Abstract
Human noroviruses are major causative agents of sporadic and epidemic gastroenteritis both in children and adults. Currently there are no licensed therapeutic intervention measures either in terms of vaccines or drugs available for these highly contagious human pathogens. Genetic and antigenic diversity of these viruses, rapid emergence of new strains, and their ability to infect a broad population by using polymorphic histo-blood group antigens for cell attachment, pose significant challenges for the development of effective antiviral agents. Despite these impediments, there is progress in the design and development of therapeutic agents. These include capsid-based candidate vaccines, and potential antivirals either in the form of glycomimetics or designer antibodies that block HBGA binding, as well as those that target essential non-structural proteins such as the viral protease and RNA-dependent RNA polymerase. In addition to these classical approaches, recent studies suggest the possibility of interferons and targeting host cell factors as viable approaches to counter norovirus infection. This review provides a brief overview of this progress.
Introduction
Human noroviruses (HuNoVs) are the most common cause of epidemic and sporadic cases of acute gastroenteritis worldwide [1]. In the US alone, HuNoVs cause approximately 19–21 million cases of acute gastroenteritis annually in all age groups [2*,3]. HuNoV infection can be life-threatening, especially in the elderly and immunocompromised transplant patients [4,5] who are at high risk for serious and prolonged chronic illness. In recent years, with the success of rotavirus vaccination in young children, HuNoVs have replaced rotaviruses as the most common cause of gastroenteritis in this age group [6,7*]. The economic burden of HuNoV infection in the US is estimated to be ~$5.5 billion [8]. In developing countries HuNoVs are estimated to cause more than 1 million hospitalizations and 218,000 deaths in children under 5 years of age occurring annually [9].
HuNoVs belong to the genus Norovirus, one of the five major genera in the Caliciviridae family. These ~400 Å icosahedral viruses have a positive-sense, single-stranded RNA genome. They exhibit enormous genetic diversity and are phylogenetically divided into at least six genogroups (GI-GVI). The GI, GII and GIV genogroups contain human pathogens. Each of these genogroups is further divided into several genotypes [10]. The HuNoVs belonging to genogroup II and genotype 4 (GII.4) are the most prevalent, and account for the majority of global outbreaks [11]. Epidemiological studies suggest that the GII.4 strains undergo epochal evolution with a new variant emerging every 2–4 years [12,13]. Recent studies also show outbreaks involving GI strains are becoming increasingly prevalent worldwide, with certain GI genotypes predominating in different geographical regions. The preponderance of global HuNoV outbreaks with periodic emergence of new variants poses a major health concern. Currently, there are no effective vaccines or antivirals available to counter HuNoV infection.
Vaccines against HuNoV infections
The genetic and antigenic diversity of HuNoVs and the lack of naturally-occurring longstanding immunity are possible significant challenges for the development of effective vaccines that can offer widespread cross-protection. However, significant effort has led to development of a bivalent vaccine, based on genotype GI.1 and a consensus GII.4 recombinant virus-like particles (VLPs) [14], which is in phase II clinical trials [15–17**]. The GII.4 VLP was designed by obtaining a consensus sequence from three GII.4 variants (Henry_2001, Yerseke_2006a, and Den Haag_2006b) using the Houston virus (Henry_2001 variant) as the backbone [18]. Point mutations were made to alter the amino acids into a consensus sequence. The consensus GII.4 VLP elicits antibody responses that recognize a wide array of GII.4 variants, including those that have yet to emerge [19*]. The HuNoV VLPs are produced by the expression of the major capsid protein VP1, which as 90 dimers forms the T=3 icosahedral capsid (Fig. 1) [20,21]. VP1 is encoded by the open reading frame (ORF) 2 of the HuNoV genome. A second minor structural protein, VP2, not present in the vaccine construct, is encoded by ORF3, whereas the ORF1 encodes a polyprotein that is processed by the virally-encoded protease into 6 non-structural proteins (NSPs). The VP1 exhibits a modular domain organization consisting of an S domain, formed by the N-terminal residues, that provides a scaffold for the protruding P domain, which is further subdivided into P1 and P2 subdomains (Fig. 1A and 1B). The distally located and surface-exposed P2 subdomain, which can be considered as a large insertion in the P1 subdomain, harbors the most sequence variations across the genogroups and genotypes and is responsible for many virus-host interactions. Recombinant VLPs are morphologically and antigenically similar to the authentic HuNoV capsid and are highly immunogenic. Such VLPs can be made from any HuNoVs genotype [22], suggesting the possibility of designing multivalent vaccines from selected multiple genotypes. In addition to the VLPs, recombinant P domain by itself elicits a strong immune response and has been suggested as a possible candidate for vaccine development efforts [23–25]. Even if an effective vaccine becomes available, there is a great interest in the development of antiviral drugs [26–28]. Antiviral treatment could be useful for therapy of chronic infection in immunocompromised patients; treatment and prophylaxis in outbreak situations where ongoing transmission continues to occur, as in a nursing home outbreak. As prophylaxis in certain circumstances, such as for travelers, if the medication is safe and vaccine is unavailable; and for treatment of acute illness, particularly in the young and old and hospitalized patients in whom symptoms may last for up to a week. What are the targets for the design and development of such anti-HuNoV drugs?
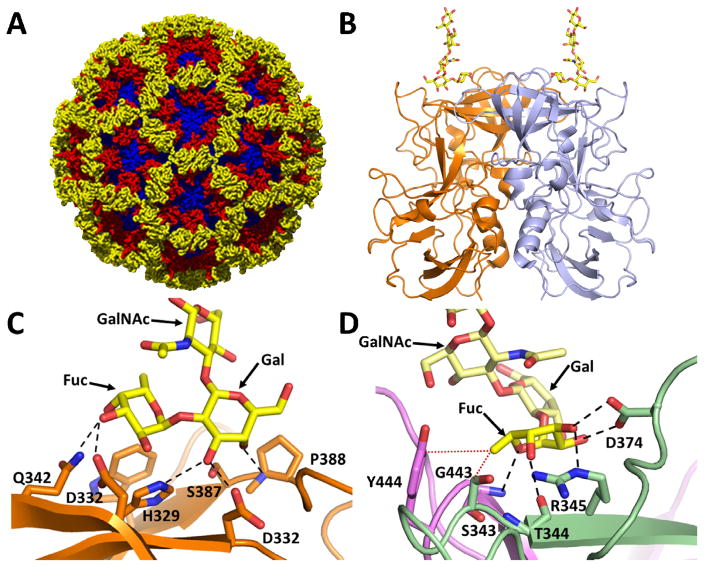
Fig. 1. NoV-HBGA binding site, a potential target for antivirals.
A) Structure of the Norwalk virus-like particle (PDB ID: 1IHM) comprised of 90 VP1 dimers. The VPI S domain, P1 and P2 subdomains are shown in blue, red and yellow respectively. B) Cartoon representation of P-domain dimer (PDB ID: 2ZL6) bound to H type HBGA. The HBGA binding site is located on the top of the P domain. The individual subunits of the dimer are shown in orange and blue, respectively, and the H type HBGA is shown in yellow as a stick model. C) Close up of HBGA binding site in GI HuNoV showing the galactose dominant nature of HBGA binding. All the residues involved in hydrogen bond interactions with H type HBGA (yellow) are contributed by the individual subunits of the dimer shown in orange stick models with oxygen (red) and nitrogen (blue) atoms shown; hydrogen bonds are shown as black dashed lines. D) Close up of the HBGA binding site in GII.4 bound to H type HBGA (PDB ID: 3SLN) showing the fucose dominant nature of HBGA binding. The HBGA binding site in GII NoVs lies on the dimeric interface with both subunits of the dimer (green and pink) contributing to HBGA binding. Residues involved in hydrogen bond interactions (dashed lines) with HBGA (yellow) are labeled and bound HBGA is shown as a stick model following the same coloring scheme as in C.
Glycan binding site as a potential target
Susceptibility to HuNoV infection is associated with expression of histo-blood group antigens (HBGAs) [1] that are found in mucosal secretions and on epithelial cells. These genetically-determined glycoconjugates function as initial cell attachment factors for HuNoVs [29,30]. The unique requirement of binding to polymorphic HBGAs may influence the evolution of NoV strains [13]. HuNoVs bind to HBGAs through the hypervariable P2 subdomain in the protruding P domain of VP1 [31–37]. Studies using VLPs or P domain constructs have shown that HuNoVs, as a result of variations in the P2 subdomain, exhibit strain-specific HBGA binding patterns. Crystallographic studies of P domain-HBGA complexes show that while the HBGA binding site is distinct between GI and GII, the sequence changes around the conserved HBGA site within each genogroup allow for modulations in the HBGA binding profiles [38] (Fig. 1C and Fig. 1D). These sequence changes result in significant alterations in the structural and electrostatic topography of the P2 subdomain consistent with strain-dependent antigenic variations [36,37]. The observation that these changes are in close proximity to the HBGA binding site is consistent with the notion that a coordinated interplay between variations in HBGA binding profiles and antigenicity are critical factors in driving the evolution of HuNoV.
Although there are differences in HBGA binding profiles between the genotypes within each genogroup, there are also conserved features. For example, all the genotypes in GI and GII genogroups primarily recognize a galactose and a fucose residue in the HBGA, respectively [38]. The structural elements including the amino acid residues in the respective P2 subdomains of these genogroups that coordinate the binding of the galactose and fucose moieties are highly conserved. In the case of a GII (GII.10) P domain, crystallographic studies have shown that fucose alone can bind the same set of P2 subdomain residues as the terminal fucose residue of HBGA [34]. Interestingly, a citrate together with a water molecule mimicking the pyranoside ring of fucose also can bind effectively to the same site [39]. Furthermore, recent studies have shown that oligosaccharides derived from human milk such as 2′-fucosyllactose and 3-fucosyllactose bind at the same site in the GII as HBGA, and they can block HBGA binding [40*]. These observations raise the possibility that HBGA binding sites can be potential targets for the design of glycomimetics or small molecules that inhibit HBGA binding. However, because these interactions with HBGA generally occur with low affinity, that is, in the low micromolar range, designing potent glycomimetics may be a challenging task.
HBGA-blocking antibodies as therapeutic agents
A more feasible approach may be to design antivirals based on monoclonal antibodies (mAbs) that inhibit HuNoV-HBGA interactions [19,41–43], particularly considering recent studies that have identified such antibodies from the sera of HuNoV infected patients [44–46]. The design and development of suitable mAbs or their derivatives, single-chain antibody fragments (also called VHH or nanobodies) and disulphide-stabilized single-chain antibody fragments that particularly target the entry mechanism, have been in consideration for prophylactic and therapeutic use to counter viruses such as influenza virus, rabies virus, Ebola virus and hepatitis B virus [47]. Adaptive humoral immunity is also involved in resistance to HuNoV infection. Serum antibodies that block interactions between the virus and HBGAs are associated with lower risks of developing infection or illness following exposure to virus [44–46*]. Such antibodies have been proposed to be functionally similar to hemagglutination inhibiting or neutralizing antibodies of influenza virus [48,49]. HBGA-blocking antibodies can vary in specificity, from genotype-specific to variant-specific and even strain-specific among the globally prevalent GII.4 NoVs, further emphasizing a correlated interplay between antigenicity and HBGA specificity in the evolution of NoVs [12,13,50]. Human monoclonal antibodies that block HBGA binding have been isolated and produced from the peripheral blood mononuclear cells of blood donors, and the few that have been characterized to date appear to be genotype-specific [51]. In addition, nanobodies that block HBGA binding in GI.1 and GII.4 VLPs, have been identified and characterized [52*]. Currently the mechanism of how these antibodies block HBGA binding is unclear. Structural studies of the P domain in complex with Fabs of these mAbs or nanobodies should provide mechanistic details of their blockade activity that then can be leveraged to design antibody-scaffolds or antibody-like molecules as HuNoV-specific therapeutic agents.
Non-structural proteins as targets
As noted earlier, the ORF1 of HuNoVs encodes a polyprotein that is proteolytically processed by the virus-encoded protease into at least six NSPs [53]. These NSPs, from N- to C-terminus of the polyprotein, include p48, whose precise function is yet to be determined; p41, an NTPase with distinct highly conserved SF3 helicase motifs similar to picornavirus 2C; p22, which shares sequence similarities with picornavirus 3A, with a possible function as an antagonist of Golgi-dependent cellular protein secretion [54]; VPg that is covalently linked to the viral RNA; a protease that is similar to picornavirus 3C; and an RNA-dependent-RNA polymerase (RdRp) orthologous to picornavirus 3Dpol [55–58]. Of these six NSPs, protease and RdRp have been considered potential antiviral targets not only because their structures and functions are well characterized, but also because their picornavirus homologues have been studied extensively for the development of antivirals.
HuNoV Protease
Proteolytic processing of the polyprotein by the virally-encoded protease is a common essential step in the replication of the (+)RNA viruses, including HuNoVs. Unlike cellular proteases that generally target one site, these viral proteases can recognize and cleave at multiple specific sites in the polyprotein. Because of their critical function in viral replication, viral proteases including HuNoV protease [56,59–64*], have been attractive targets for the design and development of small molecule drugs that inhibit proteolytic processing. Typically, such protease inhibitors are short peptides that mimic N-terminal residues preceding the cleavage site (P1–P5) of the substrates in the polyprotein and modified further at the P1 position by attaching adducts such as aldehydes, ketones, esters or bisulfite as electrophilic warhead. These inhibitors bind to the active site irreversibly and covalently modify the active-site nucleophilic residue to inhibit proteolytic activity.
The HuNoV protease, similar to picornavirus 3C, is a cysteine protease with a chymotrypsin-like fold. It is comprised of two domains separated by a cleft where the active site is located [65–67]. The active site consists of a catalytic triad with cysteine as a nucleophile, histidine as the general base catalyst, and glutamic acid as the anion to orient the imidazole ring of histidine [66,68–70]. Crystal structures of HuNoV proteases in complex with substrates bearing P1–P4 residues or substrate-mimics have shown how these residues optimally interact with the S1–S4 pockets in the protease (Fig. 2A), respectively, and how the protease accommodates the varying residue compositions in the polyprotein by undergoing suitable conformational changes in the active-site cleft [68,71,72*]. These studies have provided valuable insights into the design of peptide-mimetics that effectively inhibit protease activity for both GI.1 and GII.4 proteases [62,72*]. Crystal structures of the Norwalk virus (GI.1) protease in complex with three of these substrate-based peptide inhibitors with a terminal aldehyde showed that in addition to the formation of the covalent adducts (Fig. 2B), these inhibitors prevent the conformational change necessary for the formation of the oxyanion hole. These studies further suggest that peptido-mimetics with suitable warheads with a Glu-like chemical entity at P1 for optimal interactions with S1 pocket, and an appropriate combination of hydrophobic residues at P2 and P4 that maximizes the interactions with S2 and S4 pockets, are factors to be considered for enhancing the potency of the inhibitors. More recently, based on the observation that protease-bound peptidyl inhibitors typically adopt a β-strand conformation between the P1 and P3 positions, Weerwarna et al., [73**] have designed and synthesized a novel set of triazole-based macrocyclic inhibitors in which these two positions are linked using a suitable linker such that the P1–P3 is pre-organized into a β-strand conformation for optimal interactions with the norovirus protease as demonstrated by structural studies. In addition to potentially higher stability to metabolic enzymes these inhibitors exhibit increased cellular permeability. Further studies should be anticipated that are directed at optimizing the design strategies and improving the pharmacokinetic properties and metabolic stability of HuNoV protease inhibitors using structural analysis and cell-based assays [74–76]. Considering that the active-site residues are highly conserved between the HuNoV proteases in various genogroups and picornavirus proteases, there is a distinct possibility of designing broad-spectrum protease inhibitors as antivirals [77].
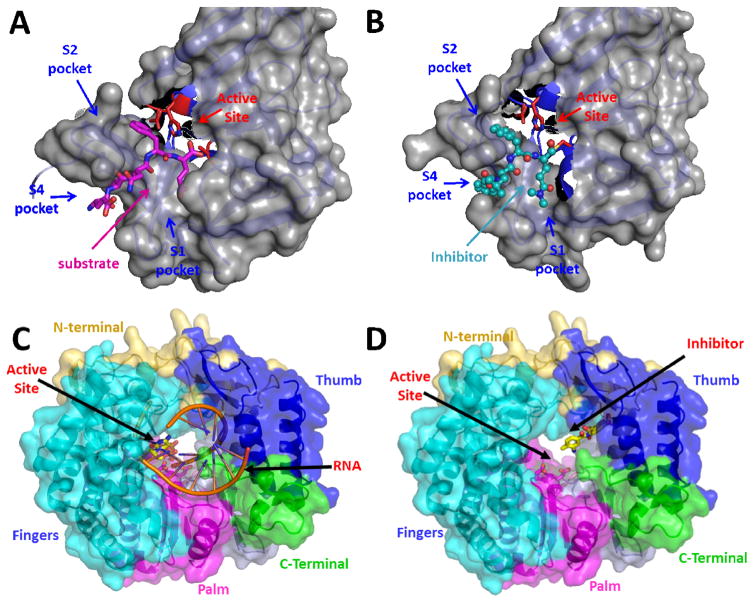
Fig. 2. HuNoV NSPs protease and RdRp as antiviral targets.
A) Structure of the Norwalk virus protease bound to its natural substrate (PDB ID: 4IN1) (PDB ID: 4IMZ syc10). The protease is depicted in both surface (grey) and cartoon model (light blue). The substrate binding pockets are labeled S1, S2 and S4 and the residues forming the catalytic triad are shown as stick model in red and labeled active site. The substrate comprising residues INFE is shown in pink as stick model with oxygen and nitrogen atoms labeled in red and blue respectively. B) Structure of the Norwalk protease bound to substrate-based peptide inhibitor syc10 (PDB ID: 4IMZ). The protease is depicted similar to Fig. 2A. The inhibitor is represented as a ball and stick model in cyan and is shown to mimic substrate binding C) Structure of the norovirus polymerase, RdRp (PDB ID:1SH2) bound to its primer-template RNA duplex. RdRp is depicted in both surface and cartoon model. The thumb (blue), fingers (cyan), palm (pink) domains, along with the N- (light yellow) and C-terminal (green) regions are indicated. The active site is labeled and is shown with bound primer-template RNA duplex (orange). D) Structure of the norovirus polymerase, RdRp (PDB ID 4NRT) bound to suramin inhibitor, which blocks RNA exit pathway, with each of the RdRp structural elements shown using the same coloring code as in Fig. 2C.
HuNoV RdRp
The NoV RdRp, similar to picornavirus 3D, is critical for synthesizing both negative-sense RNA as well as newly made positive-sense genomic RNA and has been the target for developing small molecule inhibitors. X-ray structures of RdRps from several NoV genogroups including HuNoV (GII) [78,79], sapovirus [80], and murine NoV [81,82] have been determined. As observed in all RNA/DNA polymerases, this protein exhbits a typical “right hand” configuration of palm, finger, and thumb domains [83] (Fig. 2C). The active site is located in the thumb domain. It consists of three conserved Asp residues that are critical for mediating catalysis through a two metal-ion mechanism, and other key residues such as Arg, Asn and Ser that are required for substrate binding and catalysis. Crystallographic studies have further shown that NoV RdRp can exist in two principal conformations. An ‘open’ active site conformation that represents the inactive state of the RdRp [78,80,84], and a ‘closed’ active site conformation that is primed for catalyzing nucleotidyl transfer reaction [79] by optimally positioning the nucleotide, RNA and the metal ions for catalytic reaction.
There has been significant progress in identifying small molecule inhibitors of NoV RdRp using in silico screening, and in understanding the structural basis of inhibition by analyzing co-crystal structures of RdRp with some of these inhibitors [85,86*]. Non-nucleoside inhibitors such as suramin, a drug used in the treatment of sleeping sickness caused by the protozoan Trypanosoma, and its analogue NF023, consisting of naphthalene-trisulfonic acid moiety, have been shown to be effective in inhibiting NoV RdRp with IC50s in low nanomolar range [86*]. Both these inhibitors bind RdRp along the nucleotide access pathway between the fingers and thumb domains (Fig. 2D). Nucleoside-analogs such as 2′-C-methylcytidine (2CM-C) and ribavirin, hepatitis C virus polymerase inhibitors, and 6-fluoro-3-hydroxy-2-pyrazinecarboxamide (T-705; favipiravir), a nucleoside precursor which was originally developed against influenza viruses, have been shown to be effective in inhibiting in vitro replication of murine norovirus (MNV), and also HuNoV replication using a Norwalk virus replicon model [87*–89]. More recent studies have shown that triphosphates of 2CM-C and T-705 also inhibit MNV and HuNoV RdRp activities with IC50s in the low micromolar range [90]. These studies found that 2CM-C triphosphate inhibited RdRp by directly competing with CTP during primer elongation whereas T-705 triphosphate competed mostly with ATP and GTP at the initiation and elongation steps. Further structure-based techniques coupled with high throughput screening [91] will likely lead to design and development of more potent and perhaps even broad-spectrum RdRp inhibitors with the necessary pharmacokinetic properties.
Interferons and targeting host factors as antiviral approach
Interferons are a group of peptides that have antiviral activity against a variety of viruses. In vitro studies have demonstrated that type I interferons inhibit norovirus replication in a replicon system [88] although no human studies have been reported to date. MNV replication is inhibited by both type I and II interferons [92]. Recent studies have also demonstrated that chronic infection caused by MNV can be cleared with the administration of the type III interferon, interferon λ, in the absence of an adaptive immune response [93**]. In addition, a number of cellular factors such as La, PTB, DDX3, PCPB2, and hnRNPs have been identified as critical for MNV replication using RNAi methodology, suggesting that these molecules may be targets for antiviral drug development [94**]. Small molecule inhibitors of deubiquitinases, such as WP1130, inhibit MNV replication in addition to several other RNA viruses [95]. Whether such approaches are viable for HuNoV replication needs further studies.
Conclusions
HuNoVs pose a significant global health concern. With the current lack of effective antiviral strategies, there has been an intense focus in vaccine development as well as antiviral drug discovery. Recent progress in both these fronts is encouraging. An immunogenic VLP-based candidate vaccine is in phase-II clinical trials and other candidate vaccines based on P domain, which elicit strong immune responses, also hold promise. These studies raise the possibility of designing multivalent vaccines to counter the antigenic and genetic diversity exhibited by HuNoVs. Recent progress in the isolation and characterization of HBGA-blocking human mAbs points to a distinct possibility of designing antibody-based scaffolds as immunotherapeutic agents. In parallel, several studies have focused on selected non-structural proteins such as protease and RdRp for small molecule drug discovery. Further studies are required to optimize their metabolic stability and pharmacokinetic properties. Progress made in recent years in producing human NoVs in cultured cells [96**,97**] may prove critical for robust optimization of such antiviral drugs and lead to a better understanding of the mechanisms that underlie virus replication, which in turn may pave way for discovering novel antiviral agents. Combined use of antivirals with an effective vaccine may indeed be realized in the near future to counter and control HuNoV infections.
Highlights.
Development of capsid-based vaccines – a VLP-based vaccine in phase-II clinical trials –potential for multivalent vaccines
Human monoclonal antibodies that block glycan binding - prospects for immunotherapeutic agents
Viral protease and polymerase as targets for drug discovery
Interferons and inhibitors of host cell factors as antivirals.
Acknowledgments
We acknowledge support from NIH grant PO1 AI057788 (MKE, RLA, BVVP, TP, YS), and a grant (Q1279) from the Robert Welch foundation (BVVP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ramani S, Atmar RL, Estes MK. Epidemiology of human noroviruses and updates on vaccine development. Current opinion in gastroenterology. 2014;30:25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD. Norovirus disease in the United States. Emerg Infect Dis. 2013;19:1198–1205. doi: 10.3201/eid1908.130465. This manuscript summarizes estimates of the burden of disease caused by norovirus infection in the United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Mahon BE. Foodborne Diseases Active Surveillance Network (FoodNet) in 2012: a foundation for food safety in the United States. Clin Infect Dis. 2012;54(Suppl 5):S381–384. doi: 10.1093/cid/cis257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok K, Green KY. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med. 2012;367:2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roddie C, Paul JP, Benjamin R, Gallimore CI, Xerry J, Gray JJ, Peggs KS, Morris EC, Thomson KJ, Ward KN. Allogeneic hematopoietic stem cell transplantation and norovirus gastroenteritis: a previously unrecognized cause of morbidity. Clin Infect Dis. 2009;49:1061–1068. doi: 10.1086/605557. [DOI] [PubMed] [Google Scholar]
- 6.Koo HL, Neill FH, Estes MK, Munoz FM, Cameron A, DuPont HL, Atmar RL. Noroviruses: The most common pediatric viral enteric pathogen at a large university hospital after introduction of rotavirus vaccination. J Pediatric Infect Dis Soc. 2013;2:57–60. doi: 10.1093/jpids/pis070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Payne DC, Vinje J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. This study demonstrates that noroviruses have replaced rotaviruses as the most common cause of gastroenteritis in children under 5 years of age in the United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartsch SM, Lopman BA, Hall AJ, Parashar UD, Lee BY. The potential economic value of a human norovirus vaccine for the United States. Vaccine. 2012;30:7097–7104. doi: 10.1016/j.vaccine.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroneman A, Vega E, Vennema H, Vinje J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol. 2013;158:2059–2068. doi: 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, van Duynhoven Y, Gray J, Iturriza M, Bottiger B, Falkenhorst G, Johnsen C, et al. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J Clin Microbiol. 2008;46:2959–2965. doi: 10.1128/JCM.00499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. Epochal evolution of GGII. 4 norovirus capsid proteins from 1995 to 2006. J Virol. 2007;81:9932–9941. doi: 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol Rev. 2008;225:190–211. doi: 10.1111/j.1600-065X.2008.00680.x. [DOI] [PubMed] [Google Scholar]
- 14.Richardson C, Bargatze RF, Goodwin R, Mendelman PM. Norovirus virus-like particle vaccines for the prevention of acute gastroenteritis. Expert Rev Vaccines. 2013;12:155–167. doi: 10.1586/erv.12.145. [DOI] [PubMed] [Google Scholar]
- 15.Treanor JJ, Atmar RL, Frey SE, Gormley R, Chen WH, Ferreira J, Goodwin R, Borkowski A, Clemens R, Mendelman PM. A novel intramuscular bivalent norovirus virus-like particle vaccine candidate--reactogenicity, safety, and immunogenicity in a phase 1 trial in healthy adults. J Infect Dis. 2014;210:1763–1771. doi: 10.1093/infdis/jiu337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atmar RL, Estes MK. Norovirus vaccine development: next steps. Expert Rev Vaccines. 2012;11:1023–1025. doi: 10.1586/erv.12.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, Estes MK, Graham DY, Opekun AR, Richardson C, Mendelman PM. Norovirus vaccine against experimental human Norwalk Virus illness. N Engl J Med. 2011;365:2178–2187. doi: 10.1056/NEJMoa1101245. This is a report of a proof-of-concept clinical trial that demonstrated that a VLP-based norovirus vaccine can prevent illness and infection following experimental challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra GI, Bok K, Taylor R, Haynes JR, Sosnovtsev SV, Richardson C, Green KY. Immunogenicity and specificity of norovirus Consensus GII. 4 virus-like particles in monovalent and bivalent vaccine formulations. Vaccine. 2012;30:3580–3586. doi: 10.1016/j.vaccine.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Lindesmith LC, Ferris MT, Mullan CW, Ferreira J, Debbink K, Swanstrom J, Richardson C, Goodwin RR, Baehner F, Mendelman PM, Bargatze RF, et al. Broad blockade antibody responses in human volunteers after immunization with a multivalent norovirus VLP candidate vaccine: immunological analyses from a phase I clinical trial. PLoS Med. 2015;12:e1001807. doi: 10.1371/journal.pmed.1001807. This study demonstrates that a norovirus vaccine can elicit HBGA-blocking antibodies to genotypes not included in the vaccine, suggesting possible protection against heterotypic strains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X, Wang M, Graham DY, Estes MK. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol. 2005;79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocher J, Bui T, Giri-Rachman E, Wen K, Li G, Yang X, Liu F, Tan M, Xia M, Zhong W, Jiang X, et al. Intranasal P particle vaccine provided partial cross-variant protection against human GII. 4 norovirus diarrhea in gnotobiotic pigs. J Virol. 2014;88:9728–9743. doi: 10.1128/JVI.01249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocher J, Yuan L. Norovirus vaccines and potential antinorovirus drugs: recent advances and future perspectives. Future Virol. 2015;10:899–913. doi: 10.2217/fvl.15.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang H, Tan M, Xia M, Wang L, Jiang X. Norovirus P particle efficiently elicits innate, humoral and cellular immunity. PLoS One. 2013;8:e63269. doi: 10.1371/journal.pone.0063269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufman SS, Green KY, Korba BE. Treatment of norovirus infections: moving antivirals from the bench to the bedside. Antiviral Res. 2014;105:80–91. doi: 10.1016/j.antiviral.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galasiti Kankanamalage AC, Weerawarna PM, Kim Y, Chang KO, Groutas WC. Anti-norovirus therapeutics: a patent review (2010–2015) Expert Opin Ther Pat. 2016;26:297–308. doi: 10.1517/13543776.2016.1153065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y, Galasiti Kankanamalage AC, Chang KO, Groutas WC. Recent advances in the discovery of norovirus therapeutics. J Med Chem. 2015;58:9438–9450. doi: 10.1021/acs.jmedchem.5b00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. 2002;185:1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- 30.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu W, Mamedova A, Tan M, Xia M, Jiang X, Hegde RS. Structural basis for the receptor binding specificity of Norwalk virus. J Virol. 2008;82:5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi JM, Hutson AM, Estes MK, Prasad BV. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci U S A. 2008;105:9175–9180. doi: 10.1073/pnas.0803275105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansman GS, Biertumpfel C, Georgiev I, McLellan JS, Chen L, Zhou T, Katayama K, Kwong PD. Crystal structures of GII.10 and GII. 12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J Virol. 2011;85:6687–6701. doi: 10.1128/JVI.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubota T, Kumagai A, Ito H, Furukawa S, Someya Y, Takeda N, Ishii K, Wakita T, Narimatsu H, Shirato H. Structural basis for the recognition of Lewis antigens by genogroup I norovirus. J Virol. 2012;86:11138–11150. doi: 10.1128/JVI.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII. 4 epidemic variant: implications for epochal evolution. J Virol. 2011;85:8635–8645. doi: 10.1128/JVI.00848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanker S, Czako R, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses. J Virol. 2014;88:6168–6180. doi: 10.1128/JVI.00201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad BV, Shanker S, Hu L, Choi JM, Crawford SE, Ramani S, Czako R, Atmar RL, Estes MK. Structural basis of glycan interaction in gastroenteric viral pathogens. Curr Opin Virol. 2014;7:119–127. doi: 10.1016/j.coviro.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansman GS, Shahzad-Ul-Hussan S, McLellan JS, Chuang GY, Georgiev I, Shimoike T, Katayama K, Bewley CA, Kwong PD. Structural basis for norovirus inhibition and fucose mimicry by citrate. J Virol. 2012;86:284–292. doi: 10.1128/JVI.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Weichert S, Koromyslova A, Singh BK, Hansman S, Jennewein s, Schroten H, Hansman GS. Structural basis for norovirus inhibition by human milk oligosaccharides. J Virol. 2016;90:48433–4848. doi: 10.1128/JVI.03223-15. Showed that in adition to HBGA noroviruses can recognize human milk oligoscahhrides bearing a fucose moiety. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bok K, Parra GI, Mitra T, Abente E, Shaver CK, Boon D, Engle R, Yu C, Kapikian AZ, Sosnovtsev SV, Purcell RH, et al. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc Natl Acad Sci U S A. 2011;108:325–330. doi: 10.1073/pnas.1014577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blazevic V, Malm M, Honkanen H, Knip M, Hyoty H, Vesikari T. Development and maturation of norovirus antibodies in childhood. Microbes Infect. 2015 doi: 10.1016/j.micinf.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Lindesmith LC, Debbink K, Swanstrom J, Vinje J, Costantini V, Baric RS, Donaldson EF. Monoclonal antibody-based antigenic mapping of norovirus GII. 4-2002. J Virol. 2012;86:873–883. doi: 10.1128/JVI.06200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czako R, Atmar RL, Opekun AR, Gilger MA, Graham DY, Estes MK. Serum hemagglutination inhibition activity correlates with protection from gastroenteritis in persons infected with Norwalk virus. Clin Vaccine Immunol. 2012;19:284–287. doi: 10.1128/CVI.05592-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, Graham DY, Atmar RL. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis. 2010;202:1212–1218. doi: 10.1086/656364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Czako R, Atmar RL, Opekun AR, Gilger MA, Graham DY, Estes MK. Experimental human infection with Norwalk virus elicits a surrogate neutralizing antibody response with cross-genogroup activity. Clin Vaccine Immunol. 2015;22:221–228. doi: 10.1128/CVI.00516-14. This study demonstrates that norovirus infection induces HBGA-blocking antibody responses to heterotypic strains in other genotypes and even in other genogroups. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Both L, Banyard AC, van Dolleweerd C, Wright E, Ma JK, Fooks AR. Monoclonal antibodies for prophylactic and therapeutic use against viral infections. Vaccine. 2013;31:1553–1559. doi: 10.1016/j.vaccine.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J Virol. 2002;76:12335–12343. doi: 10.1128/JVI.76.23.12335-12343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindesmith LC, Donaldson E, Leon J, Moe CL, Frelinger JA, Johnston RE, Weber DJ, Baric RS. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J Virol. 2010;84:1800–1815. doi: 10.1128/JVI.02179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol. 2010;8:231–241. doi: 10.1038/nrmicro2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindesmith LC, Beltramello M, Swanstrom J, Jones TA, Corti D, Lanzavecchia A, Baric RS. Serum immunoglobulin A cross-strain blockade of human noroviruses. Open Forum Infect Dis. 2015;2:ofv084. doi: 10.1093/ofid/ofv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Garaicoechea L, Aguilar A, Parra GI, Bok M, Sosnovtsev SV, Canziani G, Green KY, Bok K, Parreno V. Llama nanoantibodies with therapeutic potential against human norovirus diarrhea. PLoS One. 2015;10:e0133665. doi: 10.1371/journal.pone.0133665. Describes characterization of HBGA-blocking nanobodies for different HuNoV genogroups. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorne LG, Goodfellow IG. Norovirus gene expression and replication. J Gen Virol. 2014;95:278–291. doi: 10.1099/vir.0.059634-0. [DOI] [PubMed] [Google Scholar]
- 54.Sharp TM, Crawford SE, Ajami NJ, Neill FH, Atmar RL, Katayama K, Utama B, Estes MK. Secretory pathway antagonism by calicivirus homologues of Norwalk virus nonstructural protein p22 is restricted to noroviruses. Virol J. 2012;9:181. doi: 10.1186/1743-422X-9-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfister T, Wimmer E. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J Virol. 2001;75:1611–1619. doi: 10.1128/JVI.75.4.1611-1619.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sosnovtsev SV, Garfield M, Green KY. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J Virol. 2002;76:7060–7072. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hardy ME. Norovirus protein structure and function. FEMS Microbiol Lett. 2005;253:1–8. doi: 10.1016/j.femsle.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 58.Clarke IN, Lambden PR. Organization and expression of calicivirus genes. J Infect Dis. 2000;181(Suppl 2):S309–316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- 59.Blakeney SJ, Cahill A, Reilly PA. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology. 2003;308:216–224. doi: 10.1016/s0042-6822(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 60.Hardy ME, Crone TJ, Brower JE, Ettayebi K. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 2002;89:29–39. doi: 10.1016/s0168-1702(02)00114-4. [DOI] [PubMed] [Google Scholar]
- 61.Someya Y, Takeda N, Miyamura T. Characterization of the norovirus 3C-like protease. Virus Res. 2005;110:91–97. doi: 10.1016/j.virusres.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Deng L, Muhaxhiri Z, Estes MK, Palzkill T, Prasad BV, Song Y. Synthesis, Activity and Structure-Activity Relationship of Noroviral Protease Inhibitors. Medchemcomm. 2013;4 doi: 10.1039/C3MD00219E. Describes synthesis and chracterization of HuNoV protease inhbitors to GI and GII.4 HuNoVs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Tiew KC, He G, Aravapalli S, Mandadapu SR, Gunnam MR, Alliston KR, Lushington GH, Kim Y, Chang KO, Groutas WC. Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease. Bioorg Med Chem Lett. 2011;21:5315–5319. doi: 10.1016/j.bmcl.2011.07.016. Describes synthesis and chractrization of peptide-based protease inhbitors to GI Norwalk virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Mandadapu SR, Weerawarna PM, Prior AM, Uy RA, Aravapalli S, Alliston KR, Lushington GH, Kim Y, Hua DH, Chang KO, Groutas WC. Macrocyclic inhibitors of 3C and 3C-like proteases of picornavirus, norovirus, and coronavirus. Bioorg Med Chem Lett. 2013;23:3709–3712. doi: 10.1016/j.bmcl.2013.05.021. This report raises the possibility of designing broad-specturm protease-inhbitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boniotti B, Wirblich C, Sibilia M, Meyers G, Thiel HJ, Rossi C. Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J Virol. 1994;68:6487–6495. doi: 10.1128/jvi.68.10.6487-6495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeitler CE, Estes MK, Venkataram Prasad BV. X-ray crystallographic structure of the Norwalk virus protease at 1. 5-A resolution. J Virol. 2006;80:5050–5058. doi: 10.1128/JVI.80.10.5050-5058.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bazan JF, Fletterick RJ. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hussey RJ, Coates L, Gill RS, Erskine PT, Coker SF, Mitchell E, Cooper JB, Wood S, Broadbridge R, Clarke IN, Lambden PR, et al. A structural study of norovirus 3C protease specificity: binding of a designed active site-directed peptide inhibitor. Biochemistry. 2011;50:240–249. doi: 10.1021/bi1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakamura K, Someya Y, Kumasaka T, Ueno G, Yamamoto M, Sato T, Takeda N, Miyamura T, Tanaka N. A norovirus protease structure provides insights into active and substrate binding site integrity. J Virol. 2005;79:13685–13693. doi: 10.1128/JVI.79.21.13685-13693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leen EN, Baeza G, Curry S. Structure of a murine norovirus NS6 protease-product complex revealed by adventitious crystallisation. PLoS One. 2012;7:e38723. doi: 10.1371/journal.pone.0038723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leen EN, Kwok KY, Birtley JR, Simpson PJ, Subba-Reddy CV, Chaudhry Y, Sosnovtsev SV, Green KY, Prater SN, Tong M, Young JC, et al. Structures of the compact helical core domains of feline calicivirus and murine norovirus VPg proteins. J Virol. 2013;87:5318–5330. doi: 10.1128/JVI.03151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72**.Muhaxhiri Z, Deng L, Shanker S, Sankaran B, Estes MK, Palzkill T, Song Y, Prasad BV. Structural basis of substrate specificity and protease inhibition in Norwalk virus. J Virol. 2013;87:4281–4292. doi: 10.1128/JVI.02869-12. This is the first study to describe the structural basis for understanding how norovirus protease recognizes mutiple substrates and the mechanism of inhbition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73**.Weerawarna PM, Kim Y, Galasiti Kankanamalage AC, Damalanka VC, Lushington GH, Alliston KR, Mehzabeen N, Battaile KP, Lovell S, Chang KO, Groutas WC. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies. Eur J Med Chem. 2016;119:300–318. doi: 10.1016/j.ejmech.2016.04.013. Describes structure-based design and synthesis of novel set of macrocyclic inhibitors of norovirus protease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qu L, Vongpunsawad S, Atmar RL, Prasad BV, Estes MK. Development of a Gaussia luciferase-based human norovirus protease reporter system: cell type-specific profile of Norwalk virus protease precursors and evaluation of inhibitors. J Virol. 2014;88:10312–10326. doi: 10.1128/JVI.01111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006;353:463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 76.Galasiti Kankanamalage AC, Kim Y, Weerawarna PM, Uy RA, Damalanka VC, Mandadapu SR, Alliston KR, Mehzabeen N, Battaile KP, Lovell S, Chang KO, et al. Structure-guided design and optimization of dipeptidyl inhibitors of norovirus 3CL protease. Structure-activity relationships and biochemical, X-ray crystallographic, cell-based, and in vivo studies. J Med Chem. 2015;58:3144–3155. doi: 10.1021/jm5019934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim Y, Lovell S, Tiew KC, Mandadapu SR, Alliston KR, Battaile KP, Groutas WC, Chang KO. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng KK, Pendas-Franco N, Rojo J, Boga JA, Machin A, Alonso JM, Parra F. Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J Biol Chem. 2004;279:16638–16645. doi: 10.1074/jbc.M400584200. [DOI] [PubMed] [Google Scholar]
- 79.Zamyatkin DF, Parra F, Alonso JM, Harki DA, Peterson BR, Grochulski P, Ng KK. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283:7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 80.Fullerton SW, Blaschke M, Coutard B, Gebhardt J, Gorbalenya A, Canard B, Tucker PA, Rohayem J. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J Virol. 2007;81:1858–1871. doi: 10.1128/JVI.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee JH, Alam I, Han KR, Cho S, Shin S, Kang S, Yang JM, Kim KH. Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase. J Gen Virol. 2011;92:1607–1616. doi: 10.1099/vir.0.031104-0. [DOI] [PubMed] [Google Scholar]
- 82.Hogbom M, Jager K, Robel I, Unge T, Rohayem J. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J Gen Virol. 2009;90:281–291. doi: 10.1099/vir.0.005629-0. [DOI] [PubMed] [Google Scholar]
- 83.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ng KK, Cherney MM, Vazquez AL, Machin A, Alonso JM, Parra F, James MN. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J Biol Chem. 2002;277:1381–1387. doi: 10.1074/jbc.M109261200. [DOI] [PubMed] [Google Scholar]
- 85.Mastrangelo E, Pezzullo M, Tarantino D, Petazzi R, Germani F, Kramer D, Robel I, Rohayem J, Bolognesi M, Milani M. Structure-based inhibition of Norovirus RNA-dependent RNA polymerases. J Mol Biol. 2012;419:198–210. doi: 10.1016/j.jmb.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 86*.Croci R, Pezzullo M, Tarantino D, Milani M, Tsay SC, Sureshbabu R, Tsai YJ, Mastrangelo E, Rohayem J, Bolognesi M, Hwu JR. Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds. PLoS One. 2014;9:e91765. doi: 10.1371/journal.pone.0091765. Describes the discovery of non-nucleoside inhibitors of norovirus RdRp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87*.Rocha-Pereira J, Jochmans D, Dallmeier K, Leyssen P, Cunha R, Costa I, Nascimento MS, Neyts J. Inhibition of norovirus replication by the nucleoside analogue 2′-C-methylcytidine. Biochem Biophys Res Commun. 2012;427:796–800. doi: 10.1016/j.bbrc.2012.10.003. Describes first identifications of nucleoside analogues, inhbitors of norovirus RdRp, as antivirals against noroviruses. [DOI] [PubMed] [Google Scholar]
- 88.Chang KO, George DW. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J Virol. 2007;81:12111–12118. doi: 10.1128/JVI.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Costantini VP, Whitaker T, Barclay L, Lee D, McBrayer TR, Schinazi RF, Vinje J. Antiviral activity of nucleoside analogues against norovirus. Antivir Ther. 2012;17:981–991. doi: 10.3851/IMP2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin Z, Tucker K, Lin X, Kao CC, Shaw K, Tan H, Symons J, Behera I, Rajwanshi VK, Dyatkina N, Wang G, Beigelman L, Deval J. Biochemical evaluation of the inhibition properties of favipiravir and 2′-C-methyl-cytidine triphosphates against human and mouse norovirus RNA polymerases. Antimicrob Agents Chemother. 2015;59:7504–7516. doi: 10.1128/AAC.01391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eltahla AA, Lim KL, Eden JS, Kelly AG, Mackenzie JM, White PA. Nonnucleoside inhibitors of norovirus RNA polymerase: scaffolds for rational drug design. Antimicrob Agents Chemother. 2014;58:3115–3123. doi: 10.1128/AAC.02799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Changotra H, Jia Y, Moore TN, Liu G, Kahan SM, Sosnovtsev SV, Karst SM. Type I and type II interferons inhibit the translation of murine norovirus proteins. J Virol. 2009;83:5683–5692. doi: 10.1128/JVI.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93**.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. Detailed characterization of interferons as possible antiviral agents for norovirus infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94**.Vashist S, Urena L, Chaudhry Y, Goodfellow I. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J Virol. 2012;86:11977–11990. doi: 10.1128/JVI.00432-12. Identification of host cell factors in murine norovirus replication with possible implications as antiviral targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gonzalez-Hernandez MJ, Pal A, Gyan KE, Charbonneau ME, Showalter HD, Donato NJ, O’Riordan M, Wobus CE. Chemical derivatives of a small molecule deubiquitinase inhibitor have antiviral activity against several RNA viruses. PLoS One. 2014;9:e94491. doi: 10.1371/journal.pone.0094491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96**.Katayama K, Murakami K, Sharp TM, Guix S, Oka T, Takai-Todaka R, Nakanishi A, Crawford SE, Atmar RL, Estes MK. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc Natl Acad Sci U S A. 2014;111:4043–4052. doi: 10.1073/pnas.1415096111. This report describes a reverse genetics system for human noroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97**.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. This report describes the propagation of human noroviruses in a B lymphocyte cell line and the dependence of the presence of histo-blood group antigens for successful cultivation. [DOI] [PMC free article] [PubMed] [Google Scholar]