Abstract
FSH promotes maturation of ovarian follicles. One pathway activated by FSH in granulosa cells (GCs) is phosphatidylinositol-3 kinase/AKT. The AKT target FOXO1 is reported to function primarily as a repressor of FSH genes, including Ccnd2 and Inha. Based on its broad functions in other tissues, we hypothesized that FOXO1 may regulate many more GC genes. We transduced GCs with empty adenovirus or constitutively active FOXO1 followed by treatment with FSH for 24 hours, and conducted RNA deep sequencing. Results show that FSH regulates 3,772 genes ≥ 2.0-fold; 60% of these genes are activated or repressed by FOXO1. Pathway Studio Analysis revealed enrichment of genes repressed by FOXO1 in metabolism, signaling, transport, development, and activated by FOXO1 in signaling, cytoskeletal functions, and apoptosis. Gene regulation was verified by q-PCR (eight genes) and ChIP analysis (two genes). We conclude that FOXO1 regulates the majority of FSH target genes in GCs.
Keywords: FOXO1, granulosa cells, follicle-stimulating hormone, RNA deep sequencing
Introduction
Female fertility requires precise regulation of folliculogenesis to generate mature preovulatory granulosa cells (GCs§) within the follicle that can respond to the surge of luteinizing hormone (LH) that promotes ovulation and the resumption of meiosis within the oocyte. Immature, preantral follicles contain an immature oocyte arrested in prophase 1 (Chesnel and Eppig, 1995; Erickson and Sorensen, 1974). GCs that express receptors for follicle-stimulating hormone (FSH) are separated by a basement membrane from an outer layer of theca cells that express LH receptors and contain blood vessels that supply nutrients to the avascular follicle. In the intact animal, FSH is needed to promote GC proliferation and differentiation to a preovulatory phenotype as well as maturation of the oocyte (Kumar et al., 1997). The program of differentiation initiated by FSH in rat GCs in the intact animal is duplicated when GCs isolated from preantral follicles are placed into primary culture either on fibronectin or serum substrata and cultured in serum-free medium (reviewed in (Hsueh et al., 1984; Hunzicker-Dunn and Mayo, 2015)). However, induction of proliferation in primary cultures of rat GCs requires not only FSH but also a transforming growth factor (TGF)-β receptor agonist (El-Hefnawy and Zeleznik, 2001; Miro and Hillier, 1996; Park et al., 2005), such as activin or growth differentiation factor 9 derived from the oocyte (Hayashi et al., 1999; Lin et al., 2012).
We previously reported that the proliferative response of rat GCs to FSH plus activin, evidenced by the induction of Ccnd2, required relief from transcriptional repression mediated by the forkhead box O family member FOXO1 (Park et al., 2005). FOXO1 binds DNA as a monomer that can either repress or activate transcription in a gene-specific manner (reviewed in (Burgering and Kops, 2002)). FOXO1 can also function as a co-regulator, primarily of nuclear receptors, independently of its ability to bind DNA (reviewed in (Van Der Heide, Hoekman and Smidt, 2004)). FOXO1’s cellular localization and its ability to bind DNA (reviewed in (Van Der Heide et al., 2004)) are regulated by phosphorylation catalyzed primarily by AKT, the nodal kinase activated by the phosphatidylinositol-3 kinase (PI3K) pathway. The PI3K pathway is canonically activated by growth factor receptor tyrosine kinases, such as the insulin-like growth factor 1 (IGF1) receptor, via phosphorylation of insulin receptor substrate (IRS) proteins on select Tyr residues that mediate PI3K activation (reviewed in (Vanhaesebroeck et al., 2010)). In the absence of phosphorylation, FOXO1 is localized to the nucleus and functions as an active transcriptional regulator. Upon phosphorylation by AKT on Thr24, Ser256, and Ser319, FOXO1 binds 14-3-3 proteins, relocates to the cytoplasm, and is rapidly degraded by the ubiquitin-proteasome pathway (reviewed in (Van Der Heide et al., 2004; Zhao, Wang and Zhu, 2011)). The phosphorylation of all three sites is required for the relocation of FOXO1 to the cytoplasm. A commonly used tool to illuminate FOXO1 targets is an adenovirally expressed FOXO1 mutant in which the three AKT phosphorylation sites are mutated to Ala (designated FOXO1(A3)), rendering FOXO1 “constitutively active” in the nucleus.
The FOXO family of proteins regulates many biological processes, including cell proliferation, DNA repair, apoptosis, cell metabolism, glucose and fatty acid metabolism, aging, autophagy, responses to oxidative stress, and pluripotency of embryonic stem cells (reviewed in (Puthanveetil, Wan and Rodrigues, 2013; Van Der Heide et al., 2004; Zhao et al., 2011)). As a result of this broad range of functions, FOXO’s in general and especially FOXO1 regulate a large number of gene targets in a variety of tissues including skeletal and cardiac muscle, adipose tissue, and pancreatic β-cells (reviewed in (Lettieri Barbato, Aquilano and Ciriolo, 2014; Puthanveetil et al., 2013; Sanchez, Candau and Bernardi, 2014; Szydlowski, Jablonska and Juszczynski, 2014)).
In immature rat GCs, FSH signals via its G-protein coupled receptor to promote the protein kinase A (PKA)-dependent activation of the PI3K pathway (Hunzicker-Dunn et al., 2012; Law and Hunzicker-Dunn, 2016; Puri P; Zhou et al., 2013) by regulating the tyrosine phosphorylation of IRS1 by the IGF1R (Law and Hunzicker-Dunn, 2016). AKT activated downstream of PI3K in FSH-treated cells then phosphorylates FOXO1 to abrogate its transcriptional activity (Park et al., 2005), be it activating or repressing.
We showed in immature GCs transduced with the FOXO1(A3) mutant and treated with FSH and activin, that FOXO1 repressed the induction of Ccnd2, Cyp19a1, Ereg, Inha, Nr5a1, and Nr5a2 (Park et al., 2005). A more recent report extended the list of genes to 315 that were repressed and 57 genes that were up-regulated by constitutively active FOXO1(A3) in the presence of FSH (and absence activin) (Liu et al., 2009). The most significantly regulated genes were those repressed by FOXO1 and represented genes that mediate primarily lipid and steroidogenic biosynthetic pathways (Liu et al., 2009). Conditional deletion of FOXO1 and FOXO3 from GCs revealed additional genes that were up-regulated by FOXO1 in the absence of FSH, some of which are associated with the IGF pathway, that included Ctgf, Nr0b1, Igf1, Irs2, and Fbn2 (Liu et al., 2013).
However, based on the broad cellular functions of this transcriptional regulator in many tissues, we hypothesized that FOXO1 may regulate the expression of many more genes in GCs. We therefore transduced GCs with empty recombinant adenovirus or constitutively active FOXO1(A3) followed by treatment with FSH for 24 hours, and conducted RNA deep sequencing. Results show that FSH regulates 3,772 genes ≥ 2.0-fold of a total of 13,461 genes detected, and that 60% of the genes regulated by FSH are activated or repressed by FOXO1. Pathway Studio Analysis revealed enrichment of genes repressed by FOXO1 in metabolism, signaling, transport, development, and activated by FOXO1 in signaling, cytoskeletal functions, and apoptosis. Gene regulation was verified by q-PCR and ChIP analysis for select genes. Together these results show that FOXO1 regulates the majority of FSH target genes in GCs.
Materials and Methods
Materials
FSH-19 was obtained from the National Hormone and Pituitary Agency of the National Institute of Diabetes and Digestive and Kidney Diseases (Torrance, CA). The following were purchased from the indicated sources: forskolin from Sigma; human fibronectin from Corning (Bedford, MA); DMEM/F12 from Gibco, Life Technologies (Grand Island, NY); Halt Protease Inhibitor cocktail from Thermo Fisher Scientific (Rockford, IL). Antibodies were obtained from the following sources: anti-FOXO1 (FKHR H128) (rabbit, Santa Cruz; for use in western blot), anti-phospho-Ser256 FOXO1 (rabbit, Cell Signaling), anti-SHP2 (mouse, Santa Cruz), anti-Gαq (rabbit, Santa Cruz), and anti-FOXO1 (C29H4) (rabbit, Cell Signaling; for use in ChIP assay).
Animals and primary rat GC isolation
Rats were maintained under protocols approved and in accordance with the Washington State University Animal Care and Use Committee. Ovaries were harvested from immature female rats that had been injected subcutaneously once daily with 1.5 mg estradiol-17β in 0.1 mL propylene glycol on days 21 to 23 to promote preantral follicle growth. GCs were collected from harvested ovaries and plated at a density of ~3-5 ×106 cells/mL in DMEM/F12 serum-free media supplemented with 10 nM estradiol, 100 U/mL penicillin, and 100 μg/mL streptomycin onto fibronectin-coated plates, as previously described (Law et al., 2013), and treated as indicated.
Adenoviral transduction
GCs were transduced 7-10 hours after plating with control empty adenovirus (Ad-E), Ad-wild type (WT) FOXO1, or Ad-FOXO1(A3) (Park et al., 2005). The following morning, media was replaced with fresh media and after five hours, cells were treated with either vehicle (media) or 50 ng/mL FSH for 24 hours and then harvested for RNA isolation, as described below.
RNA isolation, cDNA synthesis, and q-PCR
GCs were treated with either vehicle (media) or 50 ng/mL FSH for the indicated times, rinsed with PBS, and Isol-RNA (5 Prime) was added followed by RNA isolation according to the manufacturer’s protocol. cDNA was synthesized from DNase-treated RNA (500-1,000 ng) using qScript cDNA Supermix (Quanta Biosciences) following manufacturer’s protocol. cDNA was diluted 1:5-1:15 prior to analysis by q-PCR. q-PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems) in a 20 μL final volume using a 7500 Fast Real time machine (Applied Biosystems). 2−ΔCt was determined using Rpl19 as the endogenous load control. Primer sequences are listed in Supplemental Table 1. Primers were optimized for primer concentration, primer efficiency was confirmed to be between 90-110% efficient, and melt curves analyzed. Experiments were performed three times and statistics was performed using Graphpad Prism® version 6.01. Statistical significance was evaluated by one-way ANOVA followed by Tukey’s multiple comparisons test, when multiple comparisons were made, or by unpaired Student’s t test, as indicated.
Western blotting
GCs were treated with either vehicle (media) or 50 ng/mL FSH for indicated times and harvested by rinsing with PBS and then scraping in 1X Stop buffer (Hunzicker-Dunn, 1981) followed by boiling 10-20 min to yield total cell extracts. Equal volumes of total cell extracts were loaded onto 4-20% Criterion gradient gels (BioRad). Rest of techniques are as previously described (Law et al., 2013).
ChIP assay
GCs were plated and treated the next morning for 1 hour with either vehicle (media) or 50 ng/mL FSH. ChIP assays were performed as previously described (Law et al., 2013; Weck and Mayo, 2006), except with indicated antibodies and sonication on ice 6 times for 15 sec each at setting one with a Sonic Dismembrator Model 100 (Fisher Scientific), resting cells 1.5 min on ice between sonications. The linear range for PCR reactions was determined and cycle number within the linear range was used in the analysis. Primer sequences are listed in Supplemental Table 1.
RNA sequencing(seq)
GCs were transduced with Ad-E and treated for 24 hours with vehicle or FSH, or with Ad-FOXO1(A3) and treated for 24 hours with FSH. Poly(A) mRNA was isolated from total RNA by the MicroPoly(A)Purist kit (Ambion), which excludes microRNA, rRNA, and long non coding RNA. Twenty ng of mRNA was used to construct the Ion RNA-Seq version 2 sequencing libraries by the Washington State University Molecular Biology and Genomics Core. The libraries were sequenced on an IonTorrent PGM with a single sample per Ion 318 semiconductor chip with 520 flows, generating an average 4 million 110 bp reads/sample. Mapping rate was 87%. CLC Bio genomics workbench v8.1 was used for mapping the reads from the sequencer. Results were mapped to the rat genome data base in October of 2015. Supplemental Table 2 shows genes detected in cells transduced with Ad-E and treated 24 hours with vehicle versus FSH; Supplemental Table 3 shows genes detected in cells treated with FSH for 24 hours following transduction with Ad-E versus Ad-FOXO1(A3). Regulated genes were arbitrarily designated as those with fold changes in RPKM values (Reads Per Kilobase of exon model per Million mapped reads, calculated as (total exon reads)/(mapped reads in millions) (exon length in kb)) ≥ 2.0. RPKM values of 0 for one treatment group results in fold changes of ∞. Infinity values were deleted from Supplemental Tables 2 and 3 when unique gene reads (UGRs; number of reads that match uniquely to a gene or its transcripts) for both treatment groups were < 20. Infinity values were included in counting up- or down-regulated genes: if UGRs were ≥ 20 for one treatment group, if UGRs exhibited a 3-fold increase or decrease between treatment groups, and if RPKM for one treatment group was ≥ 2.0 (such as Cyp19a1 in Supplemental Table 2). Fold change of 1.0 for treatment groups was also deleted from Supplemental Tables 2 and 3 when RPKM for both treatment groups was 0. Deleted genes were not included in gene counts. A summary of genes from Supplemental Tables 2 and 3, selected either for their known functional significance to GC maturation (such as Inha) or to other tissues (such as Cited2), is presented in Tables 1 and 2 in “Results”. An ontology enrichment analysis of the biological functions of genes up- and down-regulated by Ad-FOXO1(A3) versus Ad-E in GCs treated 24 hours with FSH was performed using Pathway Studio 9.0 (Elsevier) (see Supplemental Table 3 for the list of genes included in the analysis). Enriched groups with p values >0.01 (based on Fisher’s Exact Test) and those that contained genes highly duplicated in other groups were deleted. Gene ontology groups were then organized by functional pathways or groups (such as “angiogenesis”, “apoptosis”, etc.), as presented in Supplemental Tables 4 and 5 and summarized in Fig. 2 in “Results”.
Table 1.
Summary of genes down-regulated ≥ 2-fold 24 hours post treatment of FSH in GCs transduced with Ad-FOXO1(A3) versus Ad-E (Genes repressed by FOXO1 in the absence of FSH)
| Gene | Common Name | Fold change (a/b) |
FSH + Ad-E Unique reads |
FSH + Ad-E RPXM (a) |
FSH + Ad- FOXO1(A 3) Unique reads |
FSH + Ad- FOXO1(A3) RPXM (b) |
|---|---|---|---|---|---|---|
| P2rx7 | Purinergic receptor P2X, ligand gated ion channel, |
−14.71 | 19 | 1.74 | 4 | 0.12 |
| Nppc | Natriuretic peptide C | −10.55 | 41 | 25.2 | 7 | 2.39 |
| Cyp19a1 | Cytochrome P450, family 19, subfamily A, polypeptide 1 |
−6.37 | 278 | 27.7 | 58 | 4.35 |
| Dhh | Desert Hedgehog | −5.64 | 62 | 8.62 | 18 | 1.53 |
| Pappa | Pregnancy-associated plas- ma protein A, pappalysin 1 |
−4.95 | 174 | 7.79 | 66 | 1.57 |
| Sfrp4 | Secreted frizzled-related protein 4 |
−4.65 | 89 | 6.86 | 34 | 1.47 |
| Lhcgr | Luteinizing hormone/ choriogonadotropin receptor |
−3.92 | 419 | 50.96 | 139 | 13 |
| Scarb1 | Scavenger receptor class B, Member 1 |
−3.89 | 4486 | 573.11 | 1637 | 147.37 |
| Sgk | Serum/glucocorticoid regulated kinase |
−3.53 | 749 | 23.46 | 291 | 20.8 |
| Crabp2_2 | Cellular retinoic acid binding protein 2 |
−3.42 | 202 | 108.76 | 65 | 31.85 |
| Mdm2 | Mouse double minute 2 homolog |
−3.28 | 37 | 20.39 | 16 | 16.54 |
| Inha | Inhibin alpha | −3.25 | 14653 | 3720.95 | 6064 | 1146.18 |
| Kcnk5 | Potassium channel, two pore domain subfamily K, member 5 |
−2.97 | 22 | 2.09 | 16 | 0.7 |
| Inhba | Inhibin, beta A | −2.92 | 2225 | 299.69 | 1034 | 102.6 |
| Arf6 | ADP-ribosylation factor 6 | −2.71 | 234 | 74.18 | 115 | 27.38 |
| Ihh | Indian hedgehog | −2.51 | 48 | 7.49 | 26 | 2.98 |
| Cebpa | CCAAT/enhancer binding protein (C/EBP), alpha |
−2.47 | 200 | 27.23 | 108 | 11.04 |
| Jund | Jun D protooncogene | −2.39 | 551 | 119.25 | 310 | 49.98 |
| Inhbb | Inhibin, beta B | −2.36 | 720 | 82.23 | 418 | 35.24 |
| Wnt11 | Wingless-type MMTV integra- tion site family, member 11 |
−2.33 | 176 | 19.07 | 109 | 8.18 |
| Grem2 | Gremlin 2, DAN family BMP antagonist |
−2.23 | 767 | 65.62 | 493 | 29.48 |
| Id2 | Inhibitor of DNA Binding 2 | −2.18 | 633 | 112.51 | 484 | 51.59 |
| Ppp2r3b | Protein phosphatase 2, regulatory subunit B", Beta |
−2.08 | 70 | 9.37 | 51 | 4.51 |
| Fkbp5 | FK506 binding protein 5 | −2.07 | 726 | 71.94 | 480 | 34.77 |
| Csdc2 | Cold shock domain containing C2, RNA binding |
−2.05 | 379 | 46.38 | 275 | 22.6 |
Table 2.
Summary of genes up-regulated ≥ 2-fold 24 hours post treatment of FSH in GCs transduced with Ad-FOXO1(A3) versus Ad-E (Genes activated by FOXO1 in the absence of FSH)
| Gene | Common Name | Fold change (b/a) |
FSH + Ad- E Unique reads |
FSH + Ad- E RPXM (a) |
FSH + Ad- FOXO1(A3) Unique reads |
FSH + FOXO1(A3) RPXM (b) |
|---|---|---|---|---|---|---|
| Bcl2 | B-Cell CLL/Lymphoma 2 | 14.2 | 19 | 0.31 | 48 | 4.36 |
| Shb | Src homology 2 domain containing adaptor protein B |
9.72 | 7 | 0.24 | 44 | 2.34 |
| Adamts12 | Disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 12 |
7.51 | 30 | 0.84 | 266 | 6.33 |
| Vgll3 | Vestigial-like family member 3 |
6.98 | 65 | 15.16 | 568 | 105.79 |
| Ctgf | Connective tissue growth factor |
6.7 | 463 | 61.95 | 4151 | 415.12 |
| Cald1 | Caldesmon 1 | 6.57 | 658 | 46.55 | 6023 | 305.61 |
| Ets1 | v-ets avian erythro- blastosis virus E26 oncogene homolog |
4.36 | 44 | 2.66 | 229 | 121.62 |
| Bdnf | Brain-derived neurotrophic factor |
4.27 | 40 | 1.85 | 233 | 7.9 |
| Fn1 | Fibronectin 1 | 3.75 | 1926 | 82.23 | 9650 | 308.37 |
| Klf12 | Kruppel-like factor 12 | 3.61 | 67 | 1.4 | 206 | 5.08 |
| Klf6 | Kruppel-like factor 6 | 3.34 | 274 | 8.13 | 827 | 27.15 |
| Tgfb2 | Transforming growth factor, beta 2 |
3.03 | 342 | 24.99 | 1449 | 75.68 |
| Ahrr | Aryl-hydrocarbon receptor repressor |
2.99 | 20 | 0.62 | 48 | 1.85 |
| Cited2 | Cbp/P300-interacting transactivator, with Glu/Asp-rich carboxy- terminal domain, 2 |
2.8 | 229 | 42.54 | 861 | 119.03 |
| AKAP12 | A kinase (PRKA) anchor protein (Gravin) 12 |
2.66 | 33 | 1.37 | 104 | 3.66 |
| Fosl2 | Fos-like antigen 2 | 2.6 | 119 | 19.47 | 418 | 50.7 |
| Rnd3 | Rho family GTPase 3 | 2.56 | 309 | 34.67 | 1099 | 88.79 |
| Fos | FBJ murine osteo- sarcoma viral oncogene homolog |
2.43 | 13 | 2.73 | 42 | 6.64 |
| Per2 | Period circadian clock 2 | 2.39 | 28 | 1.31 | 86 | 3.14 |
| Gadd45a | Growth arrest and DNA- damage-inducible, alpha |
2.35 | 38 | 10.5 | 119 | 24.65 |
| Junb | Jun B proto-oncogene | 2.32 | 20 | 4.06 | 61 | 9.41 |
| Rictor | Rapamycin-insensitive companion of mTOR |
2.22 | 176 | 7.52 | 498 | 16.67 |
| Dusp1 | Dual specificity phosphatase 1 |
2.12 | 60 | 11.03 | 170 | 23.35 |
| WT1 | Wilms Tumor 1 | 2.1 | 45 | 4.28 | 122 | 8.99 |
| Prkca | Protein kinase C alpha | 2.08 | 82 | 3.04 | 244 | 6.32 |
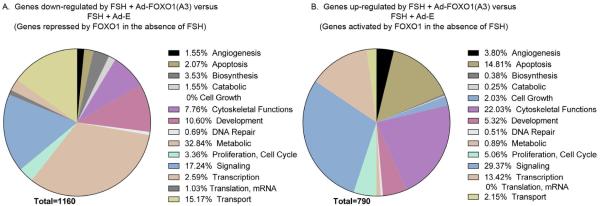
Figure 2. Summary of the predominant biological functions of genes repressed or activated by FOXO1 in the absence of FSH identified by Pathway Studio Analysis.
Panels are pie charts showing the percentage of genes down-regulated by FSH + Ad-FOXO1(A3) versus FSH + Ad-E (i.e., genes repressed by FOXO1 in the absence of FSH; Panel A) and of genes up-regulated by FSH + Ad-FOXO1(A3) versus FSH + Ad-E (i.e., genes activated by FOXO1 in the absence of FSH; Panel B). Genes identified for each biological function in Supplemental Tables 4 and 5 were counted and are reflected in Panels A and B, respectively. Biological functions that contained genes highly duplicated in other groups within the same biological function category were deleted as were enriched groups with p values >0.01 (based on Fisher’s Exact Test), as explained in Materials and Methods. A number of genes appear in different functional categories. As a result, total gene numbers do not align with results presented in Fig. 1.
Results
FSH-regulated genes
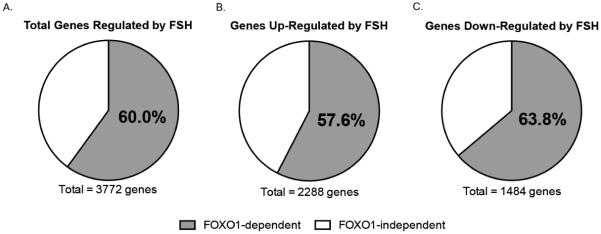
Of the 13,461 genes annotated in rat GCs by RNA Seq analysis, 2,288 (17.0%) were up-regulated ≥ 2.0-fold and 1,484 (11.0%) were down-regulated ≥ 2.0-fold by FSH; 9,689 genes (72.0%) were not regulated (i.e., regulation was < 2.0-fold) by a 24 hour treatment with FSH (Fig.1 and Supplemental Table 2). The most highly regulated genes (285- to 45-fold) included: Slc26a7, an anion exchanger; Lhcgr, the LH/choriogonadotropin G-protein coupled receptor; Cyp11a1, rate-limiting enzyme in progesterone biosynthesis; Ass1, argininosuccinate synthase involved in the urea cycle; Chst1, a carbohydrate sulfotransferase that catalyzes the sulfation of the proteoglycan keratin; Acsbg1, an acyl-CoA synthetase involved in fatty acid metabolism; Mrap, melanocortin receptor-interacting protein; Nppc, natriuretic peptide C, the precursor agonist for the natriuretic peptide receptor B (NPR2 receptor); Map2k6, a kinase that activates p38 MAP kinase; Rasd1, a member of the Ras GTPase super family; Kcnq1, a voltage-gated potassium channel; and Rxrg, retinoid X receptor gamma.
Figure 1. Summary of FSH genes regulated by FOXO1.
Panels are pie charts showing the percentage of all FSH target genes activated and repressed by FOXO1 (Panel A); the percentage of genes up-regulated by FSH that are repressed by FOXO1 (Panel B); and the percentage of genes down-regulated by FSH that are activated by FOXO1 (Panel C).
FSH-regulated genes down-regulated by constitutively active FOXO1
Of the genes up-regulated by FSH, we identified 1,317 genes (from a total of 2,288) whose expression in response to FSH was diminished (i.e., down-regulated) ≥ 2.0-fold by constitutively active Ad-FOXO1(A3) (Fig. 1 and Supplemental Table 3). This group of genes is expected to be repressed by FOXO1 in the absence of FSH; FOXO1 phosphorylation in response to FSH is expected to relieve this repression, allowing transcriptional activators to stimulate mRNA expression. (See results below for verification of these expectations.) These results suggest that 57.6% of GC genes activated by FSH are repressed by FOXO1 (Fig. 1). A summary of 25 of these genes is shown in Table 1. This group of genes encodes proteins that function as an RNA binding protein (Csdc2), regulators of intracellular signaling (Fkbp5, Ppp2r3b, Crabp2_2, Sgk, Arf6), ion channels (Kcnk5, P2rx7), a G-protein coupled receptor (Lhcgr), receptor agonists (Wnt11, Ihh, Dhh, Nppc) and antagonists (Grem2, Sfrp4), transcriptional regulators (Id2, Jund, Cebpa), hormone subunits (Inhbb, Inha), proteins involved in steroidogenesis (Cyp19a1, Scarb1), ubiquitin ligase (Mdm2), and a metalloproteinase (Pappa). In general, these genes that are repressed by FOXO1 in the absence of FSH are expected to contribute to the differentiation and proliferation responses of FSH that result in a preovulatory GC capable of responding to LH.
Predominant biological functions of enriched genes, identified by Pathway Studio Analysis (Supplemental Table 4 and Fig. 2A), whose expression is enhanced by FSH and down-regulated by constitutively active FOXO1 include: metabolism (especially small molecule, oxidation-reduction, carbohydrate, lipid, and protein metabolic processes), signaling (synaptic signaling, that includes ion transporters, potassium channels, membrane receptors; Notch, TGFβ, ephrin, activin, and TRK receptor signaling; protein phosphorylation; responses to cAMP), transport (especially transmembrane and potassium ion), development (primarily cell differentiation and development), cytoskeletal functions, biosynthesis (primarily steroid pathways), and cell proliferation (positive regulation). Underrepresented gene enrichments of biological functions included those associated with angiogenesis, apoptosis, catabolism, cell growth, DNA repair, translation, and transcription.
FSH-regulated genes up-regulated by constitutively active FOXO1
Of the genes whose expression was diminished (i.e., down-regulated) by FSH, we identified 947 genes (from a total of 1,484; 63.8%) that were up-regulated ≥ 2.0-fold by constitutively Ad-FOXO1(A3) (Fig. 1 and Supplemental Table 3). This group of genes is expected to be activated by FOXO1 in the absence of FSH; FOXO1 phosphorylation in response to FSH is expected to abrogate this activation. (See results below for verification of these expectations.) These results suggest that 63.8% of GC genes inactivated by FSH are activated by FOXO1 in the absence of FSH. A summary of 26 of these genes is shown in Table 2. This group of genes encodes proteins that function in intracellular signaling pathways (Shb, Cald1, Akap12, Rictor), transcriptional activators (Vgll3, Ets1, Klf12, Klf6, Cited2, Fosl2, Fos, Per2, Junb, Wt1) and repressors (Ahrr), growth factors (Ctgf, Bdnf, Tgfb2), a metalloprotease (Adamts12), kinases and phosphatases (Prkca, Dusp1), cell survival proteins (Bcl2, Gadd45a), and cytoskeletal proteins (Fn1, Rnd3). In general, these genes that are expressed in the absence of FSH and whose expression is reduced by FSH are expected to contribute to the maintenance of GCs in an undifferentiated stage of development and/or to inhibit differentiation of GCs to a preovulatory phenotype.
Predominant biological functions of enriched genes, identified by Pathway Studio Analysis (Supplemental Table 5, Fig. 2B), whose expression is reduced by FSH and up-regulated by constitutively active FOXO1 include: signaling (including signaling by interleukin-1; TGFβ; tumor necrosis factor and cytokines in general; gamma-aminobutyric acid; intracellular signal transduction that includes small GTPases, kinases, and phosphatases); cytoskeletal functions (especially cell adhesion, extracellular matrix, and actin cytoskeletal organization); apoptosis; transcription (both positive and negative regulation from RNA polymerase II promoter); development (although less represented compared to genes in Supplemental Table 4, Fig. 2A); and cell proliferation (both positive and negative regulation). Underrepresented gene enrichments of biological functions included those associated with angiogenesis, biosynthesis, catabolism, cell growth, DNA repair, metabolism, translation, and transport.
Validation of effect of FSH and Ad-FOXO1(A3) on select gene targets
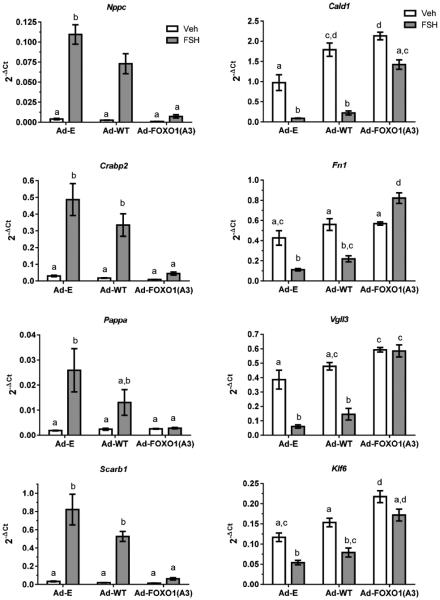
In the following experiments, we selected eight genes regulated by FSH and constitutively active FOXO1 according to RNA seq: four whose expression was up-regulated by FSH and down-regulated by Ad-FOXO1(A3), Nppc, Crabp2, Pappa, and Scarb1; and four whose expression was down-regulated by FSH and up-regulated by Ad-FOXO1(A3), Cald1, Fn1, Vgll3, and Klf6. We initially confirmed their regulation by q-PCR, as shown in Fig. 3, where cells were transduced overnight with control empty adenovirus (Ad-E), Ad-WT-FOXO1, or Ad-FOXO1(A3) and then treated for 24 hours with vehicle or FSH. Responses to Ad-WT-FOXO1 generally mirrored those of Ad-E in the absence and presence of FSH. Responses to Ad-FOXO1(A3) reversed responses to FSH in Ad-E-transduced cells. Together, these results confirm the expectation from RNA seq results: of the genes up-regulated by FSH, constitutively active FOXO1 repressed their expression in the presence of FSH (Fig. 3, left panel); of the genes whose expression is elevated in the absence of FSH and down-regulated by FSH, constitutively active FOXO1 enhanced their expression in the presence of FSH to levels seen in the absence of FSH (Fig. 3, right panel).
Figure 3. Transduction of GCs with constitutively active FOXO1 confirms that FOXO1 represses or activates FSH gene targets.
GCs were transduced overnight with Ad-E, Ad-WT-FOXO1 (Ad-WT), or constitutively active Ad-FOXO1(A3), media was replaced and 5 hours later cells were treated with vehicle or FSH (50 ng/ml) for 24 hours, as described in Materials and Methods. mRNA expression of indicated genes, relative to Rpl19 and expressed as 2−ΔCt, was determined as described in Materials and Methods. Results are means ± SEM of 3 independent experiments. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Different letter designations for each gene indicate significantly different groups (P<0.05).
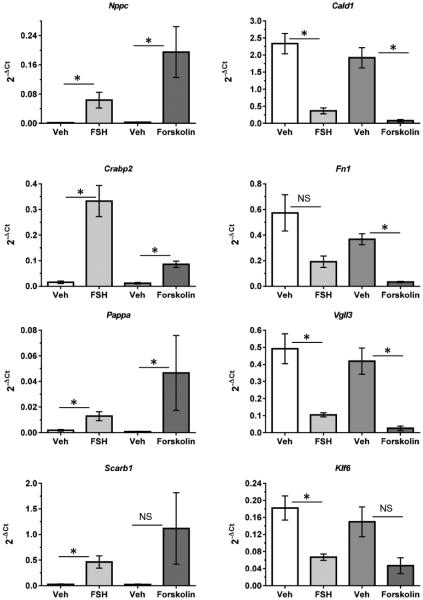
The responses of these genes to FSH were mimicked by the adenylyl cyclase activator, forskolin (Fig. 4). The ability of elevated intracellular cAMP levels to enhance expression of those genes up-regulated by FSH and to depress expression of those genes down-regulated by FSH is consistent with previous results showing the PKA-dependence of FSH signaling to activate AKT (Hunzicker-Dunn et al., 2012; Law and Hunzicker-Dunn, 2016; Puri P; Zhou et al., 2013).
Figure 4. The adenylyl cyclase activator forskolin mimics FSH to either up-regulate or down-regulate indicated genes.
GCs were treated with vehicle, FSH, or forskolin (10 μM) for 24 hours. For additional details, see legend to Fig. 3. Results are means ± SEM of 3 independent experiments. Statistical significance was determined by unpaired Student’s t test. Comparisons were made between vehicle- and FSH-, and between vehicle- and forskolin-treated samples. An asterisk indicates statistically significant differences (P<0.05); NS indicates not statistically significant.
Interaction of FOXO1 with Nppc and Cald1 promoters
We selected two genes, Nppc and Cald1, to conduct ChIP studies to show that FOXO1 indeed interacts with their gene promoters, either directly or indirectly, in the absence of FSH, and that FSH promotes the release of FOXO1. We initially compared the regulation of Nppc and Cald1 over a 72 hour time course in vehicle versus FSH-treated cells (Supplemental Fig. 1). For Nppc, a gene that is up-regulated by FSH, a maximal response was detected 24 hours post FSH treatment. mRNA levels of Nppc declined thereafter (left panel). In the absence of FSH, Nppc mRNA remained repressed over a 72 hour time course. For Cald1, a gene that is down-regulated by FSH, significant down-regulation (p<0.05) was evident only 24 hours post FSH compared to mRNA levels in vehicle-treated cells at the same time point (right panel). By 72 hours in vehicle-treated cells, levels of Cald1 had unexpectedly fallen compared to levels at 24 hours (p<0.05).
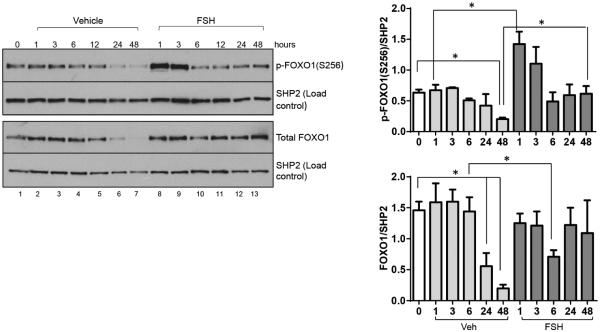
We next performed a time-course study from 1-48 hours post FSH primarily to determine the time of maximal FSH-stimulated FOXO1(Ser256) phosphorylation for subsequent ChIP analyses. Results (Fig. 5, left and top right panels) show that FOXO1 is maximally phosphorylated on Ser256 1 hour post FSH; by 6 hours, phosphorylation has returned to basal levels. We selected the 1 hour time point to conduct ChIP assays. However, we noted that total FOXO1 levels in vehicle-treated cells were unexpectedly reduced ~50% by 24 hours and ~ 80% by 48 hours while total FOXO1 levels in cells treated with FSH at these time points were not changed, except for the significant decline in total FOXO1 levels 6 hours post FSH (Fig. 5, left and lower right panels). Perhaps the large reduction in total FOXO1 protein contributes to the reduced expression of Cald1 in vehicle-treated cells cultured for 48 and 72 hours (see Supplemental Fig. 1). Based at least on the expression of this single gene (Cald1), these results suggest that over time under our in vitro culture conditions, genes expressed in the absence of FSH that may define the undifferentiated GC become compromised.
Figure 5. Time course of the phosphorylation of FOXO1 on Ser256 in GCs treated with vehicle or FSH.
GCs were plated and treated the following morning with vehicle or FSH for indicated times. Time “0” reflects cells that were not treated and was taken at the time of cell treatments. Total cell extracts were collected as described in Materials and Methods, proteins were separated by SDS-PAGE, and western blots probed with indicated antibodies (FOXO1 phosphorylated on Ser256, p-FOXO1(S256); SRC homology-2 (SH2) domain-containing tyrosine phosphatase, SHP2). Representative western blots are shown in the left panel. Results in right panels are means ± SEM of 3 independent experiments. Statistical significance was determined by unpaired Student’s t test. Comparisons for vehicle-treated cells were made relative to time 0. An asterisk indicates a statistically significant difference (P<0.05) between each vehicle treatment time and time 0 value. Comparisons for FSH-treated samples at each time point were made relative to the comparable vehicle-treated sample. An asterisk indicates a statistically significant difference (P<0.05) between vehicle- and FSH-treatment at each time point.
The Jaspar database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl) was used to identify potential FOXO1 binding sites for ChIP assays. ChIP assay results showed that FOXO1 was associated with the promoter region of both Nppc (Fig. 6A) and Cald1 (Fig. 6B) in the absence of FSH, and that FSH treatment of GCs (1 hour) reduced the detectable signal. Negative controls included both immunoprecipitation with an antibody to the G-protein Gαq, a protein localized to the plasma membrane and not the nucleus, as well as inclusion of distal primers within the coding regions of both genes. These results thus confirm that both Nppc and Cald1 are either direct or indirect targets of FOXO1.
Figure 6. FOXO1 selectively interacts with the promoter regions of Nppc and Cald1 in vehicle-treated cells; FSH promotes FOXO1 release.
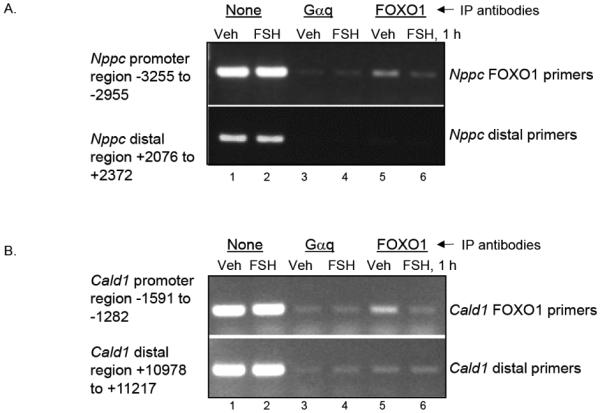
GCs were treated the morning after plating for 1 hour with vehicle or FSH. ChIP assays were conducted as described in Materials and Methods using antibodies to Gαq, as a negative control, and FOXO1. Lanes 1 and 2 reflect input DNA. Results are representative of 3 separate experiments.
Discussion
Our results show that FOXO1 is indeed a major regulator of FSH target gene expression in immature GCs. Of the 3,772 genes down- or up-regulated ≥ 2.0-fold by FSH, 60% (2,264) are regulated by FOXO1 (see Fig. 1). These results support earlier studies that concluded that FSH-dependent activation of the PI3K/AKT pathway, which leads to FOXO1 phosphorylation and nuclear exclusion, was required for follicular maturation (Liu et al., 2013; Park et al., 2005; Zeleznik, Saxena and Little-Ihrig, 2003).
Transcriptional regulators that promote gene expression in GCs in the absence of FSH are rarely investigated. Those genes expressed in the absence of FSH are expected to maintain GCs in an undifferentiated stage of development and/or to inhibit GC differentiation. We detected 1,484 genes whose expression was down-regulated ≥ 2.0 by FSH (see Fig. 1). Of those genes, the expression of 947 genes (63.8%) was enhanced ≥ 2.0 fold (in the presence of FSH) by constitutively active FOXO1 in which the three AKT sites were mutated to Ala. We validated by q-PCR that constitutively active FOXO1 in GCs treated with FSH enhanced the expression of four of these genes, Cald1, Fn1, Vgll3, and Klf6, to levels equivalent to those detected in cells cultured in the absence of FSH (see Fig. 3). Klf6 encodes a member of the Kruppel-like family of transcriptional activators. Although its gene targets in GCs are not known, KLF6 contributes to the expression of the IGF1R in some cellular models (reviewed in (Werner and Sarfstein, 2014)). The IGF1R is necessary for FSH to signal to activate PI3K (Law and Hunzicker-Dunn, 2016). Vgll3 encodes the transcriptional co-activator protein vestigial-like protein 3 that is reported to function as a tumor suppressor for epithelial ovarian cancer (Gambaro et al., 2013). Fn1 encodes the extracellular matrix protein fibronectin and is generally involved in cell adhesion and migration processes (http://www.genecards.org/cgibin/carddisp.pl?gene=FN1). Cald1 encodes the Ca2+-calmodulin and actin binding protein caldesmon that inhibits smooth muscle contraction, stabilizes actin filaments in non-muscle cells, and is reported in some contexts to function as a negative regulator of cell proliferation, cell migration, and release of metalloproteinases (Hai, 2008; Li et al., 2004; Wang, 2008). It is possible that the reduced expression of the Cald1 gene in response to FSH contributes to the ability of preovulatory GCs to enhance actin filament dynamics to promote progesterone production in response to the ovulatory surge of LH (Karlsson et al., 2010).
In view of the prominent activation by FOXO1 of genes that encode cytoskeletal proteins in the absence of FSH (see Supplemental Table 5, Fig. 2B), we selected to investigate Cald1 based on its prominent role in stabilizing actin filaments (Wang, 2008). ChIP analysis showed that Cald1 binds FOXO1 either directly or indirectly (see Fig. 6B). We noted that while a 24-hour treatment of GCs with FSH indeed reduced the expression of Cald1 mRNA ~50% (see Supplemental Fig. 1), thereafter Cald1 mRNA expression dropped an equivalent amount in vehicle-treated GCs, likely a consequence of the unexpected reduction of total FOXO1 at 48 hours under our culture conditions (see Fig. 5). We do not know if there are similar reductions in the expression of other genes that are transcriptionally activated by FOXO1 in GCs cultured greater than 24 hours in the absence of FSH.
Although there is a dearth of information on the cellular functions of undifferentiated GCs, it is expected that the genes and relevant transcriptional factors expressed in the absence of FSH function to maintain the undifferentiated state of these cells. Based on Pathway Studio Analysis of genes activated by FOXO1 and repressed by FSH, dominant genes appear to be those that enhance apoptosis (such as Gadd45, Cidea, Prune2, and Nuak2) and those that promote cell adhesion (Nedd9, Vcl, Ctgf, Fn). There is also an abundance of genes involved in signaling, including membrane receptor and receptor agonists, kinases and phosphatases, and small G-proteins, although these signaling pathways have not been investigated.
In striking contrast to the limited information on the cellular functions and transcriptional regulators that promote gene expression in GCs in the absence of FSH, the ability of FSH to enhance the expression of GC target genes that characterize the mature preovulatory phenotype has been intensely investigated. Up-regulated genes that portray the preovulatory follicle include Lhcgr, Star, Cyp11a1, Cyp19a1, Inha, Prkar2b, Egfr, and others (reviewed in (Hunzicker-Dunn and Mayo, 2015)). Indeed, our results reveal that FSH up-regulates the expression of 2,288 genes ≥ 2.0 fold (see Fig. 1). Many of the transcriptional activators that contribute to the induction of a subset of these genes in GCs, including Cyp19a1 (Bennett et al., 2012; Carlone and Richards, 1997; Parakh et al., 2006; Tremblay and Viger, 2001), Cyp11a1 (Clemens et al., 1994; Eimerl and Orly, 2002; Kurten and Richards, 1989; Oonk et al., 1990; Sher, Yivgi-Ohana and Orly, 2007), Lhcgr (Chen et al., 1999; Law et al., 2013; Shi and Segaloff, 1995), and Inha (Ito, Achermann and Jameson, 2000; Luo et al., 2012; Pei et al., 1991; Weck and Mayo, 2006), have been identified. Our results indicate that FOXO1 represses the expression of 1,317 GC genes that are up-regulated by FSH as a consequence of relief from repression in concert with transcriptional activation. We validated by q-PCR that constitutively active FOXO1 in FSH-treated GCs promoted the down-regulation of four of these genes, Nppc, Crabp2, Pappa, and Scarb1, to levels equivalent to those detected in the absence of FSH (see Fig. 3). Scarb1 encodes for scavenger receptor class B, member 1, a plasma membrane receptor for high-density lipoprotein (HDL) cholesterol. SCARB1 mediates the transfer of cholesterol from HDLs to steroidogenic pathways, has recently been shown in a proteomic analysis of lipid droplets in rat GCs to be enriched in cholesterol-ester lipid droplets (Khor et al., 2014), and is necessary for progesterone production by GCs (Kolmakova et al., 2010; Yates et al., 2011). Pappa encodes a secreted metalloproteinase which cleaves insulin-like growth factor binding proteins (http://ww.genecards.org/cgi-bin/carddisp.pl?gene=PAPPA). Its expression is linked to selection of the dominant follicle in cattle (Luo et al., 2011); deletion results in reduced fertility and steroidogenesis in mice (Nyegaard et al., 2010). Crabp2 encodes the cellular retinoic acid binding protein 2. Retinoic acid contributes to oocyte maturation (Bowles and Koopman, 2010; Tahaei et al., 2011). Nppc encodes natriuretic peptide precursor C that, upon proteolytic cleavage, generates C-type natriuretic peptide that functions as the agonist of the guanylyl cyclase membrane receptor NPR2. In ovarian follicles, mural GCs (that line the basement membrane) produce C-type natriuretic peptide that binds NPR2 on the surface of mural GCs and of nearby cumulus GCs that surround the oocyte to trigger production of cGMP (Shuhaibar et al., 2016). cGMP then transits into the oocyte and maintains meiotic arrest of fully grown oocytes until the ovulatory surge of LH that reduces oocyte levels of cGMP to trigger the resumption of meiosis (Lee et al., 2013; Norris et al., 2009; Vaccari et al., 2009; Zhang et al., 2010). Previous studies have documented that FSH or an FSH receptor agonist (pregnant mare’s serum gonadotropin) enhances expression of Nppc by GCs (Kawamura et al., 2011; Lee et al., 2013) and the corresponding increase in cGMP (Gutkowska et al., 1999), although transcriptional regulators of Nppc expression have not been investigated. Our results demonstrate that Nppc expression in the absence of FSH is repressed by FOXO1, and that FSH relieves this repression (see Figs. 3 and 6). The transcriptional factors that promote Nppc expression and are activated by FSH remain to be elucidated.
It is generally recognized that the differentiation response of GCs to FSH includes but is not restricted to the following: steroid hormone biosynthesis (estrogen and progesterone); membrane receptor biosynthesis (LHCGR, prolactin, EGFR, lipoprotein receptors), protein hormone biosynthesis (inhibin, relaxin), enhanced carbodydrate metabolism (increased glucose uptake and lactate formation), enhanced junctional membrane protein expression (especially gap junctions), altered cellular shape (from flattened to spherical with formation of microvilli), inhibition of apoptosis, and antrum formation and synthesis of follicular-fluid sulfur-enriched proteoglycans (as reviewed by (Hsueh et al., 1984)). Many of the genes identified by Pathway Studio Analysis that are repressed by FOXO1 and activated by FSH (see Supplemental Table 4 and Fig. 2A), are consistent with these responses, such as those associated with steroid and amino acid biosynthesis, carbohydrate metabolic processes, and cell-cell junctions as well as an abundance of solute carrier genes involved in membrane transport of small molecules. Unexpected gene expression includes a number of potassium channel genes, ephrins and ephrin receptor genes as well as phosphatases, cyclases, and kinases that have not been reported in GCs. Cell proliferation genes are likely under-represented in this analysis as cultured rat GCs do not proliferate in the absence of an exogenous TGF-β agonist (El-Hefnawy and Zeleznik, 2001; Miro and Hillier, 1996; Park et al., 2005).
A previous microarray analysis of genes up-regulated by FSH and repressed by constitutively active FOXO1 (315 genes) included primarily genes involved in lipid, sterol, and steroidogenic biosynthesis, transcriptional regulators of these genes, and Lhcgr (Liu et al., 2009). All of the same genes were identified in our RNA seq analysis, although the fold regulation in our analysis was generally less (often < 2.0-fold). Our RNA seq analysis also identified many of the same genes previously identified that were up-regulated by constitutively active FOXO1 (57 genes), such as Klf5, Fgf13, Nr0b1, and Irs2 (Liu et al., 2009).
In conclusion, our results reveal that FSH regulates many more genes than was previously appreciated (Escamilla-Hernandez et al., 2008; Law et al., 2013; Liu et al., 2009). Moreover, 60% (2,264) of the genes whose expression is reduced or enhanced by FSH are transcriptionally regulated by FOXO1, based on the presence of mRNA transcripts that are up- or down-regulated ≥ 2.0 fold in GCs transduced with constitutively active FOXO1(A3) compared to empty adenovirus. FOXO1-regulated genes function not only in lipid biosynthesis and steroidogenesis, as previously reported (Liu et al., 2009), but also in signaling, development, metabolism, transport, apoptosis, cytoskeletal functions, and transcription. FOXO1 thus functions in GCs, as in many other cells, to broadly regulate cellular functions. We conclude that FOXO1 can be viewed as a master regulator of FSH responsive genes.
Supplementary Material
Highlights.
RNA deep sequencing detected 13,461 genes in primary rat ovarian granulosa cells.
RNA deep sequencing showed that FSH regulates 3,772 genes ≥ 2.0-fold.
Sixty % of the genes regulated by FSH are activated or repressed by FOXO1.
Pathway Studio Analysis revealed enrichment of genes repressed by FOXO1 in metabolism, signaling, transport, and development.
Pathway Studio Analysis revealed enrichment of genes activated by FOXO1 in signaling, cytoskeletal functions, and apoptosis.
Acknowledgments
We gratefully acknowledge the contributions of Dr. John H. Nilson for his helpful suggestions. We also sincerely thank Mark R. Wildung of the Molecular Biology and Genomics Core at Washington State University for preparing mRNA sequencing libraries, for conducting RNA sequencing, and for updating results to the rat genome data base.
This research was supported by the National Institutes of Health grants R01HD065859 (to M.H.D.), R01HD62503 (to M.H.D.), T32GM083864 (to N.C.L., E.M.D., and B.K.), and the Poncin Fellowship Fund (to N.C.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
M.K.H , B.K., E.M.D., and M.H.D designed experiments; B.K. transduced cells with adenoviruses, collected RNA for RNA seq, and performed initial Pathway Studio Analysis of results (2012); M.H.D. evaluated updated Pathway Studio Analysis of results (2015); M.K.H. performed q-PCR; M.K.H. and E.M.D. performed western blots; M.K.H. and N.C.L. performed ChIP analyses; M.H.D and M.K.H. wrote the manuscript. All authors critically read the manuscript and provided constructive feedback.
Additional Information
The authors have no conflicts of interest.
- CALD1
- caldesmon 1
- FOXO1
- Forkhead Box O member
- FSH
- follicle-stimulating hormone
- GCs
- granulosa cells
- IGF1
- insulin-like growth factor 1
- IRS
- insulin receptor substrate
- LH
- luteinizing hormone
- NPPC
- natriuretic peptide precursor C
- NPR2
- natriuretic peptide receptor B
- PI3K
- phosphatidylinositol-3 kinase
- PKA
- protein kinase A
- RNA seq
- RNA sequencing
- WT
- wildtype
References
- Bennett J, Wu YG, Gossen J, Zhou P, Stocco C. Loss of GATA-6 and GATA-4 in granulosa cells blocks folliculogenesis, ovulation, and follicle stimulating hormone receptor expression leading to female infertility. Endocrinology. 2012;153:2474–2485. doi: 10.1210/en.2011-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Koopman P. Sex determination in mammalian germ cells: extrinsic versus intrinsic factors. Reproduction. 2010;139:943–958. doi: 10.1530/REP-10-0075. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Kops GJ. Cell cycle and death control: long live Forkheads. Trends Biochem.Sci. 2002;27:352–360. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- Carlone DL, Richards JS. Functional interactions, phosphorylation, and levels of 3',5'-cyclic adenosine monophosphate-regulatory element binding protein and steroidogenic factor-1 mediate hormone-regulated and constitutive expression of aromatase in gonadal cells. Mol Endocrinol. 1997;11:292–304. doi: 10.1210/mend.11.3.9900. [DOI] [PubMed] [Google Scholar]
- Chen S, Shi H, Liu X, Segaloff DL. Multiple elements and protein factors coordinate the basal and cyclic adenosine 3',5'-monophosphate-induced transcription of the lutropin receptor gene in rat granulosa cells. Endocrinology. 1999;140:2100–2109. doi: 10.1210/endo.140.5.6722. [DOI] [PubMed] [Google Scholar]
- Chesnel F, Eppig JJ. Synthesis and accumulation of p34cdc2 and cyclin B in mouse oocytes during acquisition of competence to resume meiosis. Mol Reprod Dev. 1995;40:503–508. doi: 10.1002/mrd.1080400414. [DOI] [PubMed] [Google Scholar]
- Clemens JW, Lala DS, Parker KL, Richards JS. Steroidogenic factor-1 binding and transcriptional activity of the cholesterol side-chain cleavage promoter in rat granulosa cells. Endocrinology. 1994;134:1499–1508. doi: 10.1210/endo.134.3.8119192. [DOI] [PubMed] [Google Scholar]
- Eimerl S, Orly J. Regulation of steroidogenic genes by insulin-like growth factor-1 and follicle-stimulating hormone: differential responses of cytochrome P450 side-chain cleavage, steroidogenic acute regulatory protein, and 3beta-hydroxysteroid dehydrogenase/isomerase in rat granulosa cells. Biol Reprod. 2002;67:900–910. doi: 10.1095/biolreprod.101.002170. [DOI] [PubMed] [Google Scholar]
- El-Hefnawy T, Zeleznik AJ. Synergism between FSH and activin in the regulation of proliferating cell nuclear antigen (PCNA) and cyclin D2 expression in rat granulosa cells. Endocrinology. 2001;142:4357–4362. doi: 10.1210/endo.142.10.8438. [DOI] [PubMed] [Google Scholar]
- Erickson GF, Sorensen RA. In vitro maturation of mouse oocytes isolated from late, middle, and pre-antral graafian follicles. J Exp Zool. 1974;190:123–127. doi: 10.1002/jez.1401900112. [DOI] [PubMed] [Google Scholar]
- Escamilla-Hernandez R, Little-Ihrig L, Orwig KE, Yue J, Chandran U, Zeleznik AJ. Constitutively active protein kinase A qualitatively mimics the effects of follicle-stimulating hormone on granulosa cell differentiation. Mol Endocrinol. 2008;22:1842–1852. doi: 10.1210/me.2008-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaro K, Quinn MC, Wojnarowicz PM, Arcand SL, de Ladurantaye M, Barres V, Ripeau JS, Killary AM, Davis EC, Lavoie J, Provencher DM, Mes-Masson AM, Chevrette M, Tonin PN. VGLL3 expression is associated with a tumor suppressor phenotype in epithelial ovarian cancer. Mol Oncol. 2013;7:513–530. doi: 10.1016/j.molonc.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkowska J, Jankowski M, Sairam MR, Fujio N, Reis AM, Mukaddam-Daher S, Tremblay J. Hormonal regulation of natriuretic peptide system during induced ovarian follicular development in the rat. Biol Reprod. 1999;61:162–170. doi: 10.1095/biolreprod61.1.162. [DOI] [PubMed] [Google Scholar]
- Hai CM. Caldesmon as a therapeutic target for proliferative vascular diseases. Mini Rev Med Chem. 2008;8:1209–1213. doi: 10.2174/138955708786140981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, McGee EA, Min G, Klein C, Rose UM, van Duin M, Hsueh AJ. Recombinant growth differentiation factor-9 (GDF-9) enhances growth and differentiation of cultured early ovarian follicles. Endocrinology. 1999;140:1236–1244. doi: 10.1210/endo.140.3.6548. [DOI] [PubMed] [Google Scholar]
- Hsueh AJW, Adashi EY, Jones PBC, Welsh TH., Jr. Hormonal regulation of the differentiation of cultured ovarian granulosa cells. Endocrine Reviews. 1984;5:76–110. doi: 10.1210/edrv-5-1-76. [DOI] [PubMed] [Google Scholar]
- Hunzicker-Dunn M. Selective activation of rabbit ovarian protein kinase isozymes in rabbit ovarian follicles and corpora lutea. The Journal of biological chemistry. 1981;256:12185–12193. [PubMed] [Google Scholar]
- Hunzicker-Dunn M, Mayo KE. Gonadotropin Signaling in the Ovary. Elsevier Academic Press; New York: 2015. [Google Scholar]
- Hunzicker-Dunn ME, Lopez-Biladeau B, Law NC, Fiedler SE, Carr DW, Maizels ET. PKA and GAB2 play central roles in the FSH signaling pathway to PI3K and AKT in ovarian granulosa cells. Proc Natl Acad Sci U S A. 2012;109:E2979–2988. doi: 10.1073/pnas.1205661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Achermann JC, Jameson JL. A naturally-occurring steroidogenic factor-1 (SF-1) mutation exhibits differential binding and activation of target genes. Journal of Biological Chemistry. 2000;275:31708–31714. doi: 10.1074/jbc.M002892200. [DOI] [PubMed] [Google Scholar]
- Karlsson AB, Maizels ET, Flynn MP, Jones JC, Shelden EA, Bamburg JR, Hunzicker-Dunn M. Luteinizing hormone receptor-stimulated progesterone production by preovulatory granulosa cells requires protein kinase A-dependent activation/dephosphorylation of the actin dynamizing protein cofilin. Mol Endocrinol. 2010;24:1765–1781. doi: 10.1210/me.2009-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Cheng Y, Kawamura N, Takae S, Okada A, Kawagoe Y, Mulders S, Terada Y, Hsueh AJ. Pre-ovulatory LH/hCG surge decreases C-type natriuretic peptide secretion by ovarian granulosa cells to promote meiotic resumption of pre-ovulatory oocytes. Hum Reprod. 2011;26:3094–3101. doi: 10.1093/humrep/der282. [DOI] [PubMed] [Google Scholar]
- Khor VK, Ahrends R, Lin Y, Shen WJ, Adams CM, Roseman AN, Cortez Y, Teruel MN, Azhar S, Kraemer FB. The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One. 2014;9:e105047. doi: 10.1371/journal.pone.0105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmakova A, Wang J, Brogan R, Chaffin C, Rodriguez A. Deficiency of scavenger receptor class B type I negatively affects progesterone secretion in human granulosa cells. Endocrinology. 2010;151:5519–5527. doi: 10.1210/en.2010-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nature Genetics. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- Kurten RC, Richards JS. An adenosine 3',5'-monophosphate-responsive DNA element confers forskolin sensitivity on gene expression by primary rat granulosa cells. Endocrinology. 1989;125:1345–1357. doi: 10.1210/endo-125-3-1345. [DOI] [PubMed] [Google Scholar]
- Law NC, Hunzicker-Dunn ME. Insulin Receptor Substrate 1, the Hub Linking Follicle-stimulating Hormone to Phosphatidylinositol 3-Kinase Activation. J Biol Chem. 2016;291:4547–4560. doi: 10.1074/jbc.M115.698761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law NC, Weck J, Kyriss B, Nilson JH, Hunzicker-Dunn M. Lhcgr expression in granulosa cells: roles for PKA-phosphorylated beta-catenin, TCF3, and FOXO1. Mol Endocrinol. 2013;27:1295–1310. doi: 10.1210/me.2013-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Zhang M, Sugiura K, Wigglesworth K, Uliasz T, Jaffe LA, Eppig JJ. Hormonal coordination of natriuretic peptide type C and natriuretic peptide receptor 3 expression in mouse granulosa cells. Biol Reprod. 2013;88:42. doi: 10.1095/biolreprod.112.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettieri Barbato D, Aquilano K, Ciriolo MR. FoxO1 at the nexus between fat catabolism and longevity pathways. Biochim Biophys Acta. 2014;1841:1555–1560. doi: 10.1016/j.bbalip.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Lin JL, Reiter RS, Daniels K, Soll DR, Lin JJ. Caldesmon mutant defective in Ca(2+)-calmodulin binding interferes with assembly of stress fibers and affects cell morphology, growth and motility. J Cell Sci. 2004;117:3593–3604. doi: 10.1242/jcs.01216. [DOI] [PubMed] [Google Scholar]
- Lin JY, Pitman-Crawford JL, Bibby AH, Hudson NL, McIntosh CJ, Juengel JL, McNatty KP. Effects of species differences on oocyte regulation of granulosa cell function. Reproduction. 2012;144:557–567. doi: 10.1530/REP-12-0267. [DOI] [PubMed] [Google Scholar]
- Liu Z, Castrillon DH, Zhou W, Richards JS. FOXO1/3 Depletion in Granulosa Cells Alters Follicle Growth, Death and Regulation of Pituitary FSH. Mol Endocrinol. 2013;27:238–252. doi: 10.1210/me.2012-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Rudd MD, Hernandez-Gonzalez I, Gonzalez-Robayna I, Fan HY, Zeleznik AJ, Richards JS. FSH and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol Endocrinol. 2009;23:649–661. doi: 10.1210/me.2008-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Viste K, Urday-Zaa JC, Senthil Kumar G, Tsai WW, Talai A, Mayo KE, Montminy M, Radhakrishnan I. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc Natl Acad Sci U S A. 2012;109:20865–20870. doi: 10.1073/pnas.1219028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Gumen A, Haughian JM, Wiltbank MC. The role of luteinizing hormone in regulating gene expression during selection of a dominant follicle in cattle. Biol Reprod. 2011;84:369–378. doi: 10.1095/biolreprod.110.085274. [DOI] [PubMed] [Google Scholar]
- Miro F, Hillier SG. Modulation of granulosa cell deoxyribonucleic acid synthesis and differentiation by activin. Endocrinology. 1996;137:464–468. doi: 10.1210/endo.137.2.8593790. [DOI] [PubMed] [Google Scholar]
- Norris RP, Ratzan WJ, Freudzon M, Mehlmann LM, Krall J, Movsesian MA, Wang H, Ke H, Nikolaev VO, Jaffe LA. Cyclic GMP from the surrounding somatic cells regulates cyclic AMP and meiosis in the mouse oocyte. Development. 2009;136:1869–1878. doi: 10.1242/dev.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyegaard M, Overgaard MT, Su YQ, Hamilton AE, Kwintkiewicz J, Hsieh M, Nayak NR, Conti M, Conover CA, Giudice LC. Lack of functional pregnancy-associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female fertility. Biol Reprod. 2010;82:1129–1138. doi: 10.1095/biolreprod.109.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oonk RB, Parker KL, Gibson JL, Richards JS. Rat cholesterol side-chain cleavage cytochrome P-450 gene. Journal of Biological Chemistry. 1990;265:22392–22401. [PubMed] [Google Scholar]
- Parakh TN, Hernandez JA, Grammer JC, Weck J, Hunzicker-Dunn M, Zeleznik AJ, Nilson JH. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires beta-catenin. Proc Natl Acad Sci U S A. 2006;103:12435–12440. doi: 10.1073/pnas.0603006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Maizels ET, Feiger ZJ, Alam H, Peters CA, Woodruff TK, Unterman TG, Lee EJ, Jameson JL, Hunzicker-Dunn M. Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J Biol Chem. 2005;280:9135–9148. doi: 10.1074/jbc.M409486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Dodson R, Schoderbek WE, Maurer RA, Mayo KE. Regulation of the alpha inhibin gene by cyclic adenosine 3',5'-monophosphate after transfection into rat granulosa cells. Molecular Endocrinology. 1991;5:521–534. doi: 10.1210/mend-5-4-521. [DOI] [PubMed] [Google Scholar]
- Puri P, Chandran U, Law NC, Hunzicker-Dunn M, Zeleznik AJ. Protein kinase A: The master kinase of granulosa cell differentiation. Scientific Reports. L.-I.L. in revision. [DOI] [PMC free article] [PubMed]
- Puthanveetil P, Wan A, Rodrigues B. FoxO1 is crucial for sustaining cardiomyocyte metabolism and cell survival. Cardiovasc Res. 2013;97:393–403. doi: 10.1093/cvr/cvs426. [DOI] [PubMed] [Google Scholar]
- Sanchez AM, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci. 2014;71:1657–1671. doi: 10.1007/s00018-013-1513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N, Yivgi-Ohana N, Orly J. Transcriptional regulation of the cholesterol side chain cleavage cytochrome P450 gene (CYP11A1) revisited: binding of GATA, cyclic adenosine 3',5'-monophosphate response element-binding protein and activating protein (AP)-1 proteins to a distal novel cluster of cis-regulatory elements potentiates AP-2 and steroidogenic factor-1-dependent gene expression in the rodent placenta and ovary. Mol Endocrinol. 2007;21:948–962. doi: 10.1210/me.2006-0226. [DOI] [PubMed] [Google Scholar]
- Shi H, Segaloff DL. A role for increased lutropin/choriogonadotropin receptor (LHR) gene transcription in the follitropin-stimulated induction of the LHR in granulosa cells. Mol Endocrinol. 1995;9:734–744. doi: 10.1210/mend.9.6.8592519. [DOI] [PubMed] [Google Scholar]
- Shuhaibar LC, Egbert JR, Edmund AB, Uliasz TF, Dickey DM, Yee SP, Potter LR, Jaffe LA. Dephosphorylation of juxtamembrane serines and threonines of the NPR2 guanylyl cyclase is required for rapid resumption of oocyte meiosis in response to luteinizing hormone. Dev Biol. 2016;409:194–201. doi: 10.1016/j.ydbio.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski M, Jablonska E, Juszczynski P. FOXO1 transcription factor: a critical effector of the PI3K-AKT axis in B-cell development. Int Rev Immunol. 2014;33:146–157. doi: 10.3109/08830185.2014.885022. [DOI] [PubMed] [Google Scholar]
- Tahaei LS, Eimani H, Yazdi PE, Ebrahimi B, Fathi R. Effects of retinoic acid on maturation of immature mouse oocytes in the presence and absence of a granulosa cell co-culture system. J Assist Reprod Genet. 2011;28:553–558. doi: 10.1007/s10815-011-9579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Viger RS. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology. 2001;142:977–986. doi: 10.1210/endo.142.3.7995. [DOI] [PubMed] [Google Scholar]
- Vaccari S, Weeks JL, 2nd, Hsieh M, Menniti FS, Conti M. Cyclic GMP signaling is involved in the luteinizing hormone-dependent meiotic maturation of mouse oocytes. Biol Reprod. 2009;81:595–604. doi: 10.1095/biolreprod.109.077768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem.J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nature reviews. Molecular cell biology. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Wang CL. Caldesmon and the regulation of cytoskeletal functions. Adv Exp Med Biol. 2008;644:250–272. doi: 10.1007/978-0-387-85766-4_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck J, Mayo KE. Switching of NR5A Proteins Associated with the Inhibin {alpha}-Subunit Gene Promoter after Activation of the Gene in Granulosa Cells. Molecular Endocrinology. 2006;20:1090–1103. doi: 10.1210/me.2005-0199. [DOI] [PubMed] [Google Scholar]
- Werner H, Sarfstein R. Transcriptional and epigenetic control of IGF1R gene expression: implications in metabolism and cancer. Growth Horm IGF Res. 2014;24:112–118. doi: 10.1016/j.ghir.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Yates M, Kolmakova A, Zhao Y, Rodriguez A. Clinical impact of scavenger receptor class B type I gene polymorphisms on human female fertility. Hum Reprod. 2011;26:1910–1916. doi: 10.1093/humrep/der124. [DOI] [PubMed] [Google Scholar]
- Zeleznik AJ, Saxena D, Little-Ihrig L. Protein kinase B is obligatory for follicle-stimulating hormone-induced granulosa cell differentiation. Endocrinology. 2003;144:3985–3994. doi: 10.1210/en.2003-0293. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science. 2010;330:366–369. doi: 10.1126/science.1193573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J Mol Cell Biol. 2011;3:276–282. doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
- Zhou P, Baumgarten SC, Wu Y, Bennett J, Winston N, Hirshfeld-Cytron J, Stocco C. IGF-I signaling is essential for FSH stimulation of AKT and steroidogenic genes in granulosa cells. Mol Endocrinol. 2013;27:511–523. doi: 10.1210/me.2012-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.