Highlights
-
•
A second ligand-independent pathway activates STING and leads to IFN-I-production in response to viral infections.
-
•
Mitochondrial stress activates STING-mediated pathway and therefore plays important role in host response to viruses.
-
•
BTK1, GSK3β, and SCAP promote activation of IRF3 and TBK1 downstream of STING.
-
•
Blocking STING-pathway is a common viral strategy to counteract production of IFN-I by host in response viral infection.
-
•
Several viral components directly interact with STING and TBK1 and block IFN-production.
Abstract
The identification and characterization of DNA-sensing pathways has been a subject of intensive investigation for the last decade. This interest, in part, is supported by the fact that the main outcome of DNA-responses is production of type I interferon (IFN-I), which, if produced in excessive amounts, leads to various pathologies. STING (stimulator of interferon genes) is positioned in the center of these responses and is activated either via direct sensing of second messengers or via interaction with upstream sensors of dsDNA. STING mediates responses to pathogens as well as host-derived DNA and is, therefore, linked to various autoimmune diseases, cancer predisposition and ageing. Recent mouse models of DNA damage showed the adaptor STING to be crucial for heightened resting levels of IFN-I. In this review, we will focus on recent advances in understanding the regulation of STING-signaling and identification of its novel components.
Current Opinion in Microbiology 2016, 32:144–150
This review comes from a themed issue on Host-microbe interactions: viruses
Edited by Jonathan C Kagan
For a complete overview see the Issue and the Editorial
Available online 11th July 2016
http://dx.doi.org/10.1016/j.mib.2016.05.014
1369-5274/© 2016 Elsevier Ltd. All rights reserved.
STING: a central player in the immune response to cytosolic DNA
In eukaryotic cells, the DNA is packaged in the nucleus or mitochondria. During infection or in response to a variety of stress signals, DNA may be exposed within the endosomal or cytoplasmic compartment, where it is recognized by several DNA sensors that ultimately activate caspase-1 and signaling mediated by STING [1]. The name implies that the production of IFN-I as well as the subsequent activation of IFN-stimulated genes (ISGs) [2] is the main outcome of STING activation.
STING (Tmem173/MPYS/MITA) is a tetraspanning membrane-resident sensor that recognizes both bacterial and host-derived cyclic di-nucleotides (CDNs) and cytoplasmic nucleic acids, either directly or via other cytosolic DNA sensors, such as nuclear resident IFI16 (interferon inducible 16) [3], a nucleotidyltransferase cGAS (cGAMP (GMP-AMP) synthase) [4], and helicase DDX41 (DExD/H-box) [5]. The amino-terminal domain (NTD) of STING, which encompasses approximately the first 130 amino acids, contains four transmembrane domains, while the final 250 amino acids comprise a globular carboxy-terminal domain (CTD), several crystal structures of which have been reported.
The STING CTD resides in the cytosol and provides a platform that directs TBK1 (tank binding kinase) [6] to phosphorylate and activate IRF3 (interferon regulatory factor 3) [7]. The phosphorylation of IRF3 appears to be a conserved mechanism that selectively recruits IRF3 to activate IFN-I [7]. The activation of STING triggers conformational changes in its CTD, which serves as a platform for the recruitment and auto-phosphorylation of the serine-threonine kinase TBK1. This kinase, in turn, phosphorylates and activates the transcription factor IRF3. IRF3 then travels to the nucleus, where it initiates IFN-I production and directly promotes the expression of multiple ISGs, many of which are activated via the IFN-I αβ receptor (IFNAR) pathway. The growing interest in STING-mediated DNA responses is based on the importance of IFN-I for the host defenses against DNA viruses, such as herpes viruses, and intracellular bacteria, such as Listeria monocytogenes. Another subject of interest is the regulation of IFN-I by STING; namely, tight regulation of IFN-I is essential, since excessive production of this cytokine leads to the development of autoimmunity [8•, 9], chronic inflammation [10••, 11], or sepsis. Not surprisingly, STING is a critical component in tissue repair and tumorigenesis [12].
While recent studies have highlighted the functional importance of the STING CTD [13], particularly in the recruitment and activation of the serine-threonine kinase TBK1 and the transcription factor IRF3, less is known about the function of the NTD, although some NTD mutations have been described in humans; for example, a potentially nonfunctional allele, referred to as HAQ (R71H-G230A-R293Q), was found to be homozygous in ∼3% of two American cohorts [14]. Additional evidence has been reported from murine studies, where a hypomorphic allele of STING with detrimental mutations confined to the NTD is seen in wild-derived mice [15]. However, the role of the STING NTD in its interaction with recently discovered additional components and pathways requires further analysis.
Conceptual advances in the definition of the STING signaling pathway
Given a central role of STING in IFN-I production and regulation, it would not be surprising if additional ligands exist that activate STING-signaling. Some of these ligands, such as mitochondrial DNA (mtDNA), easily fit into the DNA-damage based model of STING-activation [16], which is counterbalanced by caspases [17]. Other ligands, such as those activated downstream of lipid membrane [18], have yet to be identified. Finally, the STING signaling pathway could converge at some level with other pathways, downstream of STING.
Mitochondrial stress protects against viral infection
Mitochondria are known to respond to stress by DNA leakage, a process that induces innate immune responses [19]. Not surprisingly, stressed mitochondria release DNA in response to HSV-1 (herpes simplex virus) and, thus, provide protection against infection [20•]. Hemizygosity for Tfam (transcription factor a mitochondrial), which encodes an abundant mitochondrial protein responsible for nucleoid maintenance and architecture, keeps the mitochondria intact, but leads to a 50% loss of mtDNA. This results in IRF3 nuclear translocation and elevation of several IFNs and ISGs in a STING-dependent manner. IFN production is also dependent on cGAS, which is co-localized with mtDNA, suggesting that cGAS binds mtDNA.
Accordingly, Tfam-hemizygous MEFs (mouse embryonic fibroblasts) showed resistance to VSV (vesicular stomatitis virus) and HSV-1 infection and lower viral loads, whereas inhibition of release of mtDNA promoted viral replication. Although the authors identified the viral protein UL12.5 as responsible for mitochondrial stress induction, it seems counterintuitive that the virus activates an anti-viral mechanism of defense, particularly since numerous other viral homologues of UL12.5 induce mitochondrial stress. It is possible that a synergy exists between mitochondrial stress and induction of other, independent of DNA-damage, immune responses [21], that is, a for potential engagement and cross-talk with other pathways. Altogether, this work supports the theory that mitochondria serve as important sensors of particular pathogens.
Two pathways are required for resistance to the smallpox virus infection
In support of alternative and synergistic ways of IFN-production, He et al. report [22] that TLR9, MyD88, STING and IRF7 are required for resistance to mouse smallpox virus and for the production of IFN in draining lymph nodes (dLN). Either STING-deficiency or MyD88-deficiency leads to a decrease in IFN-I production and to an increase in mortality in infected mice. The authors show that inflammatory monocytes (iMos) recruited into the dLN are the main producers of IFN-I in response to infection and use STING to activate IRF7 and NF-kB for production of IFN-I. However, it remains to be determined which particular DNA sensor activates STING signaling in iMos. For example, there could be a specialized sensor that stimulates STING in an exclusive manner in these cells. It would also be interesting to see whether STING activates IRF7 directly in iMos or through TLR9-IRF7-STING interaction.
Posttranslational modifications provide additional regulation of STING signaling. For example, IFI16 is a DNA sensor that resides in the nucleus and binds to the sugar-phosphate backbone of dsDNA. The details of how IFI16 moves to the cytosol of infected cells and activates STING have been clarified in a recently published report, which shows that acetylation regulates IFI16 trafficking and DNA binding [23]. A key molecular step is the acetylation of DNA-bound nuclear IFI16 via p300, which decreases DNA-IFI16 binding with the viral genome and expels IFI16 from the DNA, changes its conformation, exposes nuclear import signals, and binds STING in the cytosol. Thus, IFI16 acetylation defines IFI16 location and allows this sensor to switch from BRCA1-assisted [24] binding the viral DNA to interacting with STING. These findings contradict the canonical paradigm that the nucleus is ‘immune privileged’ with regard to sensing foreign DNA and suggest that both cytoplasmic and nuclear IFI16 participate in viral DNA surveillance.
A second STING-mediated pathway is targeted by the influenza virus
In an intriguing extension of their previous report [25], Holm et al. now describe the interference of the influenza virus with, presumably, a second pathway that is mediated by STING in a DNA-independent manner [26••]; namely, they show that infection with enveloped RNA viruses leads to STING-dependent, but cGAS-independent, IFN-I production, a discovery that is surprising, since it suggests the existence of another (unknown) sensor located upstream of STING. In this study, STING co-localized with viral hemagglutinin (HA) and physically interacted with the NTD of the HA fusion protein (FP) via STING's173-163 region. The FP inhibited STING dimerization and translocation in response to the activation of cells with fusogenic liposomes [26••]. The same team made another exciting discovery when they identified Arg168 of STING to be important in fusion-mediated, but not ligand-induced, IFN-I production, thus functionally separating the two pathways. It is tempting to speculate that another, novel cytosolic component, possibly related to a Ca2+ related membrane perturbation [27•], transduces the activation signal from the lipid membrane to STING. It also suggests that enveloped viruses and fusogenic liposomes both mediate STING-dependent, ligand-independent IFN-I response independently of cGAS; whether this second pathway evolved for better recognition of viruses or enveloped viruses developed additional tools for interfering with the host response remains to be further investigated.
Novel mechanistic insights in the regulation of the STING-pathway
Several activation steps in STING-signaling have recently been identified, such as the interaction of STING with TRAF2 (TNF receptor associated factor 2) and the ubiquitin ligase TRAF3, as well as phosphorylation on several residues, including Ser358 [28]. Ubiquitination by TRIM56 (tripartite motif) and TRIM32 plays an important role in regulating expression of STING [29]. In addition to recruitment of TBK1 and IRF3, the STING-CTD also recruits STAT6 [30]. Tight regulation of IFN-production is achieved via inhibitory steps such as AKT-mediated inhibition of cGAS [31] or via TRM30a-mediated degradation of STING [32].
BTK1 phosphorylates DDX41 for activation of STING
The DNA-sensor DDX41 was known to activate the STING signaling pathway, but the specific mechanism of this activation was poorly understood. Now, Lee et al. report the importance of the BTK1 kinase in this activation [33]. BTK1-deficient cells lack phosphorylation of IRF3 and TBK1 and exhibit low levels of IFN-I. According to their model, the DEAD-box domain of DDX41 binds BTK's kinase domain, whereas the SH3/SH2 domain of BTK1 binds the transmembrane domain of STING. BTK1-mediated phosphorylation of DDX41 at Tyr414 increases the affinity of DDX41 towards dsDNA and STING and stabilizes DDX41-STING interactions, thus inducing oligomerization of STING and ultimate activation of TBK1/IRF3 and production of IFN-I. The proposed model suggests that TBK1 functions as an adaptor as well as a kinase. Although a second tyrosine at 364 is critical for the activation of STING, it is not phosphorylated by BTK1, thus leaving its role in STING activation for further investigations.
PPM1A (protein phosphatase Mg2+/Mn2+ dependent 1A) is a negative regulator of STING-activation
The phosphatase PPM1A directly interacts with STING and removes the phosphate groups from both STING and TBK1 [34], thereby acting as a negative regulator of STING signaling. This interaction prevents further oligomerization of STING and IFNβ production in response to HSV-1. Using exogenously expressed proteins, the authors show that TBK1-mediated phosphorylation of STING at Ser358 is a prerequisite for STING polymerization and efficient phosphorylation of IRF3 byTBK1.
The ER (endoplasmic reticulum) adaptor SCAP (SREBP cleavage activating protein) recruits IRF3 to STING
The mechanism of IRF3 recruitment to STING was poorly understood, but Hansen et al. now provide mechanistic insight into this process [26••]. Specifically, they show that silencing of SCAP, but not TBK1, abolishes IRF3 recruitment to STING and IFNβ production, but does not abolish STING translocation, thus providing evidence, for the first time, that IRF3 activation and STING translocation are mechanistically separated. The N-terminal (8-pass) transmembrane of SCAP interacts with the transmembrane of STING (1-175), and the C-terminus of SCAP binds IRF3 in an activation dependent mode. SCAP-deficient mice are more sensitive to HSV-1 infection than WT mice. A major question remains as to what initiates the association of SCAP and STING at the ER upon stimulation. It is also still a mystery what regulates the STING/SCAP exit from the ER.
Diversity of DNA-sensors that confer responses to Listeria
In many instances, DNA-sensors cooperate to activate STING signaling [28]. An example of such diverse DNA-responses is recognition of Listeria [35, 36]. Although the role of cGAS in mediating responses to Listeria in mice is well established, the nature of the DNA-sensor of Listeria in human cells was unclear, since human cells are neither dependent on cGAS, nor on a pump that transports CDNs into the host cytoplasm. Now Hansen et al. show [37] that cGAS, STING, and IFI16, but not DDX41, are important for sensing the bacterial DNA, and that it is DNA, rather than Listeria CDNs, that activates the STING-pathway in human myeloid cells. One likely explanation is that human STING is 500 times less responsive to bacterial-derived CDNs with 3′5′ linkage than 2′3’cGAMP. In addition, humans may have evolved to process DNA better. These data suggest a role for IFI16 in the amplification of cGAS-dependent DNA-driven responses.
NLRX1 sequesters STING to negatively regulate the interferon response
Several nod-like receptors (NLRs) have been characterized as negative regulators of the IFN-I in response to DNA [38]. Guo et al. now describe a mechanism by which NLRX1 blocks IFN-I responses and promotes early HIV infection [39]. NLRX1 facilitates its effect via binding the C-terminus of STING, which disrupts the STING-TBK1 interaction. The fact that NLRX1 resides in the mitochondria, whereas STING is located in the ER, challenges these findings. Furthermore, previously observed interactions between mitochondrial and ER proteins do not provide the reason for interaction between mitochondrial and ER proteins. One possibility might be that STING-signaling is specifically prohibited in MAMs (mitochondria associated membrane) and gets disrupted once STING accidently translocates to the mitochondria. Thus, the described negative regulation of STING by NLRX1 could be restricted to certain cellular compartments.
TRIM9s promotes the IFN-I response by recruiting GSK3β to TBK1
A short isoform of TRIM9, TRIM9s, was identified as a novel positive regulator of IFN-I signaling downstream of MAVS and STING signaling [40]. Knockdown of TRIM9s resulted in higher cellular viral loads of RNA and DNA viruses. It was shown that TRIM9s binds directly to GSK3β and TBK1 and facilitates the interaction between GSK3β and TBK1. Binding of TRIM9s to TBK1 enhances IRF3 activation after viral infection. Finally, the authors determined that TRIM9s has E3 ligase activity and is able to auto-ubiquitinate itself in order to recruit TBK1 and GSK3β. The negative effect of TRIM9s on IL6 and TNF-activation is likely to be linked to GSK3β and could be confirmed by analysis of IL-10 production, which is promoted by GSK3β.
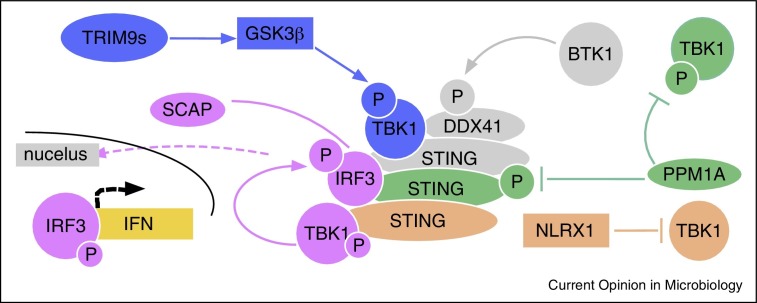
Figure 1 summarizes some of the STING-regulatory pathways that were discussed here. The effects of these pathways are confined to phosphorylation and de-phosphorylation, or direct interactions that antagonize STING-signaling or synergize with it. The specific timing and intracellular location of these interactions is not always known but, as in case with NLRX1, spatial requirement might define specificity of the effects.
Figure 1.
Recently described regulatory components of STING-mediated activation of type I IFN.
Pink — SCAP-mediated recruitment of IRF3 to STING; blue — TRIM9s-facilitated interaction of TBK1 with GSK3β; green — inhibitory effect of PPM1A on phosphorylation of TBK1 and STING; gray — BTK1-induced activation of DDX41 and its recruitment to STING; brown — inhibitory effect of NLRX1 on interaction between TBK1 and STING
Viral interference with STING signaling
Having apparently arisen as a sensor of self-DNA and stress-signals [41•], the STING-pathway has evolved to counteract viral infections. It is, therefore, not surprising that viruses have developed various mechanisms to neutralize STING signaling [42, 43]. In this section, we will discuss some of the recent advances in the characterization of viral strategies to hijack the STING-pathway.
HBV (hepatitis B virus) polymerase disrupts K63-linked ubiquitination of STING
To clarify the effect of HBV on the innate immune response, Liu et al. infected primary human hepatocytes with virus from HBV-positive sera to observe downregulation of STING-specific IFN-I production [44]. They report that viral polymerase (Pol) inhibits STING-mediated IRF3 activation and IFN-I production. They also mapped the inhibitory effect of Pol to its RT and RNAseH domains. Pol seems to physically interact with STING and does not affect expression, but rather ubiquitination, of STING via its RT domain.
KSHV (Kaposi Sarcoma Herpes Virus) targets cGAS
In the first description of viral strategy targeting cGAS, Wu et al. characterize ORF52 of Kaposi Sarcoma Herpes Virus (KSHV) as an inhibitor of cGAS. When overexpressed, ORF52 inhibited cGAS and IRF3 dimerization [45]. Because the inhibitory effect of ORF52 was attenuated by increased amounts of the DNA, this suggested that ORF52 competes with cGAS for DNA. ORF52 inhibited STING-mediated responses, but not AIM2-mediated responses. The inhibitory effect was specific for DNA, but not RNA-viruses, which altogether supported direct interaction between ORF52 and cGAS. Indeed, the authors mapped the region of ORF52 responsible for binding cGAS. Ability of ORF52 to bind both DNA and cGAS provides high specificity of the inhibitory effect. The rationale behind the strategy used by herpes viruses is that DNA accounts for 10–20% of their mass, thus making it necessary for the virus to develop a mechanism that prevents recognition of DNA. This is particularly true for KSHV, which goes through lytic cycles that release viral DNA into the cytosol. Interestingly, ORF52-deficient virus showed little difference in elicited IFN-response compared with the wild type strain, suggesting redundancy in the inhibitory effects of KSHV ORFs on IFN-I production.
KSHV protein disrupts TBK1-STING interaction
In support of the ‘redundancy model’ Ma et al. [46] searched for the KSHV ORFs responsible for the inhibitory effect and identified vIRF1 as the most efficient among the tested ORFs in disrupting the TBK1-STING interaction and attenuation of IFN-production. Concordantly, depletion of vIRF1 elevated IFN-production in response to KSHV and prevented efficient viral replication. During their lifelong persistence in the host, which includes cycles of latency and lytic activation, the herpes viruses can release their DNA into the cytoplasm or in the nucleus, or induce mitochondrial stress. Thus, KSHV viruses developed various strategies to block multiple nodes in the DNA-sensing pathway.
HCV (hepatitis C virus) protein targets STING for degradation
A similar disruption of the STING-mediated pathway has been recently reported for the proteins of HCV [47], which is, similar to Influenza virus, a ssRNA virus. Using two different viral replicons, Yi et al. identified a transmembrane domain in NS4B that inhibited responses to cGAMP. Concordantly, activation with cGAMP inhibited replication of the HCV-cassette at the mRNA and protein level in a STING-specific manner. At the mechanistic level, NS4B induced degradation of STING, which is termed ‘downregulation of STING accumulation’. Why would an RNA virus develop a strategy of interfering with STING? It is possible that STING degradation is an underlying mechanism that explains earlier reports of NS4B disrupting interaction of STING with TBK1 and MAVS.
HSV-1 serine protease VP24 inhibits DNA-response by blocking IRF3 activation
The VP24 protein is a serine protease of HSV-1 that is necessary for capsid formation of the virus. In this paper VP24 was found to dampen the interferon response to DNA by blocking the interaction between TBK1 and IRF3 and subsequent inhibition of IRF3 activation [47]. The authors show a lack of IRF3 dimerization, during HSV-1 infection that increased after the knockdown of VP24.
A summary of the discussed above viral strategies of interfering with the STING-mediates signaling is provided in Table 1 . Interestingly, viruses use direct physical interaction with the components of the STING-pathway rather than enzymatic modification. Another apparent conclusion from these data is that viral strategies cover all of the main components of STING-signaling thus suggesting that there are probably some unknown interferences with some (yet) unknown components. Therefore it seems promising to study the viral strategies of interfering with the host immune response.
Table 1.
Interference of viral proteins with components of STING signaling pathway.
| Virus | Viral protein | Pathway inhibited | Ref. |
|---|---|---|---|
| KSHV | vIRF1 | Blocks STING interaction with TBK1 | [46] |
| Adenovirus | E1a | Blocks unknown NTD STING mediated signal | [48] |
| HSV1 | VP24 | Interacts with IRF3 and blocks phosphorylation of IRF3 by TBK1 | [49] |
| IAV | HA2 fusion peptide | Inhibits STING dimerization in response to membrane fusion | [26••] |
| HTLV-1 | Tax | Inhibits TBK1 phosphorylation of IRF3 by interactions with TBK1 | [50] |
| HCV | NS4B | Inhibits STING accumulation by direct interaction with STING | [47] |
| KSHV | ORF52 | Blocks cGAS binding of DNA | [45] |
| HBV | Pol | Blocks K63 ubiquitination of STING by binding STING | [44] |
| Coronavirus | PLP2-TM | Induces STING–Beclin1 interactions to degrade STING | [51] |
| SARS | PLpro-TM | Inhibits TRAF3/TBK1/STING interactions | [52] |
In conclusion, STING is positioned as the main regulator of responses to host and pathogen-derived DNA. In addition, experimental data implicate STING in the regulation of DNA-responses in homeostasis (basal interferon), as well as in autoimmune models.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by the NIH Grants AI056234, AI119833, and Russian Science Fund project 15-15-00100 (to AP). The authors are grateful to Brigitte Huber for reading the manuscript.
References
- 1.Corrales L., Woo S.R., Williams J.B., McWhirter S.M., Dubensky T.W., Jr., Gajewski T.F. Antagonism of the STING pathway via activation of the AIM2 inflammasome by intracellular DNA. J Immunol. 2016;196:3191–3198. doi: 10.4049/jimmunol.1502538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monroe K.M., Yang Z., Johnson J.R., Geng X., Doitsh G., Krogan N.J., Greene W.C. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suschak J.J., Wang S., Fitzgerald K.A., Lu S. A cGAS-independent STING/IRF7 pathway mediates the immunogenicity of DNA vaccines. J Immunol. 2016;196:310–316. doi: 10.4049/jimmunol.1501836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewinsohn M., Brown A.L., Weinel L.M., Phung C., Rafidi G., Lee M.K., Schreiber A.W., Feng J., Babic M., Chong C.E. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127:1017–1023. doi: 10.1182/blood-2015-10-676098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freischmidt A., Wieland T., Richter B., Ruf W., Schaeffer V., Muller K., Marroquin N., Nordin F., Hubers A., Weydt P. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 7.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y., Grishin N. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015 doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 8•.Sharma S., Campbell A.M., Chan J., Schattgen S.A., Orlowski G.M., Nayar R., Huyler A.H., Nundel K., Mohan C., Berg L.J. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc Natl Acad Sci U S A. 2015;112:E710–E717. doi: 10.1073/pnas.1420217112. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses lpr model of murine systemic autoimmunity to provide evidence that STING suppresses the TLR-induced innate signaling in systemic lupus erythematosus (SLE). Mechanistically, STING-deficient macrophages failed to express negative regulators of immune activation and thus were hyperresponsive to TLR ligands. These data describe STING as a key natural regulator of SLE, which can be manipulated to provide disease-specific therapeutics.
- 9.Ahn J., Barber G.N. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 10••.Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Montealegre Sanchez G.A., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using whole exome sequencing, the authors analyzed the DNA of a patient(s) with early-onset auto-inflammatory disease to identify TMEM173, encoding STING, as the gene conferring the phenotype. Specifically, three de novo STING mutations confer gain of functions and constitutive activation of STING and hypersensitivity to ligand stimulation. In addition to being clinically relevant, this work reveals spontaneous activation of STING via the DD (dimerization domain) thus providing important mechanistic insight on STING-activation.
- 11.Crow Y.J., Casanova J.L. STING-associated vasculopathy with onset in infancy — a new interferonopathy. N Engl J Med. 2014;371:568–571. doi: 10.1056/NEJMe1407246. [DOI] [PubMed] [Google Scholar]
- 12.Ahn J., Konno H., Barber G.N. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene. 2015 doi: 10.1038/onc.2014.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lou Y.C., Kao Y.F., Chin K.H., Chen J.K., Tu J.L., Chen C., Chou S.H. Backbone resonance assignments of the 54 kDa dimeric C-terminal domain of murine STING in complex with DMXAA. Biomol NMR Assign. 2015;9:271–274. doi: 10.1007/s12104-014-9590-y. [DOI] [PubMed] [Google Scholar]
- 14.Crow Y.J. Type I interferonopathies: mendelian type I interferon up-regulation. Curr Opin Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Surpris G., Chan J., Thompson M., Ilyukha V., Liu B.C., Atianand M., Sharma S., Volkova T., Smirnova I., Fitzgerald K.A. Cutting edge: novel Tmem173 allele reveals importance of STING N terminus in trafficking and type I IFN production. J Immunol. 2015;196:547–552. doi: 10.4049/jimmunol.1501415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S., Feng M., Guan W. Mitochondrial DNA sensing by STING signaling participates in inflammation, cancer and beyond. Int J Cancer. 2016;139:736–741. doi: 10.1002/ijc.30074. [DOI] [PubMed] [Google Scholar]
- 17.White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., van Delft M.F., Bedoui S., Lessene G., Ritchie M.E. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.York A.G., Williams K.J., Argus J.P., Zhou Q.D., Brar G., Vergnes L., Gray E.E., Zhen A., Wu N.C., Yamada D.H. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rongvaux A., Jackson R., Harman C.C., Li T., West A.P., de Zoete M.R., Wu Y., Yordy B., Lakhani S.A., Kuan C.Y. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors of this paper report that mitochondrial stress is induced in response to the infection. The leaking mitochondrial DNA activates the innate immune response via cGAS-STING pathway. Thus, stressed mitochondria can be viewed as a sensor of infection.
- 21.Kemp M.G., Lindsey-Boltz L.A., Sancar A. UV light potentiates STING (stimulator of interferon genes)-dependent innate immune signaling through deregulation of ULK1 (Unc51-like kinase 1) J Biol Chem. 2015;290:12184–12194. doi: 10.1074/jbc.M115.649301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu R.H., Wong E.B., Rubio D., Roscoe F., Ma X., Nair S., Remakus S., Schwendener R., John S., Shlomchik M. Sequential activation of two pathogen-sensing pathways required for type I interferon expression and resistance to an acute DNA virus infection. Immunity. 2015;43:1148–1159. doi: 10.1016/j.immuni.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ansari M.A., Dutta S., Veettil M.V., Dutta D., Iqbal J., Kumar B., Roy A., Chikoti L., Singh V.V., Chandran B. Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-beta responses. PLoS Pathog. 2015;11:e1005019. doi: 10.1371/journal.ppat.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutta D., Dutta S., Veettil M.V., Roy A., Ansari M.A., Iqbal J., Chikoti L., Kumar B., Johnson K.E., Chandran B. BRCA1 regulates IFI16 mediated nuclear innate sensing of herpes viral DNA and subsequent induction of the innate inflammasome and interferon-beta responses. PLoS Pathog. 2015;11:e1005030. doi: 10.1371/journal.ppat.1005030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moeller H.B., Gonzalez-Dosal R., Rasmussen S.B., Christensen M.H., Yarovinsky T.O. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Holm C.K., Rahbek S.H., Gad H.H., Bak R.O., Jakobsen M.R., Jiang Z., Hansen A.L., Jensen S.K., Sun C., Thomsen M.K. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]; At the mechanistic level, this study identifies a fusion peptide in the N-terminal domain of the hemagglutinin of the dsRNA influenza virus which interacts with the dimerization domain of STING thus preventing the dimerization of STING and downstream IFN production. At the conceptual level, the data allude to existence of another pathway that is STING-dependent but cGAS-independent that interferes with the cellular membrane and provides ligand-independent activation of STING.
- 27•.Hare D.N., Collins S.E., Mukherjee S., Loo Y.M., Gale M., Jr., Janssen L.J., Mossman K.L. Membrane perturbation-associated Ca2+ signalling and incoming genome sensing are required for the host response to low-level enveloped virus particle entry. J Virol. 2015;90:3018–3027. doi: 10.1128/JVI.02642-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Experimental evidence suggests that membrane perturbation induced by fusion of the enveloped viruses with the host cells is sufficient to induce transcription of a subset of ISGs. This report has established importance of Ca2+ signaling for activation of STING and IRF3 following viral entry, suggesting that Ca2+ signaling sensitizes cells to recognize genomes within incoming virus particles. Altogether, this work further supports the second signal caused by viruses and change the cellular environment, and oxidative stress or endoplasmic reticulum stress that amplify antiviral signaling.
- 28.Barber G.N. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Sun H., You F., Sun W., Zhou X., Chen L., Yang J., Wang Y., Tang H., Guan Y. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Seo G.J., Yang A., Tan B., Kim S., Liang Q., Choi Y., Yuan W., Feng P., Park H.S., Jung J.U. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 2015;13:440–449. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O’Connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K.G., Kim S.S., Kui L., Voon D.C., Mauduit M., Bist P., Bi X., Pereira N.A., Liu C., Sukumaran B. Bruton's tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Rep. 2015;10:1055–1065. doi: 10.1016/j.celrep.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Liu G., Sun L., Teng Y., Guo X., Jia J., Sha J., Yang X., Chen D., Sun Q. PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog. 2015;11:e1004783. doi: 10.1371/journal.ppat.1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archer K.A., Durack J., Portnoy D.A. STING-dependent type I IFN production inhibits cell-mediated immunity to Listeria monocytogenes. PLoS Pathog. 2014;10:e1003861. doi: 10.1371/journal.ppat.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samba-Louaka A., Pereira J.M., Nahori M.A., Villiers V., Deriano L., Hamon M.A., Cossart P. Listeria monocytogenes dampens the DNA damage response. PLoS Pathog. 2014;10:e1004470. doi: 10.1371/journal.ppat.1004470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen K., Prabakaran T., Laustsen A., Jorgensen S.E., Rahbaek S.H., Jensen S.B., Nielsen R., Leber J.H., Decker T., Horan K.A. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crow M.K., Olferiev M., Kirou K.A. Targeting of type I interferon in systemic autoimmune diseases. Transl Res. 2015;165:296–305. doi: 10.1016/j.trsl.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H., Konig R., Deng M., Riess M., Mo J., Zhang L., Petrucelli A., Yoh S.M., Barefoot B., Samo M. NLRX1 sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV-1 and DNA viruses. Cell Host Microbe. 2016;19:515–528. doi: 10.1016/j.chom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia P., Ye B., Wang S., Zhu X., Du Y., Xiong Z., Tian Y., Fan Z. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol. 2016;17:369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- 41•.Kranzusch P.J., Wilson S.C., Lee A.S., Berger J.M., Doudna J.A., Vance R.E. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol Cell. 2015;59:891–903. doi: 10.1016/j.molcel.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper characterize the ancient origins of human cGAMP signaling by discovery of a functional cGAS-STING pathway in anemone species derived from common with humans ancestor more than 500 million years ago. These data reveal that human mixed-linkage cGAMP achieves universal signaling by exploiting a deeply conserved STING conformational intermediate, providing critical insight for therapeutic targeting of the STING pathway.
- 42.Ma Z., Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe. 2016;19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bucci M. Antivirals: alphaviruses feel the STING. Nat Chem Biol. 2016;12:53. [Google Scholar]
- 44.Liu Y., Li J., Chen J., Li Y., Wang W., Du X., Song W., Zhang W., Lin L., Yuan Z. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol. 2015;89:2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J.J., Li W., Shao Y., Avey D., Fu B., Gillen J., Hand T., Ma S., Liu X., Miley W. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Z., Jacobs S.R., West J.A., Stopford C., Zhang Z., Davis Z., Barber G.N., Glaunsinger B.A., Dittmer D.P., Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015;112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi G., Wen Y., Shu C., Han Q., Konan K.V., Li P., Kao C.C. Hepatitis C virus NS4B can suppress STING accumulation to evade innate immune responses. J Virol. 2015;90:254–265. doi: 10.1128/JVI.01720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau L., Gray E.E., Brunette R.L., Stetson D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D., Su C., Zheng C. Herpes simplex virus type 1 serine protease VP24 blocks DNA sensing signal pathway by abrogating IRF3 activation. J Virol. 2016;90:5824–5829. doi: 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuen C.K., Chan C.P., Fung S.Y., Wang P.H., Wong W.M., Tang H.M., Yuen K.S., Chan C.P., Jin D.Y., Kok K.H. Suppression of type I interferon production by human T-cell leukemia virus type 1 oncoprotein tax through inhibition of IRF3 phosphorylation. J Virol. 2016;90:3902–3912. doi: 10.1128/JVI.00129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X., Wang K., Xing Y., Tu J., Yang X., Zhao Q., Li K., Chen Z. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014;5:912–927. doi: 10.1007/s13238-014-0104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING–TRAF3–TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]