Development of rapid, highly selective and sensitive miRNA detection in a complex biological environment has attracted considerable attention. Herein, we describe a novel two step method to construct gold-nanorod functionalized polydiacetylene (PDA) microtube for miRNA detection. In PDA microtube, with a one-dimensional (1D) waveguide nature, the excitation position and emission out-coupling position are far apart, thus helpful in reducing contribution of auto-fluorescence from biological sample. The use of specially designed toehold-mediated strand displacement reaction enables the reliable and selective discrimination of miRNA sequences with high sequence homology. Based on the condensing enrichment effect, the detection limit of the proposed PDA microtube system is as low as 0.01 nM, and it can be applied directly to detect disease-specific miRNA targets in human serum. This PDA microtube waveguide system can be further integrated into the chip for the potential applications in minimally invasive, portable clinical diagnostic equipment.
MicroRNAs (miRNAs), as a kind of short, endogenous, non-coding single-stranded RNA molecules [1,2], play an important role in gene expression. MicroRNAs usually regulate and control many important biological processes such as cell death, cell differentiation, organ development chromosome segregation, the transcription level of gene expression and so on [3-6]. The variations in miRNAs expression is found to be directly related to many human diseases, including cancer, nervous system diseases, kidney and liver diseases, cardiovascular diseases, abnormal pregnancy and thus miRNA has become a key tumor biomarker for cancer diagnosis and prognosis[7-9]. Accordingly, attempts to develop methodologies for rapid, sensitive and selective detection of miRNAs has become a major aspect. Conventional detection methods, including northern blotting [10-12], molecular cloning [13-16], real-time quantitative polymerase chain reaction [17], microarray methods [18], electrochemical methods [19], magnetic nanoparticles [20], possess some limitations such as low detection sensitivity, devoid of enough analysis, imperfect specificity, large time consuming and labor, and often requiring expensive and cumbersome instruments and skilled professionals, which limited their potential application in clinical diagnostic. Fluorescence based detection methods are currently gaining increasing attention owing to its high sensitivity and easy operation [21, 22]. However, the present fluorescence based sensors have unique limitations, including small probe size, low stability, tendency of photo-bleaching, spectral cross-talk with background fluorescence, low abundance and high sequence similarity. Therefore it is still challenging to develop a novel fluorescent based sensor system for rapid, highly selective and sensitive miRNA detection in complex biological environment.
Polydiacetylene (PDA) is an interesting material for chemical and biological sensing applications, due to its fortunate conjunction of sensitive optical characteristics (including color and fluorescence) in response to external environmental stimuli such as heat, pH, mechanical stress or ligand interactions, and easy formation in self-assembled systems (liposomes, vesicles, Langmuir-Blodgett films, casting films and nanocomposites) [23-29]. A variety of PDA based liposome or vesicle probes have been utilized to detect oligonucleotides, double-stranded DNA or single-stranded DNA (ss-DNA) [30], liver cancer biomarkers and so on [31]. However, some limitations, including low stability and selectivity, the inability for precise quantitative analysis, limited their practical applications. To overcome above problems, the immobilization of PDA particles onto the solid substrate to form two-dimensional (2D) or three-dimensional (3D) networked structures has been proposed [32-34]. However, this immobilization process has resulted in intrinsic limitation of low sensitivity. As reported [34], the detection limit of the 2D and 3D PDA based microarrayed sensor chip was found to be in the range of millimole (mM) and micromole (μM) concentration, respectively, which is far higher than the typical levels of miRNA in serum (200 aM-20 pM [35]), limiting their practical applications. Moreover, most above mentioned PDA based probes lack selectivity, and often suffered from high background fluorescence interference in the complex biological environment due to that the fluorescence excitation position and emission out-coupling position are the same. Therefore, it remains challenging to develop novel PDA sensor system for the ultrasensitive and highly selective detection of miRNA in the complex biological environment including serum.
In our previous work [36, 37], a novel PDA sensor system based on single PDA microtube waveguide for selective and sensitive detection of picric acid in aqueous media has been fabricated. This approach has several advantages, including high stability and sensitivity, low background fluorescence interference, easy chemical modification, easy integration with other optical devices towards making miniaturized sensors. Herein, we fabricated gold-nanorod functionalized PDA (Au@PDA) microtubes by a novel two step method. Compared to direct hybridization, toehold-mediated recognition mechanism has the ability to enhance the specificity of nucleic-acid recognition. When the target miRNA displaces the complementary DNA within the surface of the microtube, the gold nanorods could be removed from the surface of PDA microtube, and the waveguide coupled tip emission of PDA microtube would be recovered. Further, tethering of hydrophilic PDA microtube on a hydrophobic substrate, ultratrace miRNA sensing (the detection limit is as low as 0.01 nM) could be achieved due to the condensing-enrichment effect. We expect that this work not only provides novel PDA sensor system for the selective and ultra-trace sensing of miRNA but also has great fundamental value for the rational design of novel PDA based sensor system and the understanding of their fluorimetric sensing mechanism.
2. Experimental Section
2.1 Reagents and instrumentation
All oligonucleotides used in this work were purchased from Shanghai Bio-Engineering Company (Shanghai, China), and the sequences of DNA and miRNA used in this work were shown in Table S1 (Supporting Information). 10,12-pentacosadiynoic acid were purchased from Tokyo Chemical Industry Co., Ltd. and used as received. All other solvents and reagents were of analytical grade and used as received. Milli-Q water (18.2 MΩ cm) was used in all cases. Oligonucleotides were quantified by UV-vis absorption spectroscopy (the absorbance at 260 nm), using a SHIMADZU UV-2550 PC spectrophotometer. Fluorescence spectra were measured on a JY-ihR 550 spectrophotometer. Optical microscopy images were obtained using a BX-51 fluorescence microscope. Transmission electron microscopy (TEM) characterization was performed on a JEOL-2000 microscope (operated at 200kV). X-ray photoelectron spectroscopy (XPS) characterization was obtained by using a VG ESCALAB MK-II photoelectron spectrometer. The Raman characterization was performed on a LABRAM-HR Confocal Laser Micro Raman Spectrometer with 514.5 nm radiation at room temperature. All experiments were carried out at room temperature.
2.2 The preparation of Au@PDA composite microtube
Au@PDA composite microtubes were prepared by two-step method illustrated in Figure 1a. The single-stranded DNA (5’-SH-CAGACTGATGTTGA-3’) functionalized gold nanorods (DNA@Au NR) were prepared using a similar procedure described in the literature [38]. Amine-functionalized PDA microtubes were prepared by a hierarchical self-assembly procedure as reported previously [36]. Then aqueous solution of allylglycidyl ether (200μl, 3 mmol) was added dropwise to above amine-functionalized PDA microtubes suspension solution. The vinyl-functionalized PDA microtubes were successfully prepared since the epoxy group of allylglycidyl ether has great tendency to react with amino group within the surface of PDA microtube. Then DNA (5’-SH-ACATCAACATCAGTCTGATAAGCTA-3’) modified PDA microtubes were prepared by click reaction, as described in Figure 1a. Finally, Au@PDA composite microtubes could be prepared through simple DNA hybridization process. The preparation process was monitored by XPS and Raman analysis.
Figure 1.
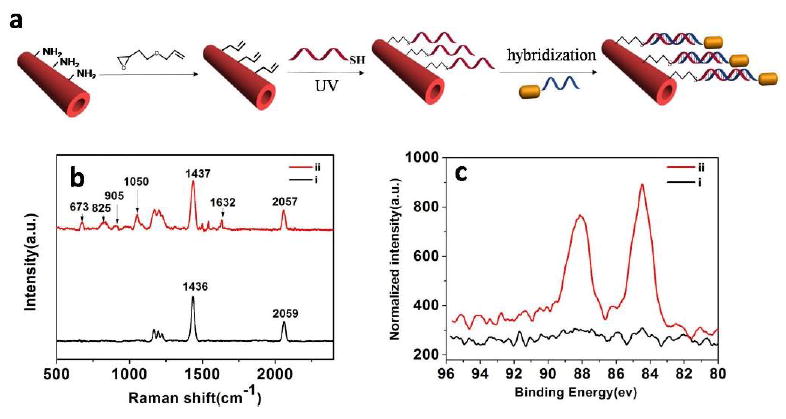
(a) Schematic illumination of the preparation process of Au@PDA microtubes. (b) Raman spectra of (i) pure PDA microtube and (ii) DNA functionalized PDA microtube. (c) Au 4f XPS spectra of PDA microtube (i) before and (ii) after modification with DNA@AuNR.
2.3 The detection experiments of miRNA-21
The detection experiments for miRNA-21 in buffer were carried out by placing as-prepared single Au@PDA composite microtube into above miRNA-21 solution at room temperature for 2 hours. Then single Au@PDA composite microtube was placed on the glass after rinsing for 4 times. A 532 nm excitation light was launched onto the body of the microtube and the out-coupled fluorescence at the tip of the microtube was measured, which was found to be dependent on the concentration of the miRNA-21. In order to test the specificity of miRNA detection, similarly sequenced miRNA including miRNA-141, miRNA-199a, single-base mismatched miRNA-21, three-base mismatched miRNA-21, were considered as ideal models to demonstrate the specificity of the assay. Moreover, single Au@PDA composite microtube was placed on hydrophobic substrate (modified with octadecyltrichlorosilane, OTS) and it was anticipated that the sensitivity of above PDA microtube probes would be enhanced based on condensing-enrichment effect. Further above PDA microtube probes were directly applied to detect the miRNA-21 in complex biological environment, e.g. the mixtures containing 1% human serum in phosphate buffered saline (PBS: 300mM NaCl, pH 7.4).
3. Results and Discussion
3.1 Fabrication of 1D Au@PDA microtube
Au@PDA composite microtubes were prepared by two-step method, which was illustrated in Figure 1a. Amine-functionalized PDA microtube, about 1-3 μm in the outer diameter (as shown in Figure S1), were prepared by a hierarchical self-assembly procedure [36]. The vinyl-functionalized PDA microtubes were prepared by incubating above amine-functionalized PDA microtube in aqueous solution of allylglycidyl ether, since the epoxy group has great tendency to react with the basic amino group of PDA microtube surface. As shown in Figure S2, two new bands (1130 cm-1 for C-O-C stretching vibration and 923 cm-1 for C=CH stretching vibration) emerged in FT-IR spectra, indicating that vinyl-functionalized PDA microtube were prepared successfully. DNA (5’-SH-ACATCAACATCAGTCTGATAAGCTA-3’) modified PDA microtubes were prepared via thiol-ene based click reaction. As shown in Figure 1b, before click reaction, there are two characteristic bands at 1436 and 2059 cm−1, which should be attributed to the C=C and C≡C stretching vibrations of PDA backbone. After incubation, new bands emerged at 673, 825, 905, 1050, 1632 cm−1, which should be attributed to the ring stretching mode in guanine, ring breathing mode in cytosine, CN bond stretching in guanine, symmetric stretching of phosphate backbone, and C=O bond stretching in thymine, respectively [39]. The XPS analysis was consistent with the Raman results (Figure S3). After click reaction, a new C 1s component was observed at 286 eV, which should be mainly assigned to the formation of C-S bond [40]. All above results confirmed the successful anchoring of the single-stranded DNA (ss-DNA) onto the surface of PDA microtube. To estimate the amount of the reacted DNA, the relative absorbance change for the reacted DNA solution was quantitatively analyzed, since the absorbance intensity at 260 nm was dependent on the concentration of DNA. The density of the modified ss-DNA within the surface of PDA microtube was calculated to be about 20 fmol • μm-2.
DNA functionalized gold nanorods (DNA@AuNR) were successfully prepared following the similar procedure [38]. The anisotropic gold nanorods (about 35 nm in length and 10 nm in width) showed the typical absorptions of a transverse SPR peak at about 515 nm and a longitudinal peak at 708 nm (Figure S4a and b). After modification with ss-DNA (5’-SH-CAGACTGATGTTGA-3’), the longitudinal peak blue-shifted to about 684 nm. As expected, in the electrophoresis analysis, DNA@Au NR run slower than that of the unconjugated ss-DNA (Figure S4c). All above results confirmed the successful anchoring of the single-stranded DNA onto the surface of Au nanorod. Using the absorption of the DNA at 260 nm in conjunction with the calculated concentration of the core Au nanorod, we estimated that the quantity of conjugated DNA to each Au nanorod was about 800, similar with that of the previous work [38]. After hybridization, the Au 4f core level region contained two peaks (84.5 and 88.1eV, respectively) could be observed, indicating that Au@PDA composite microtubes had been prepared successfully.
As an active waveguide, fluorescence could be guided within single PDA microtube through the intrinsic fluorescence of “red-phase” PDA. After hybridization, the fluorescence intensity of PDA microtube decreased quickly with increasing of the concentration of DNA@AuNR, since gold nanorods could act as the quencher to diminish the fluorescence of PDA microtube. The fluorescence quenching of PDA microtube by DNA functionalized gold nanorods could be easily discerned even at lower concentrations (0.02 nM for DNA@AuNR) and gained its maximum (about 67%) when the concentration of DNA@AuNR increased to about 0.4 nM. The Stern-Volmer quenching rate constant(Ksv) was calculated as 5.98×109M-1. Taking the fluorescence lifetime of PDA microtube in the absence of gold nanorods as 9.4 ns(Figure S5), the values of the fluorescence quenching rate constants (kq=Ksv / τ) are calculated as 6.36×1017 M−1 s−1. From the data, fluorescence resonance energy transfer and/or electron transfer processes play a major role in the fluorescence quenching of PDA microtube by gold nanorods[41].
3.2 Sensitive and selective detection of miRNA-21in buffer
The detection experiments were carried out by immersing as-prepared single Au@PDA composite microtube into miRNA-21 solution at room temperature for 2 hours. When the target miRNA displace the complementary DNA from the surface of Au@PDA microtube, the fluorescence enhancement is observed (Figure 3a). This is due to the quenching gold nanorods from the PDA surface are removed by miRNA-21 binding (Figure S6). Then single Au@PDA composite microtube was placed on to the glass and its optical waveguiding performance was studied by single-tube photoluminescence imaging method (Figure 3b). A 532 nm excitation light was launched onto the microtube body and the out-coupled tip emission of the microtube was characterized in detail. As shown in Figure 3b and c, the tip emission of Au@PDA microtube was very sensitive to the concentration of target miRNA-21. The enhancement in tip emission of the microtube could be easily discerned at low concentration (5 nM, for the target miRNA-21) and it gained maximum when the concentration of target miRNA-21 increased to 10 μM (Figure 3c). The monotonic increase of the tip emission could be directly used for miRNA detection. A linear relationship between the relative fluorescence change at 640 nm and the concentration of target miRNA-21(0-20 nM) was observed, indicating that above microtube could be applied to accurately detect the concentration of target miRNA-21 in aqueous solution (Figure 3d). The detection limit (3σ/slope) was about 2 nM, which is comparable to that of the PDA vesicle sensor system [30]. However, the present detection limit was still far higher than the typical levels of miRNA in serum, and further efforts should be made to enhance the sensitivity of PDA based sensor system. It was reasonable to use the chronological development of the relative change in tip emission at 640 nm (ΔI/I0) to determine the response time of the probe (Figure S7). The relative change of the tip emission intensity at 640 nm reached approximately 30% within 30 min, and gained its maximum over 120 min.
Figure 3.
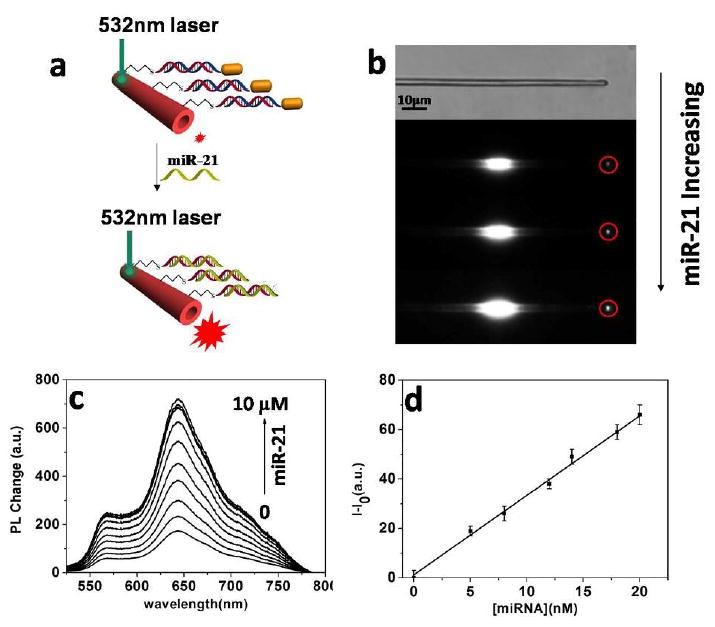
(a) Schematic illumination of single Au@PDA microtube waveguide system for miRNA-21 detection. (b) White-field image of the single PDA mictrotube and the images of single Au@PDA microtube waveguide sensor upon gradual addition of miR-21. (c) Changes in the out-coupled emission at the tip of PDA microtube with increasing concentrations of miRNA-21. (d) Plot of fluorescence-enhancement of the out-coupled tip emission of PDA microtubes upon gradual addition of miR-21 (ranged from 0 to 20nM). I0 and I represent the tip emission intensity of PDA microtube at 640 nm before and after displacement reaction with miRNA-21, respectively.
The selective sensing of miRNA-21 is highly desirable for practical applications. Compared to direct hybridization, toehold-mediated recognition mechanism has the ability to enhance the specificity of nucleic-acid recognition. As shown in Figure 4a, no obvious fluorescence enhancement could be detected upon addition of other RNA with high sequence similarity including miRNA-144, miRNA-199a, single-base mismatched miRNA-21, three-base mismatched miRNA-21 as high as 1μM. However, upon addition of only 0.2 μM miR-21, a significant change in tip emission for the microtube is observed, which confirmed that Au@PDA microtube could act as highly selective probe for miRNA-21. Another essential requirement was minor or no interference from other RNA with high sequence similarity. As shown in Figure 4b, upon the initial addition of the mixture of other four RNA (miR-144, miR-199a, single-base mismatched miRNA-21, three-base mismatched miRNA-21, each 1μM), the relative change in tip emission for the probe only reached about 3%. However, upon addition of 0.2 μM miRNA-21, remarkable enhancement in tip emission could be observed (Figure 4b). Minor interference from other RNA confirmed that Au@PDA microtube waveguide probes could detect miRNA-21 selectively even in the presence of other RNA with high sequence similarity.
Figure 4.
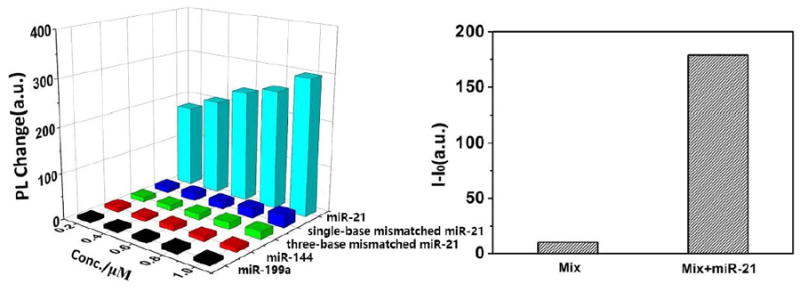
(a) Corresponding out-coupled emission change at the tip of PDA microtube upon addition of miRNA-21 or other analytes. b) The fluorescence enhancement in tip emission at 640 nm with a mixture of other RNA (mix: miR-21, single-base mismatched miR-21, three-base mismatched miR-21, miR-144, miR-199a, each 1μM) before and after addition of miR-21 (0.2 μM) to above mixture solution.
3.3 Highly sensitive detection of miRNA-21 based on condensing enrichment effect
The combination of enrichment with detection may provide an opportunity for the highly sensitive detection of miRNA. As reported in the literature [41], the analyte droplet can be concentrated to a small area after solvent evaporation due to the small interface between the hydrophobic substrate and the droplet. Au@PDA microtube placed on the hydrophobic substrate was used for the detection experiments. It is anticipated that the analytes would be enriched and then anchored onto the surface of the Au@PDA microtube due to the wettability difference between hydrophilic microtube (water contact angle of 31.3° ± 0.4°) and the hydrophobic substrate (modified with OTS, water contact angle of 103.2° ± 0.3°). Based on the condensing-enrichment effect, the effective contact frequency between the target miRNA and Au@PDA microtube would be enriched, thus allowing for highly sensitive detection of miRNA. A drop of miRNA-21 solution was placed on the sample; the droplet would not spread out and could be concentrated into a small area (Figure S8, the droplet of target was dried at 25 °C and 40% relative humidity). After rinsing for 3 times, a 532 nm excitation light was launched onto the body of the microtube and the tip emission of the microtube was characterized in detail. As shown in Figure 5b, Au@PDA microtube on the hydrophobic substrate exhibit more sensitive to the target miRNA-21, and the robust enhancement in tip emission could be easily discerned even at low concentration (0.05 nM). A linear relationship between the relative fluorescence change at 640 nm and the concentration of target miRNA-21(0.05-1nM) was observed for the microtube placed on the hydrophobic substrate, and the detection limit (3σ/slope) was about 0.01 nM, comparable to the typical levels of miRNA in serum. Thus, at least a two order increase in the sensitivity could be obtained with the sample placed on hydrophobic substrate. For comparison, the detection experiments for miRNA-21 were also carried out with Au@PDA microtube that placed on commercial hydrophilic glass slides (water contact angle of 15.6° ± 0.3°). When a drop of miRNA-21 solution was placed onto the sample, the solution spread over the whole substrate and no obvious fluorescence change could be detected even with higher concentration (1 nM). No obvious fluorescence change could be detected when single Au@PDA microtube was directly immersed into the miRNA-21 solution even with a far higher concentration (1 nM). All above results demonstrated that, by putting hydrophilic PDA microtube on a hydrophobic substrate, highly sensitive detection of miRNA could be achieved based on the condensing-enrichment effect (the detection limit is as low as 0.01 nM). Further, to investigate the reproducibility of the Au@PDA microtube probes, a series of measurements were carried out upon addition of 1 nM miRNA-21(as shown in Figure S9). It showed a good reproducibility, which was calculated to have a relative standard deviation of 2.3%.
Figure 5.
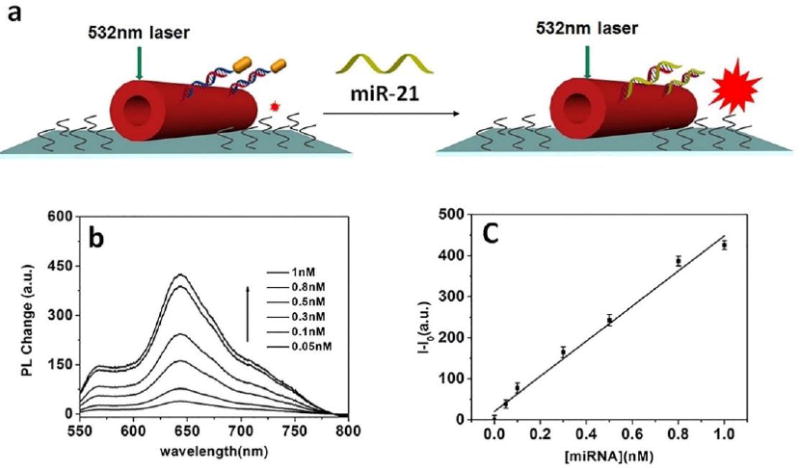
Figure 5(a) Schematic illumination of single Au@PDA microtube waveguide system placed on hydrophobic substrate for highly sensitive detection of miR-21 based on condensing-enrichment effect. (b) Fluorescence change of the out-coupled tip emission of PDA microtubes placed on hydrophobic substrate upon gradual addition of miR-21. (c) Plot of fluorescence-enhancement of the out-coupled tip emission of PDA microtubes placed on hydrophobic substrate upon gradual addition of miR-21.
For the target miRNA detection, one of the essential requirements was to minimize the interference from the complex biological environment, e.g., human serum. To prove above idea, the detection experiments based on Au@PDA microtube waveguide probes placed on hydrophobic substrate were even carried out in the miRNA-21 mixture solution containing 1%human serum. As shown in Figure 6, the robust enhancement in tip emission (480%, relative to the control test) could be easily discerned even at low concentration (0.1 nM), and it gained maximum when the concentration of target miRNA-21 increased to about 100 nM. The monotonic increase of the tip emission indicated that above Au@PDA microtube waveguide probes placed on hydrophobic substrate could be directly used for miRNA detection even in the complex biological environment.
Figure 6.
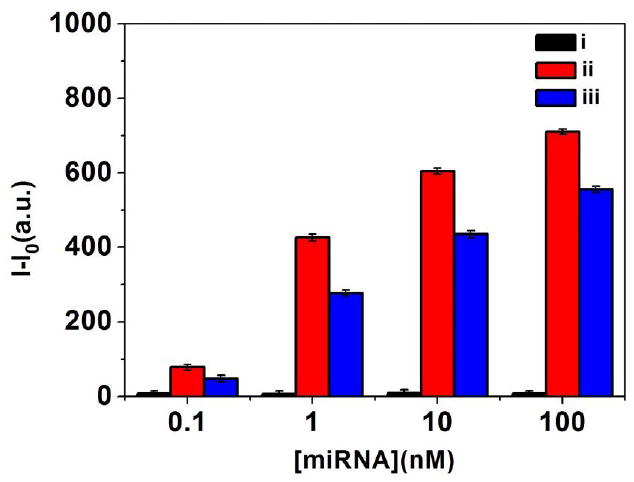
The fluorescence enhancement of tip emission for single PDA microtube after replacement reaction in (i) control solution, (ii) in PBS buffer solution and (iii) in the mixture solution containing 1% diluted human serum.
We must note here that a robust decrease in signal intensity (especially at lower concentration, about 50%) in mixture containing 1% diluted human serum could be observed compared to that in PBS buffer (Figure 6). Future work will be focused on optimizing the modified functional moieties and the structure of the composite microtube waveguide for minimizing the interference from the complex biological environment and further improving the sensitivity and selectivity of the probes.
3. Conclusion
In summary, we developed novel Au@PDA microtube and present its utility as a highly selective and sensitive probe for miRNA-21 detection in buffer. The detection limit of the proposed sensor system could be promoted to 0.01 nM based on condensing-enrichment effect, when Au@PDA microtube was placed on the hydrophobic substrate. Above PDA sensor system exhibited excellent reproducibility and could be applied directly to detect disease-specific miRNA targets even in mixture containing human serum. It is anticipated that the DNA based recognition units and the structure of the composite microtube waveguide could be optimized to further improve their sensitivity and selectivity, and this single microtube probes might be integrated into the chip for early clinical diagnosis in complex biological environment.
Supplementary Material
Figure 2.
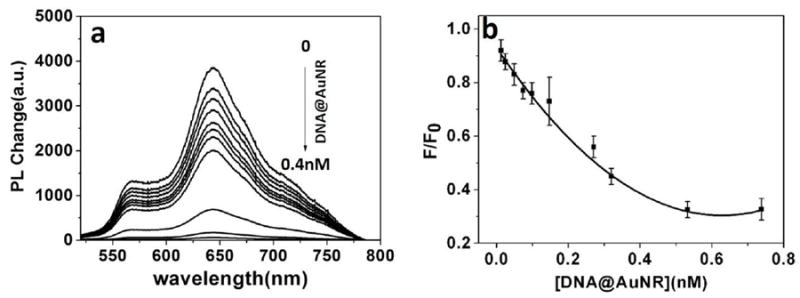
(a) PDA microtubes fluorescence quenching upon gradual addition of DNA@Au NR. (b) Plot of fluorescence-quenching ratios at 640 nm for the microtube upon gradual addition of DNA@AuNR. F0 and F represent the emission intensity at 640 nm from the body of PDA microtube before and after hybridization.
Acknowledgments
This research was carried out with funding from the National Natural Science Foundation of China (51273186, 21574120, and 91027024), the Chinese Academy of Sciences (kjcx2-yw-m11), the Basic Research Fund for the Central Universities (WK2060200012) and Science and Technological Fund of Anhui Province for Outstanding Youth (1608085J01, 1608085J02).
Contributor Information
Douguo Zhang, Email: dgzhang@ustc.edu.cn.
Yue Yu, Email: yuyuemd@yahoo.com.cn.
Gang Zou, Email: gangzou@ustc.edu.cn.
References
- 1.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croce CM. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rana TM. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Seitz H, Zamore PD. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Chen DB, Wang W. Biol Reprod. 2013;88:130. doi: 10.1095/biolreprod.113.107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trionfini P, Benigni A, Remuzzi G. Nat Rev Nephrol. 2015;11:23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- 9.McDaniel K, Herrera L, Zhou T, Francis H, Han Y, Levine P, Lin E, Glaser S, Alpini G, Meng F. J Cell Mol Med. 2014;18:197–207. doi: 10.1111/jcmm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. Nucleic Acids Res. 2010;7:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau NC, Lim LP, Weinstein EG, Bartel DP. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 16.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DZ, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Nucleic Acid Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson JM, Parker J, Perou CM, Hammond SM. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Song SP, Fan CH. Acc Chem Res. 2010;43:631–641. doi: 10.1021/ar900245u. [DOI] [PubMed] [Google Scholar]
- 20.Liang G, Xiao L, Chen H, Liu Q, Zhang S, Li F, Kong J. Biosens Bioelectron. 2013;41:78–83. doi: 10.1016/j.bios.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Chung CH, Kim JH, Jung J, Chung BH. Biosens Bioelectron. 2013;41:827–832. doi: 10.1016/j.bios.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Zhu G, Zhang C. Anal Chem. 2014;86:6703–6709. doi: 10.1021/ac501645x. [DOI] [PubMed] [Google Scholar]
- 23.Yoon B, Lee S, Kim JM. Chem Soc Rev. 2009;38:1958. doi: 10.1039/b819539k. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Zhou G, Peng X, Yoon J. Chem Soc Rev. 2012;41:4610. doi: 10.1039/c2cs35055f. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Hong L, You X, Wang Y, Zou G, Su W, Zhang QJ. Chem Commun. 2009:1356. doi: 10.1039/b820894h. [DOI] [PubMed] [Google Scholar]
- 26.Pan XJ, Wang YL, Jiang H, Zou G, Zhang QJ. J Mater Chem. 2011;21:3604. [Google Scholar]
- 27.Chen XQ, Kang S, Kim MJ, Kim J, Kim YS, Kim H, Chi B, Kim SJ, Lee JY, Yoon J. Angew Chem, Int Ed. 2010;49:1422. doi: 10.1002/anie.200905041. [DOI] [PubMed] [Google Scholar]
- 28.Sun X, Chen T, Huang S, Li L, Peng HS. Chem Soc Rev. 2010;39:4244. doi: 10.1039/c001151g. [DOI] [PubMed] [Google Scholar]
- 29.Peng H, Sun X, Cai F, Chen X, Zhu Y, Liao G, Chen D, Li Q, Lu Y, Zhu Y, Jia Q. Nature Nanotechnol. 2009;4:738. doi: 10.1038/nnano.2009.264. [DOI] [PubMed] [Google Scholar]
- 30.Jung Yun Kyung, Park Hyun Gyu. Biosens Bioelectron. 2015;72:127–132. doi: 10.1016/j.bios.2015.04.093. [DOI] [PubMed] [Google Scholar]
- 31.Hoang Hoa Thi, Lee Taemin, Kim Byeong-Su, Han Ki-Cheol, Ahn Dae-Ro. Bioorg Med Chem Lett. 2013;23:2675–2678. doi: 10.1016/j.bmcl.2013.02.087. [DOI] [PubMed] [Google Scholar]
- 32.Park M-K. Biosens Bioelectron. 2012;35:44–49. doi: 10.1016/j.bios.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 33.Kim J-M, Lee YB, Yang DH, Lee J-S, Lee GS, Ahn DJ. J Am Chem Soc. 2005;127:17580. doi: 10.1021/ja0547275. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Lee J, Lee DW, Kim J, Lee H. Chem Commun. 2016;52:926–929. doi: 10.1039/c5cc08566g. [DOI] [PubMed] [Google Scholar]
- 35.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Br J Cancer. 2010;102(7):1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu WL, Chen YK, Jiang H, Li JG, Zou G, Zhang QJ, Zhang DG, Wang P, Ming H. Adv Mater. 2014;26:3136–3141. doi: 10.1002/adma.201305712. [DOI] [PubMed] [Google Scholar]
- 37.Yang Guang, Hu Wenlong, Xia Hongyan, Zou Gang, Zhang Qijin. J Mater Chem A. 2014;2:15560–15565. [Google Scholar]
- 38.Pekcevik Idah C, Poon Lester CH, Wang Michael CP, Gates Byron D. Anal Chem. 2013;85:9960–9967. doi: 10.1021/ac4027737. [DOI] [PubMed] [Google Scholar]
- 39.Vasudev Milana, Wu Tsai-Chin, Biswas Sushmita, Dutta Mitra, Fellow, IEEE, Stroscio Michael A, Fellow, IEEE, Guthrie Stan, Reed Mark, Fellow, IEEE, Burris Kellie P, Stewart C Neal., Jr IEEE TRANSACTIONS ON NANOTECHNOLOGY. 2011;10(1) [Google Scholar]
- 40.Escorihuela Jorge, †, Bañuls María-José, †, Grijalvo Santiago, ‡, Eritja Ramón, ‡, Puchades Rosa, †, Maquieira Ángel., *, † Bioconjugate Chem. 2014;25:618–627. doi: 10.1021/bc500033d. [DOI] [PubMed] [Google Scholar]
- 41.El-Daly Samy A, Salem Ibrahim A, Hussein Mahmoud A, Asiri Abdullah M. J Fluoresce. 2015;25:379–385. doi: 10.1007/s10895-015-1523-3. [DOI] [PubMed] [Google Scholar]
- 42.Xu Li-Ping, Chen Yanxia, Yang Gao, Shi Wanxin, Dai Bing, Li Guannan, Cao Yanhua, Wen Yongqiang, Zhang Xueji, Wang Shutao. Adv Mater. 2015;27:6878–6884. doi: 10.1002/adma.201502982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


