Abstract
Hierarchical processes spanning several orders of magnitude of both space and time underlie nearly all cancers. Multi-scale statistical, mathematical, and computational modeling methods are central to designing, implementing and assessing treatment strategies that account for these hierarchies. The basic science underlying these modeling efforts is maturing into a new discipline that is close to influencing and facilitating clinical successes. The purpose of this review is to capture the state-of-the-art as well as the key barriers to success for multi-scale modeling in clinical oncology. We begin with a summary of the long-envisioned promise of multi-scale modeling in clinical oncology, including the synthesis of disparate data types into models that reveal underlying mechanisms and allow for experimental testing of hypotheses. We then evaluate the mathematical techniques employed most widely and present several examples illustrating their application as well as the current gap between pre-clinical and clinical applications. We conclude with a discussion of what we view to be the key challenges and opportunities for multi-scale modeling in clinical oncology.
Key terms: cancer, mathematical modeling, predictive oncology, numerical modeling, computational modeling, agent-based modeling, cancer screening, epidemiology
Introduction
Cancer involves spatial scales ranging from RNA to gene networks to patients to entire populations, and mathematical modeling has provided valuable insights at each level. Successes include uncovering the impact of genetic regulatory circuits on spatial dynamics at cell and tissue levels1, identifying molecular targets for therapeutic interventions2,3, establishing tumor angiogenesis as a therapeutic target4–6, and uncovering the potential of cancer immunology7. Cancer also involves diverse temporal scales ranging from gene expression and receptor-ligand interactions (minutes to hours) to tumor growth and metastasis (months to years). Multi-scale modeling that transforms diagnosis and treatment by bridging these diverse spatio-temporal scales has long been pursued, and is now maturing to the point where it shows strong promise for solving critical problems in biology in general8, and clinical oncology in particular.
The review begins with an overview of the role that multi-scale modeling can play in oncology, before discussing several of the common mathematical techniques used to attack those problems. The overarching goal of these models is to make a prediction which can then be tested against experiment (either in silico, in vitro, or in vivo) which can lead to model improvement and, eventually, clinical application. We then examine the various areas that are currently barriers to success in applying the methods of multi-scale modeling to clinical oncology. This review is designed for members of the cancer biology and oncology communities who are interested in learning more about multi-scale modeling, as well as those in the modeling community who have recently become interested in using their skills to study cancer.
The role of multi-scale modeling in cancer
Dissecting the multiscale character of cancer to identify therapeutic targets
The long-envisioned promise of multi-scale modeling in clinical oncology revolves around quantification of the hierarchical disease process and the multi-scale feedback structures that enable genetic abnormalities to manifest at the levels of tissues, organs, and systems (see Figure 1 for an overview and Figure 2 for a specific example). A goal of multi-scale modeling is to identify how perturbations at each level can affect the system as a whole, and then to exploit this to attack cancers. Well-known genetic mutations can initiate oncogenesis as these genes translate to the scale of molecular processes, which in turn translate to cellular behavior. These molecular processes can be seen as functional consequences of genetic abnormalities, and involve changes in cell signaling, receptor status activation, genetic regulation, cellular movement and interactions with the extracellular milieu. A key role of multi-scale modeling at this level has been developing knowledge of how “normal” molecular control structures function and, by contrast, how genetic abnormalities produce dysfunction.
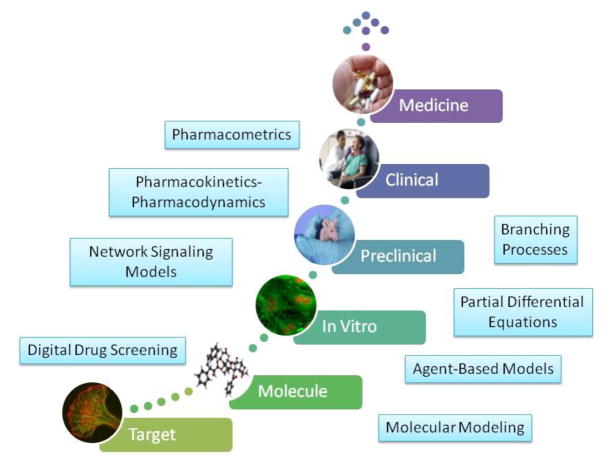
Figure 1.
The continuum of multi-scale components of cancer pharmacological therapy, and the role of each of the modeling techniques described in the text. The process starts with discovery and characterization of a target, followed by drug lead optimization and extensive in vitro and pre-clinical testing. A new medicine will also require successful testing in the clinical setting. Public domain image credits (bottom to top): NCI Center for Cancer Research (Luana Scheffer, Stephen Lockett, Jairaj Acharya); Wikimedia Commons; NCI Center for Cancer Research (Thomas Ried); National Cancer Institute (Leidos Biomedical Research, Inc.); National Cancer Institute (Rhoda Baer); photo courtesy photos-public-domain.com.
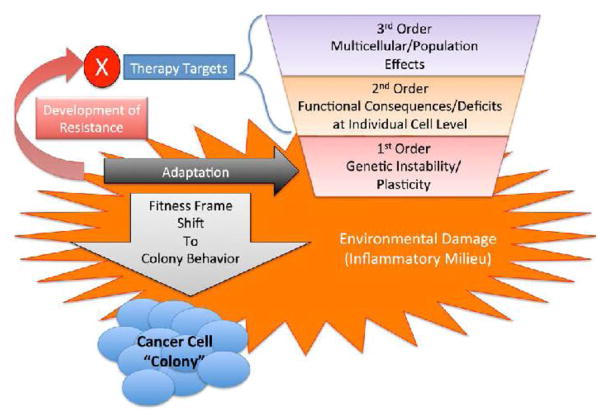
Figure 2.
Proposed Generative Hierarchies for Cancer and the Effect of Inflammation. A depiction of an example of the multi-scale effects of inflammation on the development and subsequent behavior of a tumor, incorporating evolutionary and selection effects across the scales from DNA to cellular populations. This paradigm posits that increased and accumulating genetic damage in an inflammatory milieu leads to a progressive loss of the cellular and molecular control structures that govern stable multi-cellular organization. The loss of these control structures in the tumor leads a more “colony-like” behavior, where the genetic plasticity of the increasingly disordered tumor cells provides a potential selection benefit when subjected to therapeutic interventions, akin to how microbial colonies utilize genetic heterogeneity as an adaptive strategy (as seen in antibiotic resistance). The incorporation of these concepts into a multi-scale computational model allows the exploration of various fundamental processes and behaviors involved in this hypothesis. Reprinted with permission from ref. 81.
At the cellular level, tumors are complex, non-clonal cell populations with their own internal dynamics arising from multi-scale interactions between mutated and normal cells exposed to an abnormal signaling environment. This manifests inappropriately in contextual cell-cell interaction, and is both a byproduct and further enhancer of intra-tumor heterogeneity9; cells housing mutations can “hijack” healthy neighbors into propagating a tumor. This in turn fosters selection within tumor cell populations, bringing evolutionary and ecological factors into the behavior of cancers, and defines the context by which tumors interact with their surrounding host tissue. Multiscale models hold the potential to reveal therapeutic targets arising from the ways that cells interact with their neighbors and with their physical environment during these processes.
At the tissue level, all tumors start to develop within normal tissue, and therefore have access to an “interaction space” at the border with the host that presents a potential area for “hijacking” normal processes and cellular populations. This interaction space not only directly affects the growth and selection within the tumor, but also selects for tumor processes best able to release cells into the blood stream and initiate the process of metastasis. The modeling of interactions across the cell, tumor and tissue hierarchy therefore holds the potential for unlocking additional therapeutic targets.
The progression of a tumor from a local to a system-level phenomenon represents another key point in the control of cancer, and motivates a desire to individualize the representation of tumor characteristics to better determine the specific factors involved in a particular tumor’s progression. Personalized tumor modeling might integrate genetic profiling of the primary tumor cells or tumor stem cells with the set of possible behavioral trajectories in a multi-cellular tumor that is actively interacting with its host. These latter characteristics might be represented by tumor-level properties determined by different modalities, such as histology or imaging, which would provide calibration targets for the lower-level mechanisms incorporated into such a model, and then be used to project multiple possible outcomes based on other personal factors, such as health status or interventions.
Finally, at the patient population level, the ability to potentially “simulate” an individual tumor provides a pathway to the generation of computer-generated populations of cancer patients. These simulated populations would provide a more sophisticated accounting for mechanisms than traditional population modeling. They would be key to represent and explain the observed “rarity” of cancer events, thereby accounting for stochastic processes involved in mutational events and allowing the generation of finer grained data sets for identification of subtler patterns in the pre-cancerous and early stage conditions. Ultimately, these models would form the basis of in silico clinical trials for potential therapeutic regimens, and provide another potential pathway for the design and development of cancer therapeutics.
Characterizing drug targets
Molecular targets that are cancer drivers are ultimately part of a mechanistic cascade10. Antitumor effects can be caused by many pharmacological interventions, both direct (e.g., kinase inhibition11) or indirect (e.g., immune-mediated therapy12). Given the broad landscape of potential pharmacological agents, modeling and simulation has a fundamental role in facilitating the investigation of potential targets. Systems pharmacology13 is an emerging and powerful tool in the quantitative modeler’s toolbox for guiding the early stages of discovery14, especially when tool compounds are unavailable and information is sparse about target properties such as abundance in target tissues and turnover15. Pharmacokinetic-pharmacodynamic (PK-PD) models incorporate compartmental16, or physiologically-based17, models of drug distribution and empirical or semi-mechanistic models of drug action18. They are most suited for investigating the effects of drugs on molecular targets when tool molecules are available to probe disease pathways. At the other end of the scale, pharmacometric19 models, which incorporate statistical and mechanistic features of the patient population being studied, can be used to quantify the effects of a particular treatment on populations. The statistical technique of mixed effects modeling can be applied to find explanatory variables (covariates) and, eventually, correlates of clinically significant endpoints such as overall or progression-free survival20. All of these modeling approaches ultimately characterize drug targets across the spectrum21 of target qualification (cell and tissue), pharmacology (nonclinical models and humans) and disease effect (populations).
Adverse side-effects and lack of efficacy are the two major sources of attrition in the field of drug design22. Substantial efforts have been devoted to addressing this challenge, and modeling approaches have been playing increasingly important roles in addressing the lack of efficacy and undesired off-targets effects22,23. Recent advances in structural bioinformatics have enabled the reliable prediction of drug off-target binding sites across the proteome24. Large-scale network models have also been widely applied to predict the functional effects of various therapeutics22. These two approaches have been integrated to provide a framework for assessing drug responses in silico25. A recent effort incorporated signaling pathway information for evaluating side effects on primary human hepatocytes, and obtained insightful results with clinical implications26. Collectively, the traditional experiment-based screening strategy to reduce drug off-target effects is becoming time- and resource-consuming, while a variety of recently developed computational models have begun to make significant contributions to rational drug design. In addition, many computational tools based on ordinary differential equations (ODEs) systems have been widely used to model and predict the effects of therapeutics on intracellular signaling pathways27–30. These model tools were developed based on protein-protein interaction networks with applications on exploring optimal therapeutic strategies. Collectively, ODE-based models with perturbations can be used to explore in silico candidate conditions, screen out critical factors, and guide biological experiments, by investigating drug combination effects with well-known evaluation indexes such as Loewe additivity31 and Bliss independence32. Finally, agent-based modeling techniques can be used to integrate multiple biological scales together, especially including intracellular signaling pathways33–35.
Computing the design of anticancer drugs
It is increasingly clear that there must be an extension from the “rational discovery” of potential drug candidates, often based on molecular-level assumptions of effect, to a “rational design” process, that moves beyond target identification towards characterizing the larger scale consequences of interfering with a particular target gene. This necessarily incorporates recognition of the multi-scale nature of cancer, where there are higher-order properties that involve accounting for the behavior of multi-cellular populations within a tumor, as well as the interactions of that tumor with its host environment. Given this understanding, in any attempt to recognize the potential downstream consequences of a molecular level intervention (as is the case with many anti-cancer drugs), it is critical to account for compensatory processes that remain in either the tumor or adjacent host tissue. Quantitative models that can contextualize the multi-scale processes involved in the development and behavior of cancer have an important role to play in this line of investigation36.
Digital screening of anticancer drugs
Traditional drug discovery relies upon high-throughput screening using a library that contains millions of compounds selected for and then screened for efficacy against a target of interest. While this approach has been successfully used to discover many effective anticancer drugs, it can be enhanced through digital drug screening, a powerful drug discovery technology in the post genomic era. In addition to advances in chemoinformatics37 and the deciphering of the human genome, there has been an enormous increase in the types of chemical compounds, biological and physiological systems, and diseases that have been digitized, stored and archived in publically accessible databases, such as PubChem, ChemSpider and ChEBI38. These databases offer a platform for releasing and publishing experimental data on chemical compounds and their associated structure and functional data. They also offer user-driven search engines that allow users to define and search a particular drug molecule or a class of chemical compounds37.
Conversely, while large sets of biological and medical data are frequently generated, the validation and further standardization of these data remains a significant challenge. One of the difficulties is the poor reproducibility and reliability of these biological and clinical data due to the intrinsic complexity of biological systems and hard-to-access human samples. The development of bioartificial tissue and organ-on-a-chip39 systems could help accumulate more clinically relevant biological and physiological data for digital screening of anticancer drugs. The digitalization of cancer diagnoses offers another opportunity to deposit clinical oncology data into publically available databases. Some of these databases (e.g., Therapeutic Targets Database40 and PharmGKB41) are available today for drug screening.
Another critical element of digital drug screening is provided by computational models that link the data to a specific cancer target. Such models must be multi-scale and target-driven due to the multiple spatial and temporal scales at which the motivating biological and clinical data are collected. Algorithms also need to be developed to predict whether compounds (including proteins or peptides) can be translated into anticancer drugs. For example, BioMap® (human primary cell phenotypic profiling services developed by DiscoveRx Co.) consists of primary human cell-based assay systems, a database of reference compound profiles, and computational data mining and analysis tools. A system (ChemScore) that uses reactivity based fingerprints of compounds as filters has been developed to determine a reactant-like and a product-like score for virtual drug screening42.
Taken together, the combination of available databases and computational models offers unprecedented opportunities to build computational models for drug screening, thereby enabling a fundamental shift from traditional high-throughput screening to data-driven screening of potential anticancer drugs.
Optimizing dosage, drug combinatorics, scheduling, and safety
The classical “maximal tolerated dose” (MTD) approach currently used in early Phase 1 clinical trials of anticancer drugs can be irrelevant in many situations43. For example, some compounds simply do not present a toxicity profile appropriate for reaching the MTD in a dose finding phase. For other compounds with a well-identified MTD, the upper-limit dose may not be appropriate when the compound is given in combination with other therapeutics44. Efficacy and toxicity are central issues in design of patient-specific chemotherapy regimens, but do not lend themselves well to trial-and-error approaches. A broad range of coupled in vitro and numerical modeling techniques are becoming available that offer much promise for rapid efficacy and toxicity screening45,46. Cellular and tumor-level responses may be deduced from the responses of bioartificial tissues, organ-on-a-chip systems, and murine systems in typical screening procedures. These systems also show promise for assessing toxicity of drug regimens. A particularly promising avenue is screening of compounds on bioartificial tissues whose cells are derived from a patient’s own induced pluripotent stem cells47.
A critical challenge in the development of anti-cancer drugs is the optimization of the delivery strategy including the proper dosing, timing, and scheduling of drug administration48. Optimally selecting treatments for a particular cancer subtype, particularly treatments that involve combination therapy, is an extraordinary challenge, for the number of potentially relevant adjustable parameters is too large to adequately investigate in clinical trials. Multi-scale simulation can play a pivotal role in addressing this problem by anticipating potential synergies between compounds’ mechanisms of action, thus providing a rational method for selecting a dose that would improve efficacy without affecting safety. Progress has already been made using bioinformatics algorithms49; however, applications of multi-scale modeling to solving this very important clinical problem remain to be fully explored.
Assessing intervention, prevention, and cancer screening strategies
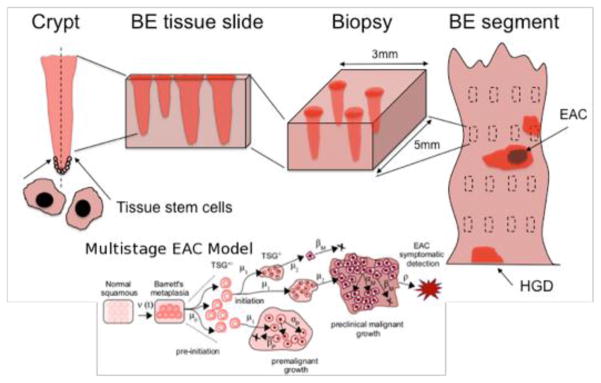
Multi-scale frameworks that explicitly model clinical outcomes in terms of underlying biological processes, in conjunction with the physical and physiological characteristics of the instrumentation used for screening or drug delivery, are likely to suggest improvements that cannot be gleaned from traditional natural history models of cancer development and drug response. Such traditional models typically ignore important biological processes and time scales in the formation of cancer and its precursors. In contrast, multi-scale models are appropriate for providing a more comprehensive understanding of the underlying mechanistic processes, as well as the spatio-temporal characteristics of cancer screening and surveillance protocols (e.g., using high-resolution imaging, or biopsies). An illustrative example of this reasoning is presented in Figure 350. Although regular biopsy-based surveillance is the standard-of-care for most Barrett’s Esophagus (BE) patients who have not progressed to dysplasia or cancer, it is not clear whether screening under current guidelines is clinically optimal and cost-effective. Curtius et al. describe a computational cell-level multiscale model for the neoplastic progression of Barrett’s metaplasia to esophageal adenocarcinoma allowing for variation in segment length, presence of dysplastic cells in the crypt-structured epithelium and their potential detection by biopsy50. Thus, multi-scale-based screening models can potentially be used to better understand the clinical performance (sensitivity/specificity) of various screening methods and the sources that limit their clinical utility.
Figure 3.
The Multi-scale nature of screening in Barrett’s esophagus (BE). The standard screening protocol for BE involves scales from stem cells in the crypt (left) to the BE cylindrical segment of the esophagus depicted (right) with rectangles representing biopsy samples taken during endoscopy. The BE segment may have dysplasia and/or malignant tissue patches that may be missed. During histological preparation, portions of each biopsy are sliced by microtome and placed on slides for pathologic assessment. A diagnosis is made by microscopic interpretation of crypt and cellular architecture, reflecting the most severe tissue grade found from all slides. Given the multi-scale nature of the problem, it is natural that a multi-scale-based screening model would have great clinical utility. (EAC = Esophageal Adenocarcinoma; HGD = High Grade Dysplsia.)
State-of-the-art multi-scale approaches to cancer
Multi-scale network signaling models
Advances in mechanistic modeling of signaling networks present opportunities for better understanding of therapeutic targets, designing therapeutic regimens (including combination therapies), as well as the de novo design of drugs. Models of growth factor signaling networks, such as vascular endothelial growth factor (VEGF) and epidermal growth factor (EGF), have been particularly well developed. The VEGF models describe molecular-detailed kinetic interactions between different splice isoforms of VEGF and their cognate receptors VEGFR1 and VEGFR2 and co-receptors neuropilins-1 and -2. The models comprise the blood, normal, and tumor compartments and they also take into account VEGF binding to the extracellular matrix, and soluble factors such as soluble VEGFR151,52. Molecular-detailed intracellular signaling models that include receptor dimerization, internalization, recycling, and degradation can potentially be used for simulating intracellular drug targeting with, for example tyrosine kinase inhibitors53. The kinetic receptor-ligand interaction model has been extended to describe the pharmacokinetics and pharmacodynamics (PK/PD) of VEGF-neutralizing anti-angiogenic drugs including the antibody bevacizumab52, and aflibercept51, a fusion of specific domains of VEGFR1 and VEGFR2. They were compared with extensive available clinical data. The difference between these molecular-detailed PK/PD models and more conventional models is in the mechanistic level of detail with which the molecular interactions are represented. Therefore, the model predicts the amounts not only of the drug in the compartments, but also the detailed distribution of the different ligands (e.g., those bound to the cell-surface receptors, extracellular matrix, and free in the interstitium)4. For the EGF system a molecular-detailed approach has been used to design and optimize an antibody for cancer applications54,55.
Pharmacometrics and nonlinear mixed effects models
A popular definition of pharmacometrics is “the science of developing and applying mathematical and statistical methods to: a) characterize, understand, and predict a drug’s PK and PD behavior, b) quantify uncertainty of information about that behavior, and c) rationalize data-driven decision making in the drug development process and pharmacotherapy”56. In practice, pharmacometrics often employs nonlinear mixed effects modeling techniques57 that combine structural models (algebraic or differential equations) that are nonlinear in their parameters with nested variability in the clinical observations (i.e., variation among and within patients) and trial execution components (i.e., patient adherence and dropout rate). There have been many applications of nonlinear mixed effects models to clinical oncology PK/PD58,59. In general, nonlinear mixed effects modeling of PK and PD benefited from the early availability of computer software60,61 and frequent application to situations where other techniques would have been difficult to deploy, such as clinical studies. Since this class of models is informed by data, mechanistic detail is a function of available information, which can be limited in clinical oncology. However useful, nonlinear mixed effects are not the only tool that can be used to understand dose-exposure-response relationships in vivo. It is through the combination and use of multiple, “fit for purpose” modeling approaches, depending on the appropriate biological scale, that we can hope to understand cancer etiology and pharmacotherapy36.
Partial differential equations
For handling clinical data that have spatial dimensions as independent variables (i.e., data that do not depend solely on time), mathematical models based on partial differential equations (PDEs) may be more appropriate than those based on ordinary differential equations (ODEs), which are more amenable to the applications described in the previous two sections. For example, medical images which are composed of rectangular, spatially-resolved voxels describing the shape, location, and texture of the tumor in addition to underlying physiological, cellular, and molecular processes62, cannot be readily handled by ODE models. More generally, PDE models are appropriate when the available data is multidimensional63. Most spatial models describe the movement of cancer cells through reaction-diffusion64 or advection65 terms. The applications are numerous, ranging from monitoring the evolution of slowly evolving tumors such as lung metastases66 to defining surgery or irradiation margins67,68, and improving the insight offered by images69. (See Figure 4). PDE models have also been used to evaluate the spatio-temporal distribution of metastases over time70, as well as mutations and resistance to treatment71. However, most applications require being able to recover the parameters of the model from clinical data in order to perform patient-specific simulations or predict patient-specific outcome.
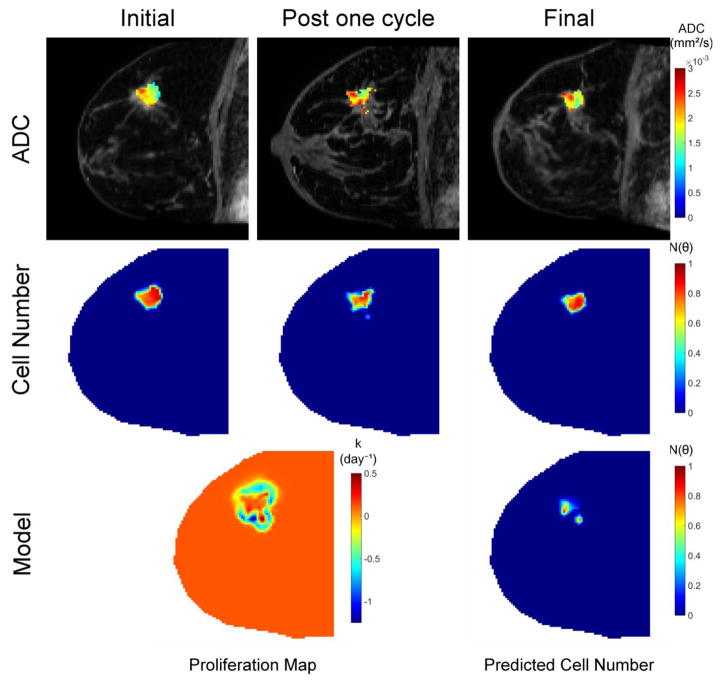
Figure 4.
This illustrative example uses serial diffusion weighted magnetic resonance imaging data to estimate tumor cellularity before and after the first cycle of neoadjuvant therapy. The top row indicates the apparent diffusion coefficient (ADC) obtained from diffusion weighted MRI data at three time points during therapy. Given the known relationship between ADC values and cellularity, this data is then converted to tumor cell number (middle row). The cell number data from the initial and post one cycle time points are then fit to a biomechanical model of tumor growth to estimate patient-specific parameters of tumor cell proliferation and migration. The model, calibrated with the patient-specific parameters determined from the fitting procedure, is then run forward in time to predict residual tumor burden at the conclusion of neoadjuvant therapy. Model predicted cell number can be compared to cell number imaging observations at the final time point in order to assess predictive performance. Details are presented in ref. 69.
It is important to note that PDE models are (relatively) computationally expensive to solve. Classical optimization techniques can often be too expensive to be realistically used. More advanced techniques relying on reduced-order models72 or exploiting the features of the solution to a model73, may prove more successful for assimilating clinical data.
Spatially discrete/cellular agent models
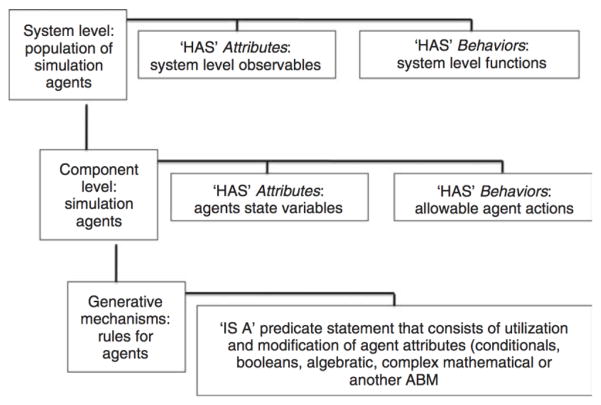
One class of biological computational modeling that has recently increased in popularity is that of spatially discrete, cell-as-agent models. Methods in this category are agent-based models (ABMs), individual based models (IBMs), cellular automata, and cellular Potts Models. For simplicity’s sake, these methods will be globally described as agent-based modeling, and can be generally described as discrete event, object oriented, rule based, and often spatially explicit methods for dynamic computer modeling that represent systems as a series of interacting components74. ABMs are programs that generate populations of discrete computational objects (or agents) that correspond to the component-level at which the reference system is being examined. These computational agents are organized into agent classes representing groupings of agents of a similar type defined by shared properties and characteristics. Agents are governed by agent rules; i.e., instructions that allow the agent to be treated as an input-output object. Individual agents incorporate the properties and rule-structures of their parent agent class, but are able to manifest diverging behavior based on local inputs. ABMs intrinsically cross scales of biological organization, utilizing behavioral rules (scale 1) to determine individual agent behavior (scale 2), and then aggregating individuals into population dynamics of the global system (scale 3). When applied to biological systems, cells form a natural agent level within this organizational structure. Subcellular components (e.g., genes, enzymes, receptors, and structural elements) are represented as state variables for the cellular agents. Their behavior and interactions (e.g., gene transcription, intracellular signaling, protein synthesis) can be represented by a wide range of mathematical and computational formulations. Individual cellular agents interact with each other by manipulating state variables of their neighbors or their shared environment. Figure 5 depicts an ontological description of an agent-based model, with an emphasis on its generality that can be tailored to specific modeling tasks. Given this framework, ABMs have been extensively used to study tumor growth and behavior74–77.
Figure 5.
A schematic of an Agent-based modeling format (ABM). The structure of an ABM intrinsically incorporates at least three representational scales of the system being modeled. For most biomedical ABMs, cells are used for the middle level representation (the agent level). Figure reprinted with permission from Elsevier Ltd.
Multi-scale, agent based modeling has been used to generate high-fidelity replications of tumor structure. The spatial representation of ABMs allows them to generate “realistic” tumor structures incorporating multiple cell types and capturing cancer heterogeneity78. This permits a detailed investigation of the multi-scale consequences of genetic or molecular perturbations, and offers the promise of potentially personalizing models based on histological features79. ABMs also allow examination of fundamental processes involved in oncogenesis by facilitating a parsimonious approach that provides insight into fundamental processes involved in tumor growth and development80. ABMs can also provide linkages to the role of general biological processes, like inflammation, in the development and progression of cancer81.
As with all modeling methods, agent-based modeling is not without its limitations. Most of these stem from the fact that ABMs do not have a common formal description, which limits the ability to subject them to formal analysis. Their “similarity” to biological systems, particularly in terms of the heterogeneity and non-linearities in their behavior, makes formal, comprehensive exploration of their parameter space difficult and only able to be accomplished using very large sets of simulations. Additionally, ABMs are relatively computationally expensive and difficult to distribute across modern, distributed computing architectures, with the result that very often biomedical ABMs are treated more akin to experimental objects where their use is dependent upon finding some subset of parameters that can provide “realistic” behavior. Despite this limitation, however, ABMs can serve a very useful purpose that can bridge between biological objects/knowledge and more formal mathematical representations.
Branching processes
Disruption of normal cell proliferation and differentiation is the sine qua non of the malignant state. However, numerous experimental and clinical studies provide evidence that the proliferation and differentiation kinetics in normal and premalignant cells are also of critical importance in the carcinogenic process. This notion was further enforced by analytic findings from mathematical modeling of cancer incidence patterns82. A prototype branching process model of cancer is the two-stage clonal expansion (TSCE) model83. Initially, this model was formulated with stochastic clonal expansions of both normal and intermediate (or premalignant) stem cells. However, due to the typically very large (and highly regulated) size of the normal tissue stem cell pool, the version most frequently used assumes a deterministic number of normal tissue stem cells. The basic TSCE model is characterized by two rate-limiting events in normal tissue stem cells (premalignant tumor initiation and malignant transformation) together with a stochastic growth process of premalignant cells that can undergo malignant transformation. Various extensions of this model have been put forward (multistage clonal expansion (MSCE) models) to better explain cancer incidence patterns in registries and cohort studies84. The availability of analytical tools and likelihood expressions for population-level clinical observations (e.g., cancer incidence, prevalence of a precursor such as colonic adenoma or dysplasia in Barrett’s esophagus patients) greatly facilitates likelihood-based parameter estimation via gradient methods or Markov-Chain Monte Carlo techniques. An example of a ‘mathematical bridge’ that connects the cellular-level with the tissue-level is the filtered Poisson process.
Homogenization approaches
Linear homogenization approaches comprise the simplest techniques for estimating parameters describing cell health and function from measurements conducted on a tissue construct. From the perspective of screening for safety, the desired outcomes are parameters describing electrophysiological and mechanical functioning of individual cells. From the perspective of screening for efficacy, cancer culture models such as the Xu model85 exist, and the challenge is determining how chemotherapy agents affect the tumor periphery and factors promoting malignancy. Models estimating tissue and cellular mechanics from multiple loadings of tumor models are capable of estimating these changes86, and for estimating effects on cells, protein structures, and networks87. Although techniques are preclinical at present, the capacity to test chemotherapy regimens on both heart tissue equivalents from a patient’s own transdifferentiated cells and tumor equivalents from a patient’s own tumor cells shows much promise. Ongoing challenges relate to refinement of electrophysiological and mechanical models to account for local variations within tissues, and to include nonlinear phenomena.
Hybrid models
Multi-scale hybrid models combine different modeling methodologies; e.g., intracellular signaling described by ODEs combined with 3D distributions of oxygen, growth factors and cells described by PDEs, or oxygen and growth factor distributions described by PDEs combined with discrete cell dynamics represented by agent-based modeling. In principle, hybrid models could include all types of models. The advantage of hybrid modeling is that different parts of the system can be described using the methodologies most appropriate for the biological question to be answered, and with a spatial and temporal resolution that makes the problem tractable.
Anderson presented a hybrid model describing the invasion of a solid tumor into healthy tissue that is governed by tumor cells, extracellular matrix, matrix-degrading enzymes and oxygen88. In his formulation, the tumor cells themselves are considered as discrete objects, while the remaining three entities are considered continuous variables. His results indicated that cell–cell interactions drive the early stages of development, but it is the loss of cell–cell interaction (due to mutation) that increases the importance of cell-matrix interactions which drive tumor invasion. Many have built on Anderson’s approach. For example, Jiang et al have extended Anderson’s approach to systematically investigate tumor cell invasion within a generalized diffusion framework89. The authors found that tumor cells can migrate by a host of diffusion modes and these predictions were supported by patterns seen both in vitro and in standard-of-care images obtained in the clinical setting. In particular, they found that in the case of invasion and metastases, tumor cells display both superdiffusion and ballistic diffusion.
Barriers to and opportunities for progress
Technical/methodological issues
Mathematical complexity
As seen in fields ranging from atomistics to astrophysics, a tradeoff exists between mathematical and computational complexity in modeling oncological problems, especially when the problem involves a large number of interacting components (cell-types), non-linear signaling between components (feedback loops), and stochastic behavior (noise). A main challenge in developing useful multi-scale models is therefore the choice of mathematical abstraction (continuum, discrete, lattice) and choice of relevant (rate-limiting) processes, which may operate at different time and length scales. However, these scales may not be known a priori and the appropriate choice may require preliminary studies and/or additional bio-mechanistic information. A case in point is the problem of emerging resistance to therapy in heterogeneous tumors, specifically whether or not the ‘resistance conferring’ alterations are preexisting, a result of the tumor and its microenviroment being under selection pressure caused by the drug, or simply due to the hypermutability or genomic instability of the tumor90. Each of these causes requires a distinct mathematical description. For example, the size fluctuations of preexisting mutants may well be captured by a Luria-Delbrück type of distribution, which allows for mutational jackpots, while a drug-induced response is unlikely to do so given the much shorter time scale of treatment91–92.
Mathematical complexity in oncologic applications of multi-scale modeling arises in the formulation of the dynamics of bulk behavior and description of underlying constituent processes in the forms described above93. It is important to note that only in exceptional cases (and often only with many simplifying assumptions) are closed form solutions available and parameter identifiability issues addressed. Further adding to the mathematical complexity is the stochastic nature of many cell-level and sub-cellular processes94. Although the implementation and mathematical treatment of the stochastic process may be complex, a considerable advantage is that it often lends itself to likelihood-based methods for parameter estimation and hypothesis testing.
Linking models with clinically available data
It is important to acknowledge that while the multi-scale cancer modeling community is rich in models from the nanoscale to the macroscale, many models are framed in terms of parameters and variables that are extraordinarily difficult to obtain in the clinical setting. This a fundamental challenge facing the validation and clinical application of multi-scale modeling. This is particularly true for cases in which electrophysiological, transport, and mechanical factors of cells and a pericellular region are of interest. For the case of the screening of anti-cancer agents, the key difficulties are measuring in the mesoscale range, characterizing the pericellular region, and sampling a sufficient number of cells to overcome the high cell-to-cell variability so inherent to three dimensional culture. These challenges result in modeling approaches that require many (often heuristic) assumptions on model parameters. Consequently, application of such models to make clinically relevant predictions can be quite limited. More specifically, the field of multi-scale modeling in cancer has largely been developed independent of the data types that are typically available in the clinical setting. The community needs to acknowledge that it is not simply enough to test a myriad of modeling approaches in silico by systematically varying parameters, coupling constants, etc. Rather, to be of clinical relevance, the community needs to build multi-scale models that can be initialized and constrained with patient specific data that is readily available in the clinical setting95. Only by proceeding along this route will we be able to test hypotheses about patients that directly testable. One area that is underexplored in making progress is the utility of medical imaging data67,69,96.
The medical imaging technologies of magnetic resonance imaging, x-ray computed tomography, positron emission tomography, single-photon emission computed tomography, and ultrasound can quantify, at multiple time points and in 3D, tumor characteristics at the physiological, cellular, and molecular levels62. Furthermore, the images themselves present a natural gridding (i.e., the image pixel or voxel) that enables direct application of finite difference and finite element methods. While using such data in statistical and informatics driven approaches has launched the fields of radioomics and radiogenomics97,98, such data are only beginning to be incorporated into mechanism-based, predictive models of tumor initiation, growth, invasion, and response to treatment.
Mathematical models of tissue health and function
There is a pressing need for multi-scale modeling techniques that enable the use of in vitro tissue surrogates and organ-on-a-chip models for safety and efficacy screening of chemotherapy agents. Although well-established methods exist for applying integrated multi-scale modeling and experiment to assess subtle, drug-induced changes to the health and function of cells within simple bioartificial tissue constructs46, advances are required to account for such tissues and organ-on-a-chip systems with heterogeneous cell populations. With these models in place, the potential to, for example, test a library of anti-cancer agents on bioartificial tissues mimicking a patient’s own tissues may one day become possible, enabling rapid optimization for both efficacy against a patient’s malignant cells and for tolerance by a patient’s healthy cells through quantitative evaluation of changes to cell function.
Institutional issues
Intrinsic to trans-disciplinary endeavors, from medicine to management, are the challenges of reconciling both different views of the world, and the different rhetorical frameworks that are used to reflect those views99. Cancer modeling, in its earliest days, involved little communication between modelers and clinicians100. Although efforts to reconcile pathophysiology with principles from the physical sciences have progressed a great deal since the 19th century courtroom battles over the topic of model validation101, many cancers remain too complex for identification of abstractions that can approximate natural laws, and validation remains a central challenge102. However, dynamic computational models representing sets of mechanistic hypotheses can stand in for formal mathematical theories for a defined context and for a constrained use103. Within this context the importance of the choice of computational methods is lessened; sufficiently strong hypothesis structures should perform equally well when instantiated in a multiplicity of modeling methods. This is the principle of cross platform validation103.
High quality, validated implementations are crucial to establishing the trustworthiness of a particular computational model, and accounting for the relative strengths, weaknesses, and representational capabilities of the different methods is essential. There also needs to be some means by which the appropriate use-context of a particular model is explicitly defined and determined, in order to avoid the misapplication and misinterpretation of a particular model. Dating back to the earliest multiphysics codes and standards (e.g., the American Society of Mechanical Engineering boiler codes104), these issues have been common to the general use of models and simulations, and have been pragmatically addressed through the establishment of guidelines and standards for model credibility, testing, and reporting in a domain specific fashion. However, for multiscale modeling of cancer, this process is in its infancy. Notable efforts to advance this include, for example, the Committee on Credible Practice in Modeling and Simulation in Healthcare105, and the EFPIA MID3 Workgroup which recently published a white paper on good practices in model-informed drug discovery and development106. We view such collaborative efforts as important, as the field continues to advance towards systematically using model-aided design and testing in the regulatory arena (for testing of drugs and devices) and for personalized/precision decision support.
Conclusion
No shortage of powerful—and promising—computational techniques exists for use in the multi-scale modeling of cancer and cancer treatments. What is lacking, however, is access to relevant clinical data and practical modeling approaches to incorporating such data into relevant models. This will allow modelers, experimentalists, and oncologists to effectively close the “build-test-refine cycle” by directly testing the predictive power of a particular model and then improving upon it to the point where it can, ultimately, be applied to clinical problems. We also need a more rigorous understanding of the key components that go into the growth and response to treatment of individual tumor subtypes. Both of these issues are exacerbated by the social constructs in academia. In practice, future progress in clinically relevant multi-scale modeling in oncology requires interdisciplinary collaborations between clinicians, experimentalists (biologists, physiologists, biophysicists, bioengineers, etc.), and mathematical and computational modelers. Although this truth is clearly recognized at the level of the funding agencies, the difficulty in developing such collaborations is generally underappreciated. However, a corollary of successfully working across multiple disciplines is the education and training of a future generation of students and postdocs who are accomplished at both the bench and computer, thereby allowing them to explore and integrate their data into clinically useful models. We believe this is happening as we speak, and that such integration of disciplines through multi-scale modeling will bring great value to clinical oncology.
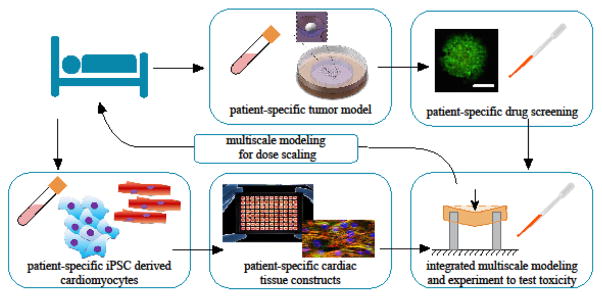
Figure 6.
Patient-specific drug and cardiotoxicity systems are available, but require integrated multi-scale modeling and experiment to apply. Tumor cells can be multiplied in systems like the Xu system (ref. 85) for patient-specific drug screening. Commercial systems exist for testing patient-specific cardiotoxicity on tissue constructs derived from induced pluripotent stem cells (iPSCs); for example. Multi-scale models are required to derive metrics of cellular health from measurements of the mechanical function of tissue constructs, and for scaling dosages. Image credits: Top center and top right: ref. 71; bottom left and bottom center: Invivosciences, LLC. All images used with permission.
Acknowledgments
The authors thank the NIH for funding through U01CA174706 (TEY), 1R01-GM-115839 and P30-DK-42086 (GA), 1U01CA166886 (XZ), French National Research Agency ANR-10-IDEX-03-02 (OS), R01CA138264 (ASP), U01CA152926 (EGL), U01EB016422 (GMG), and CPRIT RR160005 (T.E.Y.). T.E.Y. is a CPRIT Scholar in Cancer Research.
References
- 1.Rubinacci S, Graudenzi A, Caravagna G, Mauri G, Osborne J, Pitt-Francis J, Antoniotti M. CoGNaC: A Chaste Plugin for the Multiscale Simulation of Gene Regulatory Networks Driving the Spatial Dynamics of Tissues and Cancer. Cancer Inform. 2015;14(Suppl 4):53–65. doi: 10.4137/CIN.S19965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y, Powathil G, Kang H, Trucu D, Kim H, Lawler S, Chaplain MM. Strategies of eradicating glioma cells: a multi-scale mathematical model with MiR-451-AMPK-mTOR control. PLoS One. 2015;10:e0114370. doi: 10.1371/journal.pone.0114370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson-Tessi M, Gillies RJ, Gatenby RA, Anderson AR. Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res. 2015;75:1567–79. doi: 10.1158/0008-5472.CAN-14-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finley SD, Chu LH, Popel AS. Computational systems biology approaches to anti-angiogenic cancer therapeutics. Drug Discov Today. 2015;20:187–97. doi: 10.1016/j.drudis.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qutub AA, Mac Gabhann F, Karagiannis ED, Vempati P, Popel AS. Multi-scale models of angiogenesis. IEEE Eng Med Biol Mag. 2009;28:14–31. doi: 10.1109/MEMB.2009.931791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharan S, Woo S. Systems pharmacology approaches for optimization of antiangiogenic therapies: challenges and opportunities. Front Pharmacol. 2015;6:33. doi: 10.3389/fphar.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cappuccio A, Tieri P, Castiglione F. Multi-scale modelling in immunology: a review. Brief Bioinform. 2015 doi: 10.1093/bib/bbv012. [DOI] [PubMed] [Google Scholar]
- 8.Garny A, Cooper J, Hunter PJ. Toward a VPH/Physiome ToolKit. Wiley Interdiscip Rev Syst Biol Med. 2010;2(2):134–47. doi: 10.1002/wsbm.63. [DOI] [PubMed] [Google Scholar]
- 9.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Gross S, Rahal R, Stransky N, Lengauer C, Hoeflich KP. Targeting cancer with kinase inhibitors. J Clin Invest. 2015;125:1780–9. doi: 10.1172/JCI76094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavallo F, De Giovanni C, Nanni P, Forni G, Lollini PL. The immune hallmarks of cancer. Cancer Immunol Immunother. 2011;60:319–26. doi: 10.1007/s00262-010-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorger PK, Allerheiligen SRB, Abernethy DR, Altman RB, Brouwer KLR, Califano A, D’Argenio DZ, Iyengar R, Jusko WJ, Lalonde R, Lauffenburger DA, Shoichet B, Stevens JL, Subramaniam S, Van der Graaf P, Vicini P, War R. An NIH white paper by the QSP workshop group. Bethesda: NIH; 2011. Quantitative and systems pharmacology in the post-genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms. http://www.nigms.nih.gov/News/reports/Documents/SystemsPharmaWPSorger2011.pdf. [Google Scholar]
- 14.Kirouac DC, Du JY, Lahdenranta J, Overland R, Yarar D, Paragas V, Pace E, McDonagh CF, Nielsen UB, Onsum MD. Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Sci Signal. 2013;6:ra68. doi: 10.1126/scisignal.2004008. [DOI] [PubMed] [Google Scholar]
- 15.Visser SA, de Alwis DP, Kerbusch T, Stone JA, Allerheiligen SR. Implementation of quantitative and systems pharmacology in large pharma. CPT Pharmacometrics Syst Pharmacol. 2014;3:e142. doi: 10.1038/psp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang GR, Harris RZ, Lau DT. Pharmacokinetics and its role in small molecule drug discovery research. Med Res Rev. 2001;21:382–96. doi: 10.1002/med.1015. [DOI] [PubMed] [Google Scholar]
- 17.Jones HM, Chen Y, Gibson C, Heimbach T, Parrott N, Peters SA, Snoeys J, Upreti VV, Zheng M, Hall SD. Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective. Clin Pharmacol Ther. 2015;97:247–62. doi: 10.1002/cpt.37. [DOI] [PubMed] [Google Scholar]
- 18.Mager DE, Jusko WJ. Development of translational pharmacokinetic-pharmacodynamic models. Clin Pharmacol Ther. 2008;83:909–12. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JS, Fossler MJ, Cadieu KD, Gastonguay MR. Pharmacometrics: a multidisciplinary field to facilitate critical thinking in drug development and translational research settings. J Clin Pharmacol. 2008;48:632–49. doi: 10.1177/0091270008315318. [DOI] [PubMed] [Google Scholar]
- 20.Claret L, Bruno R. Assessment of tumor growth inhibition metrics to predict overall survival. Clin Pharmacol Ther. 2014;96(2):135–7. doi: 10.1038/clpt.2014.112. [DOI] [PubMed] [Google Scholar]
- 21.Lalonde RL, Kowalski KG, Hutmacher MM, Ewy W, Nichols DJ, Milligan PA, Corrigan BW, Lockwood PA, Marshall SA, Benincosa LJ, Tensfeldt TG, Parivar K, Amantea M, Glue P, Koide H, Miller R. Model-based drug development. Clin Pharmacol Ther. 2007;82(1):21–32. doi: 10.1038/sj.clpt.6100235. [DOI] [PubMed] [Google Scholar]
- 22.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 23.Tatonetti NP, Liu TY, Altman RB. Predicting drug side-effects by chemical systems biology. Genome Biology. 2009;10(9) doi: 10.1186/gb-2009-10-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie L, Li J, Bourne PE. Drug Discovery Using Chemical Systems Biology: Identification of the Protein-Ligand Binding Network to Explain the Side Effects of CETP Inhibitors. Plos Computational Biology. 2009;5(5):e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang RL, Xie L, Bourne PE, Paisson BO. Drug Off-Target Effects Predicted Using Structural Analysis in the Context of a Metabolic Network Model. Plos Computational Biology. 2010;6(9):e10000938. doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao HW, Peng T, Ji Z, Zhou X. Systematically Studying Kinase Inhibitor Induced Signaling Network Signatures by Integrating Both Therapeutic and Side Effects. Plos One. 2013;8(12):e80832. doi: 10.1371/journal.pone.0080832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng H, Wen JG, Li HW, Chang J, Zhou X. Drug Inhibition ProfilePrediction for NFκB Pathway in Multiple Myeloma. PLoS ONE. 2011;6(3):e14750. doi: 10.1371/journal.pone.0014750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng H, Wen J, Li H, Zhang L, Chang CC, Zu Y, Zhou X. A systematic modeling study on the pathogenic role of p38 MAPK activation in myelodysplastic syndromes. Molecular BioSystems. 2012;8(4):1366–1374. doi: 10.1039/c2mb05184b. [DOI] [PubMed] [Google Scholar]
- 29.Sun X, Bao J, Nelson KC, Li KC, Kulik G, Zhou X. Systems modeling of anti-apoptotic pathways in prostate cancer: psychological stress triggers a synergism pattern switch in drug combination therapy. PLoS Comput Biol. 2013;9(12):e1003358. doi: 10.1371/journal.pcbi.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng H, Peng T, Wen J, Engler DA, Matsunami RK, Su J, Zhang L, Chang CC, Zhou X. Characterization of p38 MAPK isoforms for drug resistance study using systems biology approach. Bioinformatics. 2014;30(13):1899–907. doi: 10.1093/bioinformatics/btu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3(6):285–90. [PubMed] [Google Scholar]
- 32.Fitzgerald JB, Schoeberl B, Nielson UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2(9):458–66. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 33.Tang L, Su J, Huang D-S, Lee DY, Li KC, Zhou X. An integrated multiscale mechanistic model for cancer drug therapy. ISRN Biomathematics. 2012;2012:1–12. [Google Scholar]
- 34.Sun X, Su J, Bao J, Peng T, Zhang L, Zhang Y, Zhou X. Cytokine combination therapy prediction for bone remodeling in tissue engineering based on the intracellular signaling pathway. Biomaterials. 2012;33(33):8265–76. doi: 10.1016/j.biomaterials.2012.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su J, Zhang L, Zhang W, Choi DS, Wen J, Jiang B, Chang CC, Zhou X. Targeting the biophysical properties of the myeloma initiating cell niches: a pharmaceutical synergism analysis using multi-scale agent-based modeling. PLoS One. 2014;9(1):e85059. doi: 10.1371/journal.pone.0085059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatakrishnan K, Friberg LE, Ouellet D, Mettetal JT, Stein A, Trocóniz IF, Bruno R, Mehrotra N, Gobburu J, Mould DR. Optimizing oncology therapeutics through quantitative translational and clinical pharmacology: challenges and opportunities. Clin Pharmacol Ther. 2015;97(1):37–54. doi: 10.1002/cpt.7. [DOI] [PubMed] [Google Scholar]
- 37.Begam BF, Kumar JS. A study on chemoinformatics and its applications on modern drug discovery. Procedia Engineering. 2012;38:1264–1275. [Google Scholar]
- 38.Clark AM, Williams AJ, Ekins S. Machines first, humans second: on the importance of algorithmic interpretation of open chemistry data. J Cheminformatics. 2015;7:9, 1–20. doi: 10.1186/s13321-015-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reardon S. Organs-on-chips. Nature. 2015;423:266. doi: 10.1038/523266a. [DOI] [PubMed] [Google Scholar]
- 40.http://bidd.nus.edu.sg/group/cjttd/
- 41.https://www.pharmgkb.org/
- 42.Karthikeyan M, Vyas R, Tambe SS, Radhamohan D, Kulkarni BD. Role of Chemical Reactivity and Transition State Modeling for Virtual Screening. Comb Chem High Throughput Screen. 2015;18(7):638–657. doi: 10.2174/1386207318666150703113135. [DOI] [PubMed] [Google Scholar]
- 43.Mathijssen RH, Sparreboom A, Verweij J. Determining the optimal dose in the development of anticancer agents. Nat Rev Clin Oncol. 2014;11(5):272–81. doi: 10.1038/nrclinonc.2014.40. [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Sirota M, Butte AJ, Chen B. Characteristics of drug combination therapy in oncology by analyzing clinical trial data on ClinicalTrials.gov. Pac Symp Biocomput. 2015:68–79. [PMC free article] [PubMed] [Google Scholar]
- 45.Tang J, Aittokallio T. Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles. Curr Pharm Des. 2014;20(1):23–36. doi: 10.2174/13816128113199990470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elson EL, Genin GM. Tissue constructs: platforms for basic research and drug discovery. Interface Focus. 2016;6(1):20150095. doi: 10.1098/rsfs.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirt MN, Hansen A, Eschenhagen TT. Cardiac tissue engineering state of the art. Circulation research. 2014;114(2):354–367. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 48.Sachs JR, Mayawala K, Gadamsetty S, Kang SP, de Alwis DP. Optimal Dosing for Targeted Therapies in Oncology: Drug Development Cases Leading by Example. Clin Cancer Res. 2016;22(6):1318–24. doi: 10.1158/1078-0432.CCR-15-1295. [DOI] [PubMed] [Google Scholar]
- 49.Pritchard JR, Bruno PM, Gilbert LA, Capron KL, Lauffenburger DA, Hemann MT. Defining principles of combination drug mechanisms of action. Proc Natl Acad Sci U S A. 2013;110(2):E170–9. doi: 10.1073/pnas.1210419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curtius K, Hazelton WD, Jeon J, Luebeck EG. A Multi-scale Model Evaluates Screening for Neoplasia in Barrett’s Esophagus. In: Glazier JA, editor. PLoS computational biology. 5. Vol. 11. 2015. p. e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finley SD, Angelikopoulos P, Koumoutsakos P, Popel AS. Pharmacokinetics of anti-VEGF agent aflibercept in cancer predicted by data driven, molecular-detailed model. CPT: Pharmacometrics & Systems Pharmacology. 2015;4(11):641–9. doi: 10.1002/psp4.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finley SD, Popel AS. Effect of tumor microenvironment on tumor VEGF during anti-VEGF treatment: systems biology predictions. J Natl Cancer Inst. 2013;105(11):802–11. doi: 10.1093/jnci/djt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clegg LW, Mac Gabhann F. Site-Specific Phosphorylation of VEGFR2 Is Mediated by Receptor Trafficking: Insights from a Computational Model. PLoS Comput Biol. 11(6):e1004158. doi: 10.1371/journal.pcbi.1004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoeberl B, Pace E, Howard S, Garantcharova V, Kudla A, Sorger PK, Nielsen UB. A data-driven computational model of the ErbB receptor signaling network. Conf Proc IEEE Eng Med Biol Soc. 2006;1:53–4. doi: 10.1109/IEMBS.2006.259754. [DOI] [PubMed] [Google Scholar]
- 55.Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L, Linggi B, Kalra A, Paragas V, Bukhalid R, Grantcharova V, Kohli N, West KA, Leszczyniecka M, Feldhaus MJ, Kudla AJ, Nielsen UB. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci Signal. 2009;2(77):ra31. doi: 10.1126/scisignal.2000352. [DOI] [PubMed] [Google Scholar]
- 56.Ette EI, Williams PJ. Pharmacometrics: The Science of Quantitative Pharmacology. John Wiley & Sons, Inc; Hoboken, New Jersey: 2007. [Google Scholar]
- 57.Upton RN, Mould DR. Basic concepts in population modeling, simulation, and model-based drug development: part 3-introduction to pharmacodynamic modeling methods. CPT Pharmacometrics Syst Pharmacol. 2014;3:e88. doi: 10.1038/psp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joerger M. Covariate pharmacokinetic model building in oncology and its potential clinical relevance. AAPS J. 2012;14(1):119–32. doi: 10.1208/s12248-012-9320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houk BE, Bello CL, Poland B, Rosen LS, Demetri GD, Motzer RJ. Relationship between exposure to sunitinib and efficacy and tolerability endpoints in patients with cancer: results of a pharmacokinetic/pharmacodynamic meta-analysis. Cancer Chemother Pharmacol. 2010;66(2):357–71. doi: 10.1007/s00280-009-1170-y. [DOI] [PubMed] [Google Scholar]
- 60.Kiang TK, Sherwin CM, Spigarelli MG, Ensom MH. Fundamentals of population pharmacokinetic modelling: modelling and software. Clin Pharmacokinet. 2012;51(8):515–25. doi: 10.2165/11634080-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 61.Sherwin CM, Kiang TK, Spigarelli MG, Ensom MH. Fundamentals of population pharmacokinetic modelling: validation methods. Clin Pharmacokinet. 2012;51(9):573–90. doi: 10.1007/BF03261932. [DOI] [PubMed] [Google Scholar]
- 62.Yankeelov TE, Abramson RG, Quarles CC. Quantitative multimodality imaging in cancer research and therapy. Nat Rev Clin Oncol. 2014;11(11):670–80. doi: 10.1038/nrclinonc.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brikci FB, Clairambault J, Ribba B, Perthame B. An age-and-cyclin-structured cell population model for healthy and tumoral tissues. J Mathematical Biology. 2008;57(1):91–110. doi: 10.1007/s00285-007-0147-x. [DOI] [PubMed] [Google Scholar]
- 64.Gatenby RA, Gawlinski ET. A reaction-diffusion model of cancer invasion. Cancer Research. 1996;56(24):5745–53. [PubMed] [Google Scholar]
- 65.Ambrosi D, Preziosi L. On the closure of mass balance modes for tumors growth. Mathematical Models and Methods in Applied Sciences. 2002;12(05):737–753. [Google Scholar]
- 66.Colin T, Cornelis F, Jouganous J, Palussière J, Saut O. Patient specific simulation of tumor growth, response to the treatment and relapse of a lung metastasis: a clinical case. J Computational Surgery. 2015;2:1. [Google Scholar]
- 67.Swanson KR, Rostomily RC, Alvord EC. A mathematical modelling tool for predicting survival of individual patients following resection of glioblastoma: a proof of principle. British J Cancer. 2008;98(1):113–119. doi: 10.1038/sj.bjc.6604125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konukoglu E, Clatz O, Bondiau P-Y, Delingette H, Nicholas A. Extrapolating glioma invasion margin in brain magnetic resonance images: Suggesting new irradiation margins. Medical Image Analysis. 2010;14(2):111–125. doi: 10.1016/j.media.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Weis JA, Miga MI, Arlinghaus LR, Li X, Chakravarthy AB, Abramson VV, Farley J, Yankeelov TE. A mechanically coupled reaction-diffusion model for predicting the response of breast tumors to neoadjuvant chemotherapy. Phys Med Biol. 2013;58(17):5851–66. doi: 10.1088/0031-9155/58/17/5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benzekry S, Gandolfi A, Hahnfeldt P. Global Dormancy of Metastases Due to Systemic Inhibition of Angiogenesis. PLoS ONE. 2014;9(1):e84249–11. doi: 10.1371/journal.pone.0084249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lorz A, Lorenzi T, Hochberg ME, Clairambault J, Perthame B. Populational adaptive evolution, chemotherapeutic resistance and multiple anti-cancer therapies. ESAIM: Mathematical Modelling and Numerical Analysis. 2013;47:377–399. [Google Scholar]
- 72.Colin T, Iollo A, Lombardi D, Saut O. System Identification in Tumor Growth Modeling Using Semi-empirical Eigenfunctions. Mathematical Models & Methods In Applied Sciences. 2012;22(06):1250003–1. [Google Scholar]
- 73.Konukoglu E, Clatz O, Delingette H, Ayache N. Personalization of reaction-diffusion tumor growth models in mr images: Application to brain gliomas characterization and radiotherapy planning. In: Deisboeck Thomas S, Stamatakos Georgios., editors. Multi-scale Cancer Modeling, Chapman & Hall/CRC Mathematical & Computational Biology. CRC Press; Dec, 2010. [Google Scholar]
- 74.An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based models in translational systems biology. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2009;1(2):159–71. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Z, Butner JD, Kerketta R, Cristini V, Deisboeck TS. Simulating cancer growth with multiscale agent-based modeling. Semin Cancer Biol. 2015;30:70–8. doi: 10.1016/j.semcancer.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kam Y, Rejniak KA, Anderson AR. Cellular modeling of cancer invasion: integration of in silico and in vitro approaches. J Cell Physiol. 2012;227(2):431–8. doi: 10.1002/jcp.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andasari V, Roper RT, Swat MH, Chaplain MA. Integrating intracellular dynamics using CompuCell3D and Bionetsolver: applications to multiscale modelling of cancer cell growth and invasion. PLoS One. 2012;7(3):e33726. doi: 10.1371/journal.pone.0033726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gallaher J, Anderson AR. Evolution of intratumoral phenotypic heterogeneity: the role of trait inheritance. Interface Focus. 2013;3(4):20130016. doi: 10.1098/rsfs.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Macklin P, Edgerton ME, Thompson AM, Cristini V. Patient-calibrated agent-based modelling of ductal carcinoma in situ (DCIS): From microscopic measurements to macroscopic predictions of clinical progression. J Theor Biol. 2012;301:122–40. doi: 10.1016/j.jtbi.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engelberg JA, Ropella GE, Hunt CA. Essential operating principles for tumor spheroid growth. BMC Syst Biol. 2009;2(1):110. doi: 10.1186/1752-0509-2-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.An G, Kulkarni S. An agent-based modeling framework linking inflammation and cancer using evolutionary principles: description of a generative hierarchy for the hallmarks of cancer and developing a bridge between mechanism and epidemiological data. Math Biosci. 2015;260:16–24. doi: 10.1016/j.mbs.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meza R, Jeon J, Moolgavkar SH, Luebeck EG. Age-specific incidence of cancer: Phases, transitions, and biological implications. Proceedings of the National Academy of Sciences. 2008;105(42):16284. doi: 10.1073/pnas.0801151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moolgavkar SH, Knudson AG. Mutation and cancer: A model for human carcinogenesis. J Nat Cancer Inst. 1981;66:1037–1052. doi: 10.1093/jnci/66.6.1037. [DOI] [PubMed] [Google Scholar]
- 84.Luebeck E, Moolgavkar S. Multistage carcinogenesis and the incidence of colorectal cancer. Proceedings of the National Academy of Sciences. 2002;99(23):15095. doi: 10.1073/pnas.222118199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu F, Celli J, Rizvi I, Moon S, Hasan T, Demirci U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol J. 2011;6:204–212. doi: 10.1002/biot.201000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Engler AJ, Humbert PO, Wehrle-Haller B, Weaver VM. Multi-scale modeling of form and function. Science. 2009;324(5924):208–212. doi: 10.1126/science.1170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chakrabarti A, Verbridge S, Stroock AD, Fischbach C, Varner JD. Multi-scale models of breast cancer progression. Annals Biomedical Engineering. 2012;40(11):2488–2500. doi: 10.1007/s10439-012-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson AR. A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math Med Biol. 2015;22(2):163–86. doi: 10.1093/imammb/dqi005. [DOI] [PubMed] [Google Scholar]
- 89.Jiang C, Cui C, Li L, Shao Y. The anomalous diffusion of a tumor invading with different surrounding tissues. PLoS One. 2014;9(10):e109784. doi: 10.1371/journal.pone.0109784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bae YH. Drug targeting and tumor heterogeneity. Journal of controlled release: official journal of the Controlled Release Society. 2009;133(1):2. doi: 10.1016/j.jconrel.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bozic I, Nowak MA. Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers. Proc Natl Acad Sci U S A. 2014;111(45):15964–8. doi: 10.1073/pnas.1412075111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen BB, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bellouquid A, Delitala M. Mathematical methods and tools of kinetic theory towards modelling complex biological systems. Mathematical models and methods in applied sciences. 2005;15(11):1639–1666. [Google Scholar]
- 94.Adam J, Bellomo N. A survey of models for tumor-immune system dynamics. Springer Science & Business Media; 2012. [Google Scholar]
- 95.Yankeelov TE, Quaranta V, Evans KJ, Rericha EC. Toward a science of tumor forecasting for clinical oncology. Cancer Res. 2015;75(6):918–23. doi: 10.1158/0008-5472.CAN-14-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yankeelov TE, Atuegwu N, Hormuth D, Weis JA, Barnes SL, Miga MI, Rericha EC, Quaranta V. Clinically relevant modeling of tumor growth and treatment response. Sci Transl Med. 2013;5(187):187ps9. doi: 10.1126/scitranslmed.3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin PP. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;3(5):4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278(2):563–77. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Green SE, Jr, Li Y. Rhetorical institutionalism: Language, agency, and structure in institutional theory since Alvesson. Journal of Management Studies. 1993;48(7):1662–97. [Google Scholar]
- 100.Byrne HM. Dissecting cancer through mathematics: from the cell to the animal model. Nat Rev Cancer. 2010;10(3):221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 101.Heilbron JL. The affair of the countess Görlitz. Proceedings of the American Philosophical Society. 1994;138(2):284–316. [Google Scholar]
- 102.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer. 2011;11(7):512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.An G. Closing the scientific loop: bridging correlation and causality in the petaflop age. Sci Transl Med. 2010;2(41):41ps34. doi: 10.1126/scitranslmed.3000390. [DOI] [PubMed] [Google Scholar]
- 104.Cross W. The Code: An Authorized History of the ASME Boiler and Pressure Vessel Code. New York: American Society of Mechanical Engineers; 1990. [Google Scholar]
- 105.http://www.imagwiki.nibib.nih.gov/content/committee-credible-practice-modeling-simulation-healthcare-description
- 106.EFPIA MID3 Workgroup. Marshall S, Burghaus R, Cosson V, Cheung S, Chenel M, DellaPasqua O, Frey N, Hamrén B, Harnisch L, Ivanow F, Kerbusch T, Lippert J, Milligan P, Rohou S, Staab A, Steimer J, Tornøe C, Visser S. Good Practices in Model-Informed Drug Discovery and Development: Practice, Application, and Documentation. CPT: Pharmacometrics & Systems Pharmacology. 2016;5:93–122. doi: 10.1002/psp4.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]