variant 2 (V2), encodes a 559 AA protein that contains the three DENN domains and the clathrin-binding domain, but lacks the proline-rich domain, and includes a C-terminal 33 AA sequence that differs from V1. Whole exome sequencing did not identify DENND1A coding or splicing variants in PCOS (35), but the single nucleotide polymorphisms (SNPs) in DENND1A identified by GWAS are located in introns, and lack apparent functions. Recent work has directly implicated V2 in the pathophysiology of PCOS: V2 mRNA was abundant in urinary exosomes of PCOS women but not normally-cycling women; V2 mRNA and protein were over-expressed in PCOS theca cells compared to normal theca cells; over-expression of V2 in normal theca cells increased the abundance of CYP17A1 mRNA, augmented DHEA production and increased the activity of transfected CYP11A1 and CYP17A1 promoter-reporter constructs; and siRNA knockdown of V2 in PCOS theca cells reversed the PCOS phenotype (36). Thus, we have sought to determine the mechanisms leading to production of DENND1A.V2.
2. Materials and methods
2.1 Normal and PCOS theca cells
Human theca interna tissue was obtained from follicles of women undergoing hysterectomy. All subjects gave informed consent to use tissue for research under a protocol approved by the Institutional Review Board of The Pennsylvania State University College of Medicine. All surgeries were performed during the luteal phase of the menstrual cycle. Indications for surgery were dysfunctional uterine bleeding, endometrial cancer, and/or pelvic pain. PCOS was diagnosed according to the NIH consensus guidelines (37), which include hyperandrogenemia, oligoovulation, polycystic ovaries, and the exclusion of 21-hydroxylase deficiency, Cushing syndrome, and hyperprolactinemia; insulin resistance was not considered among the diagnostic criteria.
Theca cells from follicles of normal cycling women and from PCOS follicles were isolated and grown as described in detail previously (38,39). Cells were isolated from size-matched follicles obtained from age-matched subjects. The characteristics of the normal and PCOS theca cells have been reported, including their steroidogenic phenotypes. These phenotypes result from the inherent properties of the cells, rather than the phase of the cycle at the time the cells were isolated (13,14,16). All PCOS theca cell preparations came from ovaries of women with fewer than six menses per year and elevated serum levels of total testosterone or bioavailable testosterone. All PCOS ovaries contained multiple subcortical follicles < 10 mm in diameter. Normal theca cell preparations came from ovaries of fertile women with normal menstrual histories, menstrual cycles of 21–35 days, and no clinical signs of hyperandrogenism. No subjects were receiving hormonal medications at the time of surgery. All theca cells were used at their 4th-passage (31–38 population doublings).
2.2 Genomic DNA sequencing
Genomic DNA isolated from leukocytes and from normal and PCOS theca cells (36) was prepared using the ‘Wizard’ genomic DNA purification kit (Promega), and initial direct sequencing was initiated with primers S1280 or AS1670 (Table 1). The region encompassing exons 20 to 20A was amplified with Platinum Taq DNA Polymerase (Life Technologies) using primers S1626 and AS3776 and the resulting 2150 bp fragment was cloned into pSC-A-amp/kan vector. Between 7 and 9 individual clones were selected from each sample and sequenced with primers S1280 or AS1670. In the analysis of intron 20 sequence data, visual inspection of the original trace files showed errors in calling where double peaks were found. In addition to occasional double peaks, some traces showed peaks that were out of phase, particularly after the dinucleotide repeat region. To improve quality and detect out of phase reads, the original files were reprocessed using TraceTuner with the option to allow heterozygote calls.
Table 1.
Sequences of primers used for sequencing and for amplification of genomic fragments and splicing products from minigene constructs
| Type of assay | Sequences (5′ to 3′) |
|---|---|
| PCR | |
| S1626 | AACCGAGCTTTCCAGGAGCACCC |
| AS3776 | CCAAACTGGCTCCCATGCTCCCA |
| S260 | ACGAGAGGCTGGACCTCTTCATC |
| AS715 | TATATAAGCTTCCCGGGTTGCACTAGAAATACC |
| S18977 | CCACCACCACCTGAGCACCTGAG |
| AS19828 | CAAAGGTCCCTGGCTGCTCCCTC |
| S23 | AGAGAACCCACTGCTTACTGGCT |
| AS124 | AGCGTCGACGAATTCCAGCACA |
| S74 | AGCTCGGATCCACTAGTAACGGC |
| AS813 | GGGCAAACAACAGATGGCTGGC |
| AS514 | CCCACTGGTTTATTTCAACCCCA |
| AS371 | GACACCGGCTCTTTGGCACACAC |
| –1.8dir | GAGGCCACCTGAGCAGAC |
| –0.8dir | GGTAGCTGTGATTATTGCTTCC |
| –0.6rev | TCCAACATCGAAGCCTCCTG |
| Int1rev1 | CCCACAGTTGAAGGACCATT |
| Minigene linker | |
| S-Eco-Nhe-Pci | AATTCGTCGACGCTAGCA |
| AS-Eco-Nhe-Pci | CATGTGCTAGCGTCGACG |
| Sequencing | |
| S1280 | CAGAAAATGGGAAAGAAGGTAAGG |
| AS1670 | ATAAGGGGGCAATCATAATCCATC |
| Mutagenesis | |
| S-ex21-Gmut | GCCTGCTCTGTCCCCGACAGGCCGCAGCCGT |
| AS-ex21-Cmut | ACGGCTGCGGCCTGTCGGGGACAGAGCAGGC |
Restriction sites are underlined, and mutations are in boldface. S, sense; AS, antisense.
2.3 Minigene construction
Three PCR products were generated: 1) a 2150 bp fragment amplified with primers S1626 and AS3776, which were also used to generate clones for sequencing; 2) a 456 bp fragment that spans exon 20A amplified with primers S260 and AS715; 3) a 852 bp fragment spanning exon 21 amplified with primers S18977 and AS19828. Primer sequences are given in Table 1. The 2150 bp fragment from each sequence variant was digested with PciI and BsrFI to produce a 1731 bp fragment; the 456 bp fragment was digested with BsrFI and HindIII to produce a 366 bp fragment; and the 852 bp fragment was digested with HindIII and PstI to yield a 674 bp fragment. The 2771 bp minigene construct was assembled by ligation of the 1731 bp, 366 bp and 674 bp fragments to a double-stranded linker containing EcoRI-NheI-PciI sites (Table 1) and an EcoRI-PstI digested pSC-A-amp/kan vector (Stratagene). The 1731 bp minigene insert was excised with EcoRI and XbaI, gel-purified, subcloned into EcoRI-XbaI sites in pcDNA3.1 vector and sequenced to ensure all construction junctions were correct. Mutation of the G-to-C change adjacent to exon 21 were introduced by site-directed mutagenesis using the oligonucleotides listed in Table 1, and the resulting changes were confirmed by sequencing.
2.4 Splicing analysis
DENND1A minigene constructs in pcDNA3.1 vector were individually transfected into steroidogenic human adrenal NCI-H295A cells (40) using Effectene (Qiagen). Total RNA was prepared using Trizol (Life Technologies/Thermo Fisher), digested with RNase-free DNase I (Promega), and first stand cDNA was synthesized using oligo dT primers and Superscript III reverse transcriptase. Splicing products containing exons 20 and 21 were detected using primers S74 and AS813 (Table 1); and splicing products containing exons 20 and 20A were detected using primers S74 and AS514, and primers S74 and AS371. Transfection efficiency was normalized using vector-specific primers S23 and AS124 amplifying 102 bp of vector sequences downstream from the transcriptional start site and upstream from exon 20 in the minigene constructs (Table 1). All splicing products were verified by DNA sequencing.
2.5 RNA isolation and quantitative RT-PCR
Total RNA was extracted from NCI-H295A cells using Trizol, followed by reverse transcription using Superscript II reverse transcriptase (Invitrogen). The resulting cDNAs were quantitated using the MyiQ Single Color Real-Time PCR Detection System (BioRad). Reactions were performed in 10 μL reaction buffer containing AccuPower 2X Greenstar qPCR Master Mix (Bioneer, Alameda, CA), 20 ng cDNA and 10 pmol of each specific oligonucleotide primer for each gene (Table 2).
Table 2.
Sequences of primers used for RT-PCR
| Name | Sequence | Size of product |
|---|---|---|
| CYP11A1 For | 5′- TCC AGA AGT ATG GCC CGATT -3′ | 75 bp |
| CYP11A1 Rev | 5′- CAT CTT CAG GGT CGATGA CAT AAA -3′ | |
| CYP17A1 For | 5′- TCT CTG GGC GGC CTC AA -3′ | 63 bp |
| CYP17A1 Rev | 5′- AGG CGATAC CCT TAC GGTTGT -3′ | |
| DENND1A V1 For | 5′- GGA GCA GCC AGG GAC CTT TGA CTA | 302 bp |
| DENND1A V1 Rev | 5′- CTT GGG GCC GGG GGATGGTGA TG | |
| DENND1A V2 For | 5′- AAA GAG CAA CAT CGC AGT GGA AGG | 176 bp |
| DENND1A V2 Rev | 5′- GGA AAA GCA ACG GCGTAA GTC AAT | |
| GAPDH For | 5′- CGG GGCTCT CCA GAA CAT CAT CC -3′ | 199 bp |
| GAPDH Rev | 5′- CGA CGC CTG CTT CAC CAC CTT CTT -3′ | |
| ACTIN For | 5′- AACTCCATCATGAAGTGTGACG -3′ | 234 bp |
| ACTIN Rev | 5′- GATCCACATCTGCTGGAAGG -3′ |
For RT-PCR and RNase protection assays, total RNA was isolated using Trizol (Invitrogen) or mirVana miRNA Isolation Kit (Ambion) according to manufacturers’ protocols. Using Superscript III reverse transcriptase (Invitrogen), reactions were carried out with 1μg total RNA in a final volume of 10 μL. PCR of the resulting cDNAs (0.3 μL/reaction in a final volume of 10 μL) was done with Platinum Taq DNA Polymerase (Life Technologies) using the primers listed in Table 3, and products were separated by agarose gel electrophoresis.
Table 3.
Sequences of primers used for RT-PCR of alternatively spliced products
| Name | Sequences (5′ to 3′) | Panel |
|---|---|---|
| Figures 3 and 5 | ||
| S234 | ATGGGCTCCAGGATCAAGCAGAA | B, C, E |
| AS2095 | CTCTCACTCCCGCCCAGATCCAG | B, F |
| ASex20A3’end | CCATGCTCCCACTGGTTTATT | C, D |
| Sex17 | ATCCGGCCATGAAGACTGT | D |
| AS3int20A | ATGTGCAATTGGAAGTGAGAGT | E |
| Sex20A | GCAACACCAGCTACACTCCA | F |
| S648 | CCCATCCCTGACCCAGGAGTGTC | G, H |
| Asint19B | CCAGTCCATCGCCTACATTT | G |
| ASex19-A1 | AATTTTTGAAAATTTGCCTGTCC | H |
| Sex13A | CAGCCGAAGTGTCCACTTATC | I |
| Asex21 | CTCTGGACTCTCTGCCTCGT | I |
| Sint17 | GTTTTCGACTTGATGATCCC | J |
| AS1932 | GGAAAAGCAACGGCGTAAGTCAAT | J |
| Figure 5 | ||
| S1757 | AAAGAGCAACATCGCAGTGGAAGG | A, B |
| AS2179 | ACATGGCATTTGGAAGGGGCGGA | A, B |
S, sense; AS, antisense.
2.6 Overexpression of DENND1A.V2 in NCI-H295A cells
The DENND1A.V2 adenovirus vector hDENND1A.V2-pADenoG (built by Applied Biological Materials, Vancouver, BC) has been described (36). Adenovirus particles made from hDENND1A.V2-pADenoG and from the empty pADenoG-null control were used to infect NCI-H295A cells cultured in RPMI-1640 medium supplemented with 2% FCS, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml selenium, and 50 μg/ml gentamycin. For mRNA measurement, we used 12 well plates with 600,000 cells per well; for western blots, we used 10 cm dishes with 2–3 x 106 cells. Each infection experiment was performed using 3 PFU/cell. Total cellular RNA was used for SYBR green-based quantitative RT-PCR of mRNAs for P450scc and P450c17, as described above using 40 ng cDNA per reaction. The data for the relative expression of each gene vs. the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) internal control were determined using the 2−ΔΔCT method.
2.7 Immunoblotting
NCI-H295A cells were lysed in RIPA buffer containing a proteinase inhibitor cocktail (Roche Diagnostics). Lysates were cleared by centrifugation (10,000 x g for 20 min) and Bradford protein was determined Aliquots (30 μg) of protein were separated by 8% SDS-PAGE, electrotransferred to polyvinylidene difluoride membranes (Hybond-P, Amersham-Pharmacia Biotech, Piscataway, NJ), and incubated with rabbit antibodies directed against a 75 amino acid peptide sequence of DENND1A encoded by DENND1A exons 17–19 (Sigma catalog # HPA020481); antiserum to β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a loading control. Proteins were detected using horseradish peroxidase-conjugated secondary antibodies (Thermo Scientific).
2.8 RNase protection assays
RNase protection assays were carried out essentially as described (41) except the total volume of hybridization was reduced to 5 μL, the combined RNase A/RNaseT1 digestion was carried out in 50 μL and chloroform-phenol extraction was replaced by phenol extraction. Antisense RNAs labeled in vitro with 32P were produced using T3 and T7 RNA polymerases (Fermentas, Thermo-Fisher, Boston MA). For this purpose RT-PCR products, generated with primer pairs Sex17 and ASex20A3’end and with S1757 and AS2179 (Table 1) and treated with Klenow polymerase (New England Biolabs) were cloned into the EcoRV site of pBS KS. Each run-off transcription reaction contained 20μCi 32P-UTP (3000 Ci/mmol, PerkinElmer), 10 units RNA polymerase (Fermentas) and 200 ng plasmid in a final volume of 5 μL. To ensure efficient synthesis of full-length RNAs, non-radioactive UTP was added to a final concentration of 0.2 mM. RNA-RNA hybridization was done in 5 μL and dual digestion with RNase A (Sigma) and RNase T1 (Fermentas) was done in 55 μL. Products were separated by polyacrylamide gel electrophoresis and analyzed by phosphoimager (Molecular Dynamics).
2.9 Statistics
The significance of the association of intron sequence variation with PCOS was assessed using the Pearson’s chi square test. For quantitation of RNA by RT-PCR, the expression levels of P450c17 and P450scc were normalized to the level of GAPDH, as determined from the average threshold cycle (Ct) of triplicate samples. Statistical analyses were performed using two-tailed unpaired t tests; significance was accepted for tests where P < 0.05 and data are represented as mean ± SD (n=3).
3. Results
3.1 DENND1A.V2 arises by exonization of sequences in intron 20
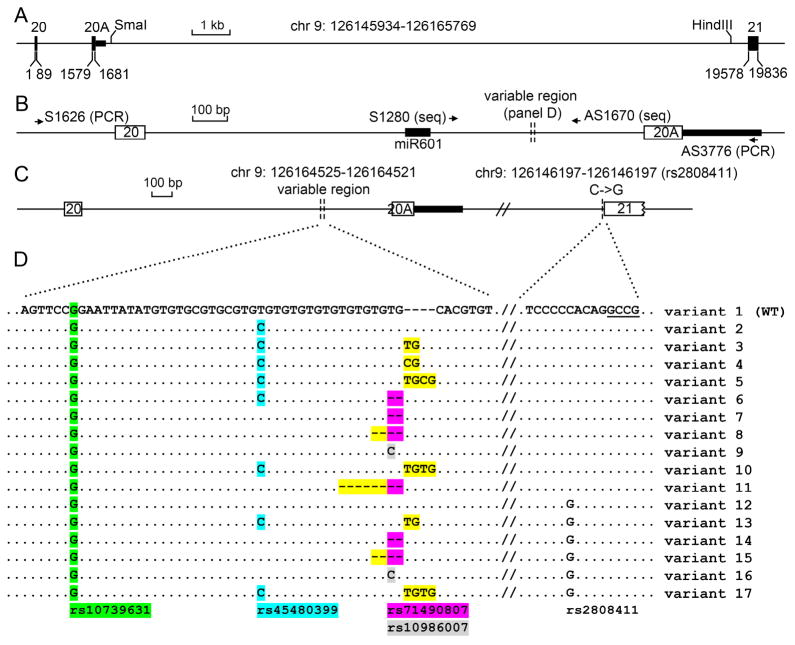
The NCBI database lists multiple DENND1A transcripts; V1 and V2 are the major ones, and are predicted to encode proteins of 1009 AA and 559 AA, respectively (34,36). Examining the protein-coding potential of the V1 and V2 mRNAs showed that they are identical through the end of exon 20 and then diverge. V1 continues with canonical exons 21 and 22 to yield a 1009 AA open reading frame, while V2 lacks the sequences encoded by exons 21 and 22 and has a different coding sequence that is responsable for a 559 AA open reading frame with a unique C-terminal 33 AA. Using this sequence in silico to probe the DENND1A gene, we found that the 33 AA C-terminus of V2 was generated by exonization of sequences located ~1.5 kb downstream from canonical exon 20 and 19.5 kb upstream from canonical exon 21 (Fig 1A). Because this is a cryptic exon, we named this exon 20A.
Fig 1. Diagram of the 3′ end of the DENND1A gene and splicing variants identified.
A. Scale diagram of ~20 kb spanning DENND1A exons 20 and 21; bases are numbered starting with the first base of exon 20 (chr 9:126145934) designated as base 1; exons are shown as boxes. The intronic regions between exons 20 and 20A and between exons 20A and 21 are 1490bp and 17897 bp, respectively. B. Magnified diagram of the region between exons 20 and 20A; note the difference in the scale bars in panels A and B. The hypervariable region between the vertical dotted lines was identified in PCR products generated with primers S1626 and AS3776; this hypervariable region is shown in detail in panel D. DNA sequencing (seq) of PCR products and cloned DNA was done using primers S1280 and AS1670. The 3′ end of miR601 (shown as a heavy black line) is 282 bp upstream of the hypervariable region. The heavy black line following exon 20A represents the DNA encoding the 3′ untranslated region of DENND1A V2 mRNA.
C. Locations of detected sequence variants. The hypervariable region was detected 330 bp upstream from exon 20A at chr 9: 126164525-126164521; and rs2808411 is adjacent to exon 21 at chr 9: 126146197-126146197.
D. Sequence variants in the hypervariable region. Known SNPs in this region are highlighted: green, rs10739631; cyan, rs45480399; purple, rs71490807; grey, rs10986007. rs2808411 (C->G) occurs 5 bp upstream of exon 21 (underlined). Newly identified variants at chr 9: 126164525-126164521 are highlighted in yellow.
3.2 Intron 20 sequence variation
To determine if there is genetic variation in intron 20 that might give rise to alternative RNA splicing that might determine whether the DENND1A gene expresses mRNA encoding DENND1A V1 vs. V2, we amplified genomic DNA from human leukocytes collected from 20 normal women and 19 women with PCOS (Table 4). Using primers S1626 (23-mer, 239 nt upstream from exon 20) and AS3776 (23-mer; 231 nt downstream from exon 20A), we amplified a 2.15 kb region, then used primers S1280 and AS1670 (24-mers 251 nt upstream and 140 nt downstream from a putative variable region of intron 20) (Fig. 1B) to sequence the PCR products. Sequencing identified a variable region 330 bp upstream from exon 20A; this region is complex with an extended dinucleotide repeat. Direct sequencing of this region in some genomic DNA samples yielded garbled sequence data, suggesting the presence of multiple sequence variants. Therefore, we cloned the 2150 bp product and then sequenced multiple clones; this yielded different sequence variants at chr 9: 126164525-126164521 (Fig. 1). Among the 10 variants (variants 2–11) identified in genomic DNA from normal and PCOS women, none appeared to be overrepresented in PCOS women. Variants 2,4,9 and 11 were found in a limited number of samples from theca cells, and variants 2 and 4 were only found in samples from PCOS patients. We detected known SNPs including rs10739631, rs45480399, rs71490807, and rs10986007 (Fig 1B). We also used primers S18977 and AS19828 to amplify a 852 bp region spanning the 3′ end of intron 20A and exon 21 (Fig 1A and Table 1); this detected SNP rs2808411 (C->G change, 5 nt upstream from exon 21) in variants 12–17. These seven variants are identical to variants 1, 3, 7, 8, 9 and 10 in the hypervariable region, but they differ by the rs2808411 SNP adjacent to exon 21. There were six samples (C31, C35, P08, P21, P26 and P29) in which we detected more than two haplotypes due to variation in the dinucleotide repeat region (Table 4); we could not determine if this represents true somatic variation or DNA sequencing artifacts.
Table 4.
Sequence variation in intron 20 (see Results, section 3.2).
| Sample | Phenotype | Variant | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 11 | ||
| C11 | Normal | 2 | 3 | ||||||||
| C12 | Normal | 1 | 4 | ||||||||
| C13 | Normal | 2 | 2 | ||||||||
| C14 | Normal | 2 | 3 | ||||||||
| C15 | Normal | 4 | |||||||||
| C16 | Normal | 4 | |||||||||
| C17 | Normal | 4 | |||||||||
| C18 | Normal | 5 | |||||||||
| C19 | Normal | 2 | 3 | ||||||||
| C20 | Normal | 4 | 1 | ||||||||
| C31 | Normal | 2 | 2 | 1 | |||||||
| C32 | Normal | 3 | 2 | ||||||||
| C33 | Normal | 1 | 4 | ||||||||
| C34 | Normal | 1 | 2 | ||||||||
| C35 | Normal | 4 | 1 | 1 | |||||||
| C36 | Normal | 1 | 4 | ||||||||
| C39 | Normal | 5 | |||||||||
| C40 | Normal | 1 | 3 | ||||||||
| C41 | Normal | 5 | |||||||||
| C42 | Normal | 4 | 1 | ||||||||
| P01 | PCOS | 2 | 2 | ||||||||
| P02 | PCOS | 2 | 3 | ||||||||
| P03 | PCOS | 5 | |||||||||
| P05 | PCOS | 1 | 3 | ||||||||
| P06 | PCOS | 4 | 1 | ||||||||
| P07 | PCOS | 2 | 3 | ||||||||
| P08 | PCOS | 5 | 1 | 1 | |||||||
| P09 | PCOS | 2 | |||||||||
| P10 | PCOS | 2 | 1 | ||||||||
| P21 | PCOS | 1 | 2 | 1 | |||||||
| P23 | PCOS | 2 | 2 | 1 | |||||||
| P24 | PCOS | 4 | 1 | ||||||||
| P25 | PCOS | 1 | 3 | ||||||||
| P26 | PCOS | 1 | 3 | 1 | |||||||
| P27 | PCOS | 1 | 2 | 1 | |||||||
| P28 | PCOS | 1 | 3 | ||||||||
| P29 | PCOS | 1 | 1 | 3 | |||||||
| P30 | PCOS | 5 | |||||||||
| VCU014 | PCOS | 4 | 1 | ||||||||
The sequence of the 330 bp variable region of intron 20 was assessed in multiple clones of genomic DNA from theca preparations from 20 control and 19 PCOS individuals; the table shows the number of clones carrying each variant sequence, defined as in Fig. 1.
3.3 Minigene analysis of alternative splicing
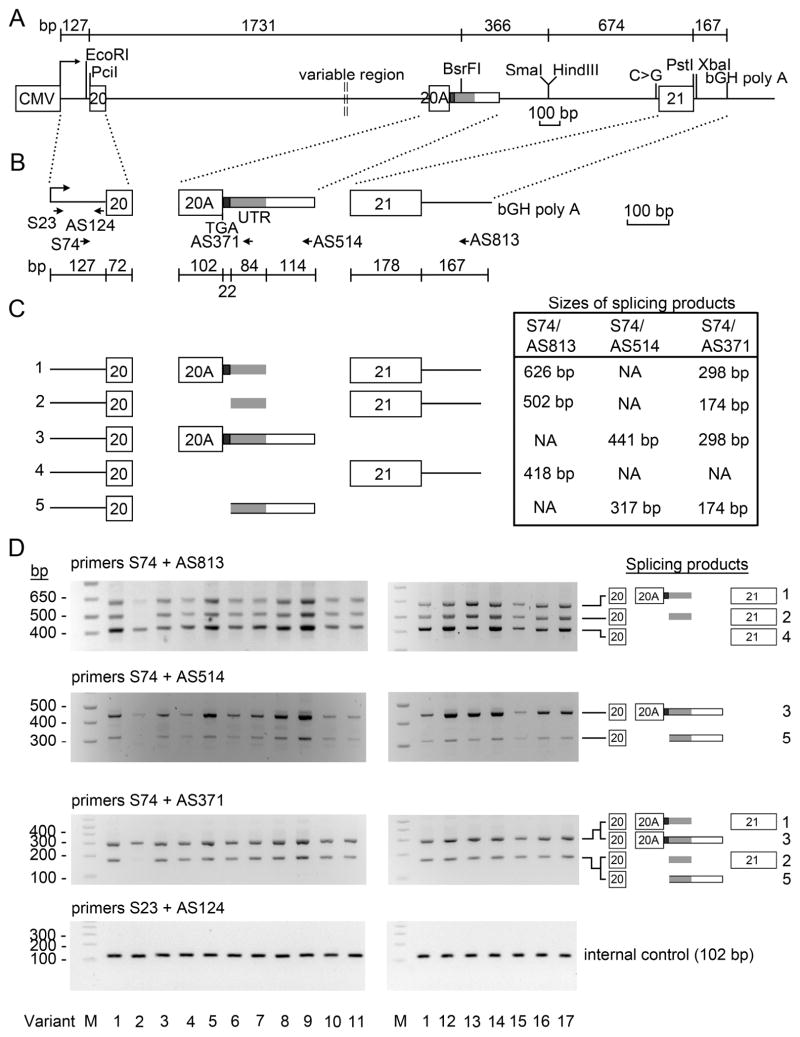
To determine the potential effects of the intron 20 sequence variants identified in Fig. 1 on splicing of DENND1A mRNA, we built a series of minigene expression constructs using the strategy shown in Fig 2A to see what forms of RNA were produced from each variant. The design retains exons 20, 20A and 21 and all of the intronic DNA between exons 20 and 20A, but excludes the 16978 bp between the SmaI and HindIII sites in the intronic DNA between exons 20A and 21. Each of the intron 20 sequence variants identified in Fig. 1D were created by ligation of a 1731 bp PciI-BsrFI fragment, 366 bp BsrFI-HindIII fragment, and 674 bp HindIII-PstI fragment (Fig 2A), and verified by sequencing. Because there are no immortalized differentiated lines of human theca cells, the constructs were transfected into human adrenal NCI-H295A cells, as this is the only human androgen-producing cell line presently available. After transfection, RNA splicing was assessed by RT-PCR followed by electrophoresis in 2% agarose (Fig. 2B–2D). To distinguish the splicing products arising from the minigene constructs vs. those endogenously expressed in NCI-H295A cells, we used the vector-specific 5′ primer S74 for RT-PCR (Fig. 2B–2D).
Fig 2. Design and expression of the minigenes.
A. Design of minigenes: upper line, scale diagram showing sizes of component fragments; lower line, diagram of construction strategy, placing DENND1A sequences under the control of the promoter from cytomegalovirus (CMV). The construct contains 72 bp of exon 20, 1490 bp of intron 20, 322 bp of exon 20A, 201 bp of the 5′-most segment of intron 20A, 498 bp of 3′-most segment of intron 20A, and 178 bp of exon 21; the construct was created by ligation of a 1731 bp PciI-BsrFI fragment, 366 bp BsrFI-HindIII fragment, and 674 bp HindIII-PstI fragment. The construct lacks 16978 bp of intronic sequences from the SmaI to HindIII sites between exon 20A and exon 21. The minigene vector contains a cytomegalovirus promoter (CMV) and bovine growth hormone polyadenylyation signal (bGH poly A). SNP rs2808411 is shown as C>G upstream of exon 21.
B. Strategy for identifying splicing products. PCR was performed using vector-specific primers S74 and AS813, and exon 20A-specific primers AS371 and AS514. Transfection efficiency was normalized using vector-specific primers S23 and AS124 in the 5′ end of the minigene transcripts. Alternative splicing of exon 20A can terminate transcription; when this occurs, its 3′UTR consists of 22 bp that contains a cryptic splice acceptor (black bar), 84 bp that contains a 5′ splice donor (grey bar) and 114 bp of downstream 3′ UTR DNA (open bar). The relative positions of the TGA stop codon and 3′ UTR in exon 20A are shown.
C. Transcripts arising from the minigene constructs. The minigenes produced the five splicing products diagrammed. The sizes of the PCR products are shown in the box to the right of the diagrams of the five PCR products.
D. Agarose gels of PCR products. The identified splicing products are indicated to the right of the figure, and are diagrammed and numbered as in Panel C. The 17 different minigene constructs analyzed, corresponding to the variant numbers in Fig 1D, are indicated at the bottom.
We detected five splicing products arising from the minigene constructs (Fig. 2C). Product 1 contains exon 20, exon 20A with 106 nt of its 3′ UTR, plus exon 21; this pattern of RNA splicing would yield a transcript encoding V2 but not V1 due to the TGA stop codon in exon 20A. Product 2 contains exon 20, 84 nt of 3′ UTR of exon 20A (skipping exon 20A and its 22 immediate 3′ bases) and exon 21; the 84 nt segment contains a translational stop codon, so that the resulting mRNA would encode a transcript of 551 AA containing the first 526 residues common to V1 and V2, but with 25 unique C-terminal residues; product 2 has 98.9% identity with a 551-residue hypothetical DENND1A isoform predicted in the olive baboon (NCBI database reference sequence XP_009186392.1). Product 3 contains exon 20, exon 20A and the complete 3′ UTR of exon 20A, but lacks exon 21 and would be predicted to encode DENND1A.V2. Product 4 contains exons 20 and 21 while lacking exon 20A, corresponding to the mRNA expected to encode V1. Product 5 contains exon 20 and the 3′ UTR of exon 20A but lacks exon 20A, the 22 nt immediately 3′ to exon 20A, and exon 21; this mRNA would encode the same 551 AA protein encoded by Product 2. Agarose gels showing these RT-PCR products are shown in Fig. 2D; the minigene constructs correspond to the sequence variants shown in Fig. 1B. In two independent experiments with all constructs, none of these consistently favored splicing to favor the mRNA encoding the V2 isoform.
3.4 Other forms of alternative splicing of DENND1A
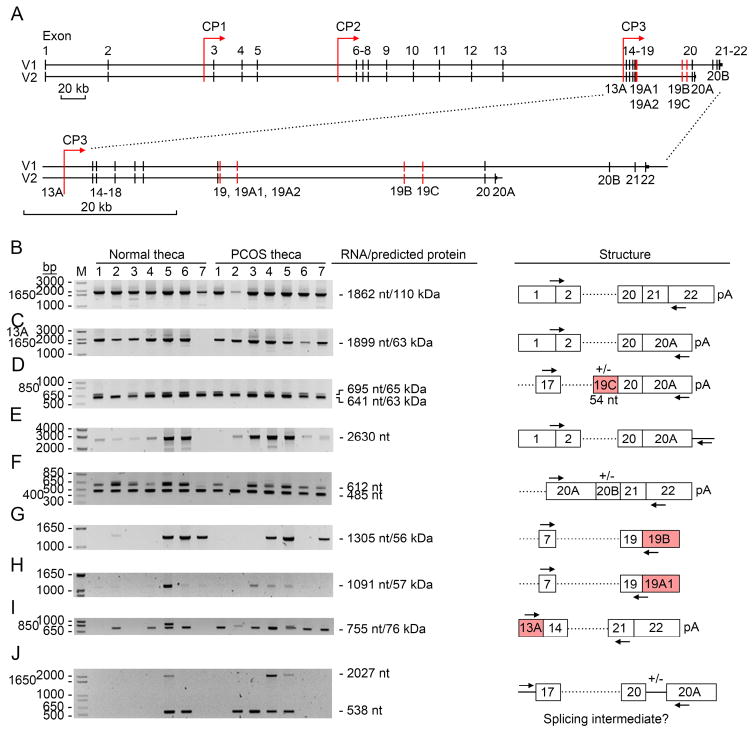
To search for other potential modes of alternatively spliced DENND1A mRNAs in human theca cells from normal and PCOS women, we performed a systematic series of RT-PCR experiments (Fig. 3); the sequences and locations of the PCR primers are given in Table 3. A series of potential alternatively spliced mRNAs was selected from the UCSC Genome Browser (GRCh37/hg19; Feb. 2009); we sought fragments (ESTs) or complete sequences derived from DENND1A.V1 and V2 mRNAs (exons 1-22 and 1-20A, respectively), exonization of introns 18, 19 and 20A, and retention of introns 17 and 20 (Fig. 3A). Total RNA was prepared from theca cells and subjected to RT-PCR (Fig.3B–3J). The sizes of the PCR products and predicted encoded protein products were calculated from the first in-frame ATG in exon 1 (Fig. 3B–D and G–H) or from the first in-frame ATG in exonized intron 13 (Fig. 3I). For V1 and V2 mRNAs there was some quantitative variability among samples but V1 was more abundant than V2 in only one of seven samples; internal exon skipping was not seen (Fig. 3B, C). Other splice variants are also seen with exonization in intron 19 (Fig. 3D, G, H); if translated, these RNAs are predicted to yield proteins of 63 and 65 kDa, or 56 and 57 kDa, respectively. A 295 bp region downstream from exon 20A was retained in properly spliced pre-V2 mRNA (Fig. 3E), suggesting inefficient transcriptional termination and/or polyadenylylation. Additional assays revealed properly processed V2 mRNA (Fig 4). When the alternate splicing mechanism choses exon 20A, the resulting mRNA may have one of two different 3′ ends (Fig. 2C, splicing forms 1 and 3). In some cases an additional segment of intron 20 (3.8 kb upstram from exon 21) may be exonized yielding ‘exon 20B’, but the stop codon in exon 20A would result in this mRNA also encoding V2 (Fig. 3F). A transcript terminating with exon 20A including or excluding intron 20 was probably a splicing intermediate of V2 mRNA (Fig. 3I). There were no consistent differences in the repertoire of splice variants from normal and PCOS theca cells and all of the RT-PCR products detected in theca cells in Figs. 3B–I were also detected in NCI-H295A cells (data not shown).
Fig 3. Alternative splicing of DENND1A in theca cells.
A. Upper paired bars: Intron/exon organization of the entire DENND1A gene (redrawn from Genome Browser). Exons are shown as vertical lines; the organization of the genetic segments encoding DENND1A V1 and V2 are shown in the two lines designated V1 and V2. Locations of cryptic promoters (CP’s) and exonizations of intronic sequences are shown. The lower paired V1 and V2 lines show an expanded view of the 3′ region of the gene encompassing exons 13A to 22 (not difference in scale bar).
B–J. Alternatively spliced DENND1A mRNAs. For each panel, autoradiograms of the RT-PCR products from theca cell RNAs from seven normal and seven PCOS subjects are shown on the left. The sizes of the PCR products and predicted encoded protein products, calculated from the first in-frame ATG in exon 1 (panels B–D and G–H) are shown to the right of each autoradiogram, and the responsible splicing structures are diagrammed on the right. The locations of the rightward and leftward primers used are indicated with arrows above and below the splicing diagrams; the primer sequences are listed in Table 3.
Fig 4. Detection of processed V2 transcripts.
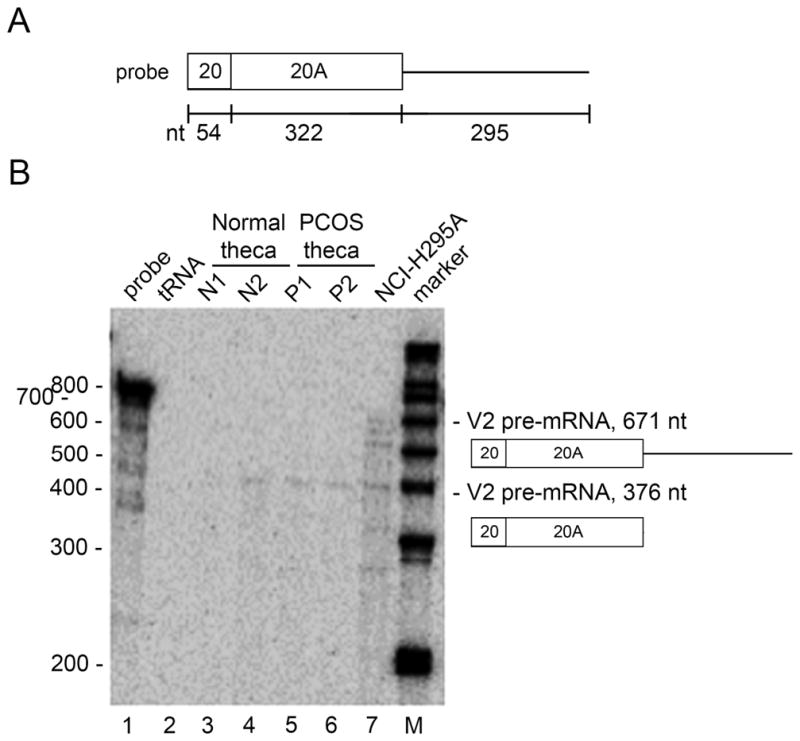
A. Diagram of 32P-labeled antisense RNA probe used for RNase protection assays, with sizes (in nt) of each exon and the intronic region downstream from exon 20A shown below.
B. RNase protection assays. Each lane contains the probe shown in Panel A plus 3 μg of each of the following RNAs: Lane 1, probe alone. Lane 2, tRNA control. Lanes 3 and 4, normal theca cells from two individuals. Lanes 5 and 6, Theca cells from two PCOS individuals. Lane 7 total RNA from NCI-H295A cells. Lane M, 32P-labeled 1 kb DNA ladder (Invitrogen). The additional bands seen with NCI-H295A RNA may represent additional alternatively spliced forms. Sequences of primers used for probe preparation are shown in Table 3.
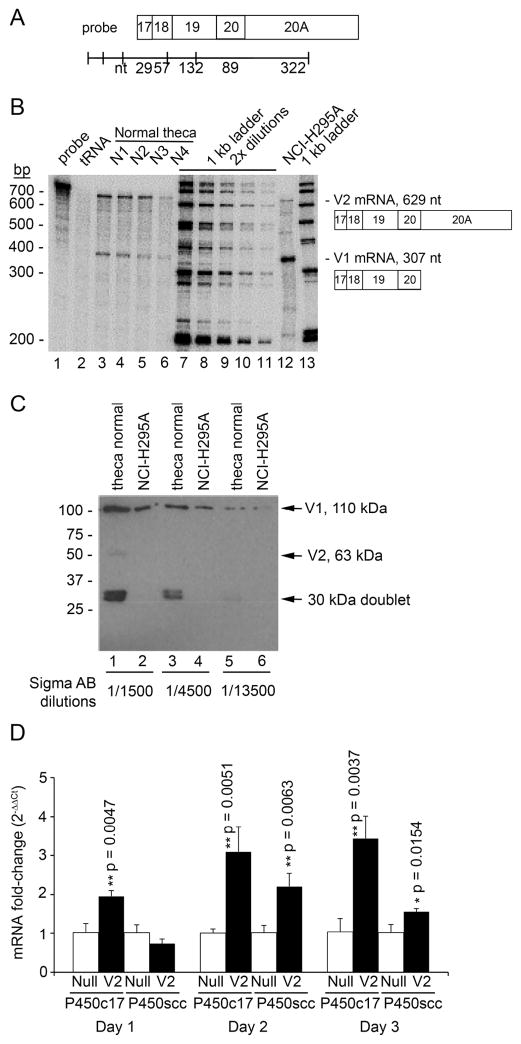
3.5 Relative expression of V1 and V2 in NCI-H295A cells
To study adrenal expression of DENND1A, we used RNase protection assays to determine the amounts of V1 and V2 mRNAs in human adrenal NCI-H295A cells. The ratio of the mRNAs for V1 and V2 in NCI-H295A cells was about 10:1, compared to a ratio of ~1:1 in normal human theca cells (when signal intensity is adjusted for the length of the protected fragment) (Fig. 5A and 5B). Western blotting showed that the V1 isoform predominates in both normal theca and NCI-H295A cells (Fig 5C), but that neither form is expressed appreciably in fetal heart, liver or kidney (not shown). A 30 kDa doublet not seen in previous work (36) was apparent in the theca cell extract, possibly reflecting translation of another splice variant or partial proteolysis of V1 or V2. When NCI-H295A cells were infected with a virus expressing human V2, expression of the mRNAs encoding CYP17A1 and CYP11A1 were significantly increased (Fig. 5D), confirming a role for V2 in the control of steroidogenesis and replicating responses previously observed in normal human theca cells (36).
Fig 5. Quantitation of V1 and V2 mRNAs by solution hybridization.
A. Diagram of 32P-labeled antisense RNA probe used for RNase protection assays, with sizes (in nt) of each exon shown below. Boxes represent DENND1A exons.
B. RNase protection assays. Each lane contains the probe shown in Panel A plus 3 μg of each of the following RNAs: Lane 1, Probe alone. Lane 2, tRNA, showing complete digestion of probe. Lanes 3–6, RNA from normal theca cells. Lanes 7–11 32P- labeled 1 kb DNA ladder (Invitrogen) with two-fold serial dilutions for semi-quantitative estimation of the amounts of V1 and V2 mRNA. Lane 12, total RNA from NCI-H295A cells. Lane 13, 32P- labeled 1 kb DNA ladder. A diagram of the protected exons and their sizes are shown on the right, distinguishing the V1 and V2 mRNAs. Sequences of primers used to prepare probe are shown in Table 3.
C. Immunoblotting of protein lysates from normal theca and NCI-H295A cells using different antibody dilutions of polyclonal antiserum (from Sigma) directed against DENND1A residues 410–484, which is found in both V1 and V2.
D. Overexpression of DENND1A.V2 increases adrenal expression of the mRNAs for P450c17 and P450scc. NCI-H295A cells were infected with adenovirus expressing DENND1A.V2 or an empty null vector for 1, 2 and 3 days. Total cDNA was prepared by reverse transcription using random hexamers, and cDNAs corresponding to P450c17 and P450scc were quantitated by qPCR, normalized using GAPDH as an internal control. Each bar represents the mean +/− SD from three experiments, each performed in duplicate (n=3); p-values comparing cells transfected with empty vector (null) vs. cells transfected with the vector expressing V2, were calculated using two-tailed unpaired t tests.
4. Discussion
We report that the V2 isoform of DENND1A is produced by exonization of sequences in intron 20, which generates a unique exon 20A, encoding the C-terminus of V2. We report that human adrenal NCI-H295A cells express V1 and V2 mRNAs, and that, similarly to prior findings in theca cells (36), forced expression of V2 in NCI-H295A cells resulted in increased abundance of the mRNAs for CYP17A1 (P450c17) and CYP11A1 (P450scc). We also found genetic variation in the intronic region 330 bp upstream from exon 20A, but found no clear association of these variants with PCOS. This variable region did not consistently favor splicing of the V2 transcript. Thus increased V2 expression in PCOS theca cells does not appear to result from genomic sequence variation in intron 20.
Multiple GWAS studies (29–33) have linked PCOS to SNPs rs10818854 (A>G at 125486599), rs2479106 (G>A at 125565033) and rs10986105 (C>A at 125589776) on chromosome 9q33.3. These three SNPs lie within the DENND1A gene: rs10818854 is in intron 5, 7.7 kb upstream from exon 6; rs2479106 is in intron 4, 5.1 kb upstream from exon 5; rs10986105 is in intron 3, 4.9 kb downstream from exon 3. Thus DENND1A has emerged as a prominent candidate gene for PCOS. The DENND1A gene is over 500 kb long, and its encoded RNA is alternatively spliced into mRNA for DENND1A.V1, DENND1A.V2 and other forms. DENND1A isoforms V1 and V2 are differentially expressed in theca cells from normal and PCOS women (36). That work found that: i) V2 is overexpressed in PCOS theca compared to normal theca; ii) V2 mRNA was found in the urinary exosomes of PCOS women in greater abundance than in normal women; iii) forced expression of V2 in normal theca cells induced a PCOS-like steroidal phenotype; and iv) knockdown of V2 in PCOS theca cells reversed the PCOS steroidal phenotype (36). Those data suggested that a key event in PCOS is the alternative splicing of DENND1A mRNA to its V2 variant rather than its V1 variant, and that V2 directly drives androgen synthesis through as-yet undetermined mechanisms.
Examining the AA sequences of V1 and V2 and comparing these to the protein-coding potential of DENND1A sequences shows that these isoforms are identical through the end of exon 20, and then diverge, with V1 continuing with AAs encoded by canonical exons 21 and 22, while V2 has a unique 33 AA carboxyl terminus. Searching downstream from exon 20 for DNA that might encode these 33 AAs identified exon 20A, encoding the complete 33 AA C-terminal segment of V2. As alternative splicing may be mediated by genetic variations in intronic sequences that direct the processing of pre-mRNAs, we sought DNA sequence variants in the intronic DNA lying between exons 20 and 20A as a potential mechanism driving the choice of splicing to the V2 vs. V1 isoform, and found a hypervariable region 330 bp upstream from exon 20A. However, when each of these sequence variants is tested in a minigene system, none consistently favored splicing to the mRNA encoding the V2 rather than V1 isoform. As there are no immortalized, differentiated lines of human theca cells, we analyzed splicing of the DENND1A minigenes in human adrenal NCI-H295A cells, as this is the only human androgen-producing cell line presently available.
In addition to the ovarian hyperandrogenism of PCOS, the adrenals also overproduce androgens in PCOS. As adrenal tissue from normal and PCOS women is not available, we initiated studies with NCI-H295 cells, which are the only available human adrenal steroidogenic cell line. These cells make C19 steroids (42) and express the genes for all the steroidogenic enzymes needed to make androgens (43). We found that these cells express abundant V1 but very little V2 mRNA. However, over-expresion of V2 in NCI-H295A cells increased the mRNAs for CYP17A1 and CYP11A1, which is consistent with a role for V2 in regulating androgen synthesis.
DENND1A isoforms V1 and V2 are not the only forms of DENND1A produced. Using RT-PCR and RNase protection assays, we detected at least six additional forms of DENND1A mRNA. Transcripts containing unprocessed 3′ sequences downstream from exon 20A were common (Fig. 3, panel E); such transcripts have also been detected in two ESTs listed in GenBank: AI671592 (pooled germ cell tumors) and CD365540 (alveolar macrophages) suggesting that this processing defect is relatively common. Exon 19C, which is in frame with the DENND1A reading frame, was seen in almost all theca cells, (Fig. 3, panel D), but no similar ESTs are listed in GenBank, suggesting that this splicing variation may be unique to theca. Exon 20B, also in frame with the DENND1A reading frame, is also frequently included (Fig. 3, panel F); similar ESTs listed in GenBank (AB046828, AK299867, AK295710, and others) are found in human brain. Exonization in intron 19 (Fig. 3, panel G) also appears to be unique to theca cells, as only one EST (BC028061, pooled from adult colon, kidney, and stomach) has been found. The extent to which of these alternatively spliced mRNAs encode variants of the DENND1A protein remains under investigation.
HIGHLIGHTS.
Polycystic ovary syndrome (PCOS) includes hyperandrogenism and metabolic dysfunctions
Genome-wide association studies have identified DENND1A as a candidate gene for PCOS
PCOS theca cells overexpress DENND1A variant 2 (V2; 559 AA), but not V1 (1009 AA)
V2 arises by exonization within intron 20 generating a unique exon 20A
Overexpression of V2 in human adrenal cells increases CYP11A1 and CYP17A1 mRNAs
Genetic variation 330 bp upstream of exon 20A is not associated with expression of V2
Acknowledgments
This work was supported by philanthropic gifts to the WLM lab; BL was supported by grant P2BSP3 148469 from the Swiss National Science Foundation and MS was supported by a grant from the Baltic-American Friendship Foundation; BM, JMM, and JFS were supported by National Institutes of Health Grants U54HD344449 (to JFS & JMM), R01HD058300 (to JMM), and R01HD33852 (to JMM) and by a cooperative agreement between Pennsylvania State University (PSU) and Virginia Commonwealth University (VCU) Clinical and Translational Science Awards, UL1T000058 (to VCU) and UL1T0000127 (to PSU).
Footnotes
Disclosure statement: The authors have nothing to disclose
Author contributions
WLM, MKT, MS and JFS designed research; MKT, MS, BL, BM and MET performed research; JMM and JFS provided research materials MKT, MS, JFS and WLM analyzed data; and MKT, MS, JFS and WLM wrote the paper.
Conflict of interest statement
JMM and JFS have patents pending related to this material. JFS is a consultant to Takeda on topics of infertility treatment.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knochenhauer ES, Key TJ, Kahsar-Miller M, Waggoner W, Boots LR, Azziz R. Prevalence of the polycystic ovary syndrome in unselected black and white women of the southeastern United States: a prospective study. J Clin Endocrinol Metab. 1998;83:3078–3082. doi: 10.1210/jc.83.9.3078. [DOI] [PubMed] [Google Scholar]
- 2.Diamanti-Kandarakis E, Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev. 2012;33:981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legro RS, Arslanian SA, Ehrmann DA, Hoeger KM, Murad MH, Pasquali R, Welt CK. Diagnosis and treatment of polycystic ovary syndrome: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2013;98:4565–4592. doi: 10.1210/jc.2013-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran LJ, Misso ML, Wild RA, Norman RJ. Impaired glucose tolerance, type 2 diabetes and metabolic syndrome in polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod Update. 2010;16:347–363. doi: 10.1093/humupd/dmq001. [DOI] [PubMed] [Google Scholar]
- 5.Fauser BCJM 29 co-authors: Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19:41–47. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 6.Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, Witchel SF. Criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an Androgen Excess Society guideline. J Clin Endocrinol Metab. 2006;91:4237–4245. doi: 10.1210/jc.2006-0178. [DOI] [PubMed] [Google Scholar]
- 7.Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7:219–231. doi: 10.1038/nrendo.2010.217. [DOI] [PubMed] [Google Scholar]
- 8.Miller WL, Bose HS. Early steps in steroidogenesis: intracellular cholesterol trafficking. J Lipid Res. 2011;52:2111–2135. doi: 10.1194/jlr.R016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lachelin GC, Judd HL, Swanson SC, Hauck ME, Parker DC, Yen SSC. Long term effects of nightly dexamethasone administration in patients with polycystic ovarian disease. J Clin Endocrinol Metab. 1982;55:768–773. doi: 10.1210/jcem-55-4-768. [DOI] [PubMed] [Google Scholar]
- 11.Rittmaster RS, Thompson DL. Effect of leuprolide and dexamethasone on hair growth and hormone levels in hirsute women: the relative importance of the ovary and the adrenal in the pathogenesis of hirsutism. J Clin Endocrinol Metab. 1990;70:1096–1102. doi: 10.1210/jcem-70-4-1096. [DOI] [PubMed] [Google Scholar]
- 12.Gilling-Smith C, Willis DS, Franks S. Oestradiol feedback stimulation of androgen biosynthesis by human theca cells. Hum Reprod. 1997;12:1621–1628. doi: 10.1093/humrep/12.8.1621. [DOI] [PubMed] [Google Scholar]
- 13.Nelson VL, Legro RS, Strauss JF, 3rd, McAllister JM. Augmented androgen production is a stable steroidogenic phenotype of propagated theca cells from polycystic ovaries. Mol Endocrinol. 1999;13:946–957. doi: 10.1210/me.13.6.946. [DOI] [PubMed] [Google Scholar]
- 14.Wickenheisser JK, Quinn PG, Nelson VL, Legro RS, Strauss JF, 3rd, McAllister JM. Differential activity of the cytochrome P450 17α-hydroxylase and steroidogenic acute regulatory protein gene promoters in normal and polycystic ovary syndrome theca cells. J Clin Endocrinol Metab. 2000;85:2304–2311. doi: 10.1210/jc.85.6.2304. [DOI] [PubMed] [Google Scholar]
- 15.Jakimiuk AJ, Weitsman SR, Navab A, Magoffin DA. Luteinizing hormone receptor, steroidogenesis acute regulatory protein, and steroidogenic enzyme messenger ribonucleic acids are overexpressed in thecal and granulosa cells from polycystic ovaries. J Clin Endocrinol Metab. 2001;86:1318–1323. doi: 10.1210/jc.86.3.1318. [DOI] [PubMed] [Google Scholar]
- 16.Nelson VL, Qin KN, Rosenfield RL, Wood JR, Penning TM, Legro RS, Strauss JF, 3rd, McAllister JM. The biochemical basis for increased testosterone production in theca cells propagated from patients with polycystic ovary syndrome. J Clin Endocrinol Metab. 2001;86:5925–5933. doi: 10.1210/jc.86.12.5925. [DOI] [PubMed] [Google Scholar]
- 17.Yildiz BO, Azziz R. The adrenal and polycystic ovary syndrome. Rev Endocr Metab Disord. 2007;8:331–342. doi: 10.1007/s11154-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 18.Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, Witchel SF. The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril. 2009;91:456–488. doi: 10.1016/j.fertnstert.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 19.Ehrmann DA, Rosenfield RL, Barnes RB, Brigell DF, Sheikh Z. Detection of functional ovarian hyperandrogenism in women with androgen excess. N Engl J Med. 1992;327:157–162. doi: 10.1056/NEJM199207163270304. [DOI] [PubMed] [Google Scholar]
- 20.Legro RS, Driscoll D, Strauss JF, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legro RS, Kunselman AR, Demers L, Wang SC, Bentley-Lewis R, Dunaif A. Elevated dehydroepiandrosterone sulfate levels as the reproductive phenotype in the brothers of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:2134–2138. doi: 10.1210/jc.87.5.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibanez L, Potau N, Virdis R, Zampolli M, Terzi C, Gussinye M, Carrascosa A, Vicens-Calvet E. Postpubertal outcome in girls diagnosed of premature pubarche during childhood: increased frequency of functional ovarian hyperandrogenism. J Clin Endocrinol Metab. 1993;76:1599–1603. doi: 10.1210/jc.76.6.1599. [DOI] [PubMed] [Google Scholar]
- 23.Oppenheimer E, Linder B, DiMartino-Nardi J. Decreased insulin sensitivity in prepubertal girls with premature adrenarche and acanthosis nigricans. J Clin Endocrinol Metab. 1995;80:614–618. doi: 10.1210/jc.80.2.614. [DOI] [PubMed] [Google Scholar]
- 24.Ibañez L, Potau N, Zampolli M, Prat N, Virdis R, Vicens-Calvet E, Carrascosa A. Hyperinsulinemia in postpubertal girls with a history of premature pubarche and functional ovarian hyperandrogenism. J Clin Endocrinol Metab. 1996;81:1237–1243. doi: 10.1210/jc.81.3.1237. [DOI] [PubMed] [Google Scholar]
- 25.Ibañez L, Potau N, Marcos MV, de Zegher F. Exaggerated adrenarche and hyperinsulinism in adolescent girls born small for gestational age. J Clin Endocrinol Metab. 1999;84:4739–4741. doi: 10.1210/jc.84.12.4739. [DOI] [PubMed] [Google Scholar]
- 26.Ibañez L, Diaz R, Lopez-Bermejo A, Marcos MV. Clinical spectrum of premature pubarche: Links to metabolic syndrome and ovarian hyperandrogenism. Rev Endocr Metab Disord. 2009;10:63–76. doi: 10.1007/s11154-008-9096-y. [DOI] [PubMed] [Google Scholar]
- 27.Miller WL. Androgen synthesis in adrenarche. Rev Endocr Metab Disord. 2009;10:3–17. doi: 10.1007/s11154-008-9102-4. [DOI] [PubMed] [Google Scholar]
- 28.Strauss JF, 3rd, McAllister JM, Urbanek M. Persistence pays off for PCOS gene prospectors. J Clin Endocrinol Metab. 2012;97:2286–2288. doi: 10.1210/jc.2012-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, Liang X, Zhao X, Zhao J, Sun Y, Zhang B, Jiang H, Zhao D, Bian Y, Gao X, Geng L, Li Y, Zhu D, Sun X, Xu JE, Hao C, Ren CE, Zhang Y, Chen S, Zhang W, Yang A, Yan J, Li Y, Ma J, Zhao Y. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, Zhang B, Liang X, Li T, Chen J, Shen J, Zhao J, You L, Gao X, Zhu D, Zhao X, Yan Y, Qin Y, Li W, Yan J, Wang Q, Zhao J, Geng L, Ma J, Zhao Y, He G, Zhang A, Zou S, Yang A, Liu J, Li W, Li B, Wan C, Qin Y, Shi J, Yang J, Jiang H, Xu JE, Qi X, Sun Y, Zhang Y, Hao C, Ju X, Zhao D, Ren CE, Li X, Zhang W, Zhang Y, Zhang J, Wu D, Zhang C, He L, Chen ZJ. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 31.Goodarzi MO, Jones MR, Li X, Chua AK, Garcia OA, Chen YD, Krauss RM, Rotter JI, Ankener W, Legro RS, Azziz R, Strauss JF, 3rd, Dunaif A, Urbanek M. Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. J Med Genet. 2012;49:90–95. doi: 10.1136/jmedgenet-2011-100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welt CK, Styrkarsdottir U, Ehrmann DA, Thorleifsson G, Arason G, Gudmundsson JA, Ober C, Rosenfield RL, Saxena R, Thorsteinsdottir U, Crowley WF, Stefansson K. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab. 2012;97:E1342–1347. doi: 10.1210/jc.2011-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louwers YV, Stolk L, Uitterlinden AG, Laven JS. Cross-ethnic meta-analysis of genetic variants for polycystic ovary syndrome. J Clin Endocrinol Metab. 2013;98:E2006–2012. doi: 10.1210/jc.2013-2495. [DOI] [PubMed] [Google Scholar]
- 34.Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksen MB, Nielsen MF, Brusgaard K, Tan Q, Andersen MS, Glintborg D, Gaster M. Genetic alterations within the DENND1A gene in patients with polycystic ovary syndrome (PCOS) PLoS One. 2013;8:e77186. doi: 10.1371/journal.pone.0077186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAllister JM, Modi B, Miller BA, Biegler J, Bruggeman R, Legro RS, Strauss JF., 3rd Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci USA. 2014;111:E1519–1527. doi: 10.1073/pnas.1400574111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legro RS, Arslanian SA, Ehrmann DA, Hoeger KM, Murad MH, Pasquali R, Welt CK. Diagnosis and treatment of polycystic ovary syndrome: an endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2013;98:4565–4592. doi: 10.1210/jc.2013-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson-Degrave VL, Wickenheisser JK, Hendricks KL, Asano T, Fujishiro M, Legro RS, Kimball SR, Strauss JF, 3rd, McAllister JM. Alterations in mitogen-activated protein kinase kinase and extracellular regulated kinase signaling in theca cells contribute to excessive androgen production in polycystic ovary syndrome. Mol Endocrinol. 2005;19:379–390. doi: 10.1210/me.2004-0178. [DOI] [PubMed] [Google Scholar]
- 39.Wickenheisser JK, Biegler JM, Nelson-DeGrave VL, Legro RS, Strauss JF, 3rd, McAllister JM. Cholesterol side-chain cleavage gene expression in theca cells: augmented transcriptional regulation and mRNA stability in polycystic ovary syndrome. PloS One. 2012;7:e48963.6. doi: 10.1371/journal.pone.0048963. doi.org/10.1371/journal.pone.0048963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez H, Hum DW, Staels B, Miller WL. Transcription of the human genes for cytochrome P450scc and P450c17 is regulated differently in human adrenal NCI-H295 cells than in mouse adrenal Y1 cells. J Clin Endocrinol Metab. 1997;82:365–371. doi: 10.1210/jc.82.2.365. [DOI] [PubMed] [Google Scholar]
- 41.Kaer K, Branovets J, Hallikma A, Nigumann P, Speek M. Intronic L1 retrotransposons and nested genes cause transcriptional interference by inducing intron retention, exonization and cryptic polyadenylation. PLoS One. 2011;6(10):e26099. doi: 10.1371/journal.pone.0026099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gazdar AF, Oie HK, Shackleton CH, Chen TR, Triche TJ, Myers CE, Chrousos GP, Brennan MF, Stein CA, La Rocca RV. Establishment and characterization of a human adrenocortical carcinoma cell line that expresses multiple pathways of steroid biosynthesis. Cancer Res. 1990;50:5488–5496. [PubMed] [Google Scholar]
- 43.Staels B, Hum DW, Miller WL. Regulation of steroidogenesis in NCI-H295 cells: a cellular model of the human fetal adrenal. Mol Endocrinol. 1993;7:423–433. doi: 10.1210/me.7.3.423. [DOI] [PubMed] [Google Scholar]