Abstract
Insects represent over 70% of all animal species. Recent virome analyses reveal unprecedented genetic diversity of insect viruses, which appears to match that of their hosts. Thus, insect-virus interactions may provide information on a vast repertoire of antiviral immune mechanisms. Tapping into this diversity is challenging because of several constraints imposed by the uniqueness of each insect model. Nevertheless, it is clear that many conserved and divergent pathways participate in the control of viral infection in insects. Co-evolution between hosts and viruses favors the development of immune evasion mechanisms by the pathogen. Viral suppressors can offer unique perspective on host pathways and emphasize the importance of RNA interference, apoptosis, but also NF-κB pathways and translation control in insect antiviral immunity.
Introduction
Viruses are an important burden for all living organisms. These obligate intracellular pathogens are intimately associated with host cells, hijacking their machineries to replicate. As a result, viruses exert great selective pressure on their host to evolve resistance pathways. These, in turn, favor the adaptation of viruses to escape antiviral mechanisms. This arms race favors the diversification of host-defense and virus escape mechanisms. It is therefore instructive to investigate virus-host interaction in a range of animals, to sample in depth the diversity of antiviral strategies. Insects represent the largest and most diverse group of animals, with over 70% of all species [1]. Although the number of known insect viruses does not outnumber that of other animals, it is becoming apparent that their diversity is unprecedented [2,3]. Besides tapping into biodiversity, there are other reasons to specifically study antiviral immunity in insects. For example, several important human diseases, such as dengue and zika fevers, are caused by insect-borne viruses. In addition, viruses infecting beneficial insects such as bees or silkworms can cause important economic losses. Finally, viruses can be used as biological control agents against pest insects. Here, we give a broad overview of pathways involved in insect antiviral immunity. We discuss the challenges associated with studies on insect-virus interactions and illustrate how the identification of evasion mechanisms encoded by viruses can validate antiviral pathways.
General overview of antiviral pathways in insects
RNA interference (RNAi) is often described as the major antiviral pathway in insects (e.g. mosquitoes, flies, bees, lepidopterans and even non-insect arthropods such as ticks) [4–12]. RNAi refers to a series of mechanisms of gene regulation mediated by small RNAs associated with proteins of the Argonaute family that drive degradation of viral RNA in a sequence specific manner [13]. In insects, two distinct types of virus-derived small RNAs, small interfering RNAs (siRNAs) and piwi-interacting RNAs (piRNAs), have been detected in vivo [14,15].
Programmed cell death is another broad strategy to control viruses in insects by eliminating infected cells [16–19]. Cell death can stop viral replication before it is completed and also promote clearance of infected cells by phagocytes thus preventing dissemination. Indeed, in Drosophila, control of FHV and picorna-like viruses, Drosophila C virus (DCV) and Cricket paralysis virus (CrPV), requires blood cells (hemocytes) and phagocytosis in vivo [20,21]. Hemocytes may also be important to initiate systemic inducible responses.
Insects, like most organisms, mount a transcriptional response to virus infection. Indeed, the transcriptome of virus-infected insects such as mosquitoes, flies, bees and silkworms reveals that expression of large sets of genes is upregulated (e.g. [22–26]). This requires, at least in part, evolutionary conserved pathways such as Jak-STAT or NF-κB. In fruit flies, Aedes and Culex mosquitoes, the cytokine activated Jak/STAT pathway controls the expression of antiviral genes in response to infection with DCV, Dengue virus (DENV) and West Nile virus (WNV) [23,26–28]. Moreover, the Toll and IMD pathways, which regulate different members of the NF-κB family of transcription factors, have also been reported to participate in the antiviral response against DCV, CrPV, Sindbis virus (SINV) and DENV in Drosophila and mosquitoes [24,29–31]. Although many examples of transcriptional responses have been described, the mechanism of activation and the function of induced genes remain poorly characterized.
In addition, several other pathways have been reported to play a role in insect antiviral defense, such as the ubiquitin proteasome pathway, autophagy or heat-shock response [12,20,26,32]. This plethora of insect strategies to counteract viruses may reflect the selective pressure these pathogens impose on a very diverse group of hosts. There is still is a large diversity of pathways that remain unknown, but studies of insect antiviral immunity need to take into account the restricted tools available and their inherent limitations.
Constraints on studies in insect antiviral immunity
Many studies on insect antiviral immunity are based on cell lines, which offer the advantage to work on a more homogenous cell population where it is easier to control the timing and multiplicity of infection. However, the interpretation of these results need to take into account some limitations of cell lines such as (i) unclear origin, (ii) changes due to immortalization, (iii) lack of input from other cell types that are present in vivo and (iv) presence of persistent virus infections, which can interfere with studies of antiviral defense. Furthermore, insect cell lines are not available for most species, and when they exist, their number is limited.
Cell lines are well suited to perform genome-wide RNAi screens based on dsRNA-mediated gene silencing. These have been instrumental to dissect different antiviral pathways both in vivo and in cell culture. One caveat with RNAi is the risk to silence off-target genes, which complicate data interpretation [33]. Therefore, phenotypes observed upon gene silencing, both in vivo and in cell culture, need to be confirmed with real mutants, if possible, or with independent dsRNAs targeting different regions of the target gene. The availability of gene editing mediated by CRISPR/Cas systems can greatly help in future studies on insect immunity, although it also requires validation due to possible off-target effects.
Even the strong genetic evidence provided by in vivo studies with mutants, need to take into account for the heterogeneity of the genetic background in non-isogenic insect strains. Even in the most common insect model, Drosophila melanogaster, polymorphisms in host restriction factors can significantly affect the outcome of viral infection (e.g. [34,35]). This needs to be addressed, ideally by performing rescue experiments in the same genetic background. For in vivo experiments, the route of infection also becomes an important issue. Direct injection of viruses into the animal body cavity (hemocoele) is often utilized, as it provides an efficient way to control timing and multiplicity of infection. However, direct hemocole injection is probably not the most common route of infection during an insect life cycle, even though there are natural examples such as when mites act as vectors for the systemic delivery of Deformed Wing virus in bees [36]. Oral infections are likely more prevalent and may trigger different systemic responses compared to direct hemocoele infection (e.g. [37]). Control of the timing and multiplicity of infection is a major challenge for the oral delivery of viruses. In vivo, there are also other issues such as the relevant developmental stages. For example, adult mosquitoes get exposed to viruses when blood feeding, whereas Lepidoteran insects do not feed as adults, and only get exposed at the larval stage.
Finally, the virus utilized will influence the results on insect antiviral immunity. Insect viruses that have been used can be divided into opportunistic or natural pathogens, both of which can cause acute or persistent infections [38]. Opportunistic or non-natural pathogens are less likely to be well adapted to the host and thus more likely to reveal antiviral mechanisms [12]. Natural viruses, on the other hand, might not activate these mechanisms because they either encode suppressors or avoid recognition. In fact, the presence of specific suppressors of a host pathway in a virus can be used as a validation of the importance of this pathway for antiviral immunity. For example, studying viral evasion proteins, such as many encoded by Vaccinia virus, has provided valuable information about the antiviral response not only in mammals but also insects [12]. Below, we focus on antiviral pathways that are targeted by suppressors proteins encoded by insect viruses (Figure 1).
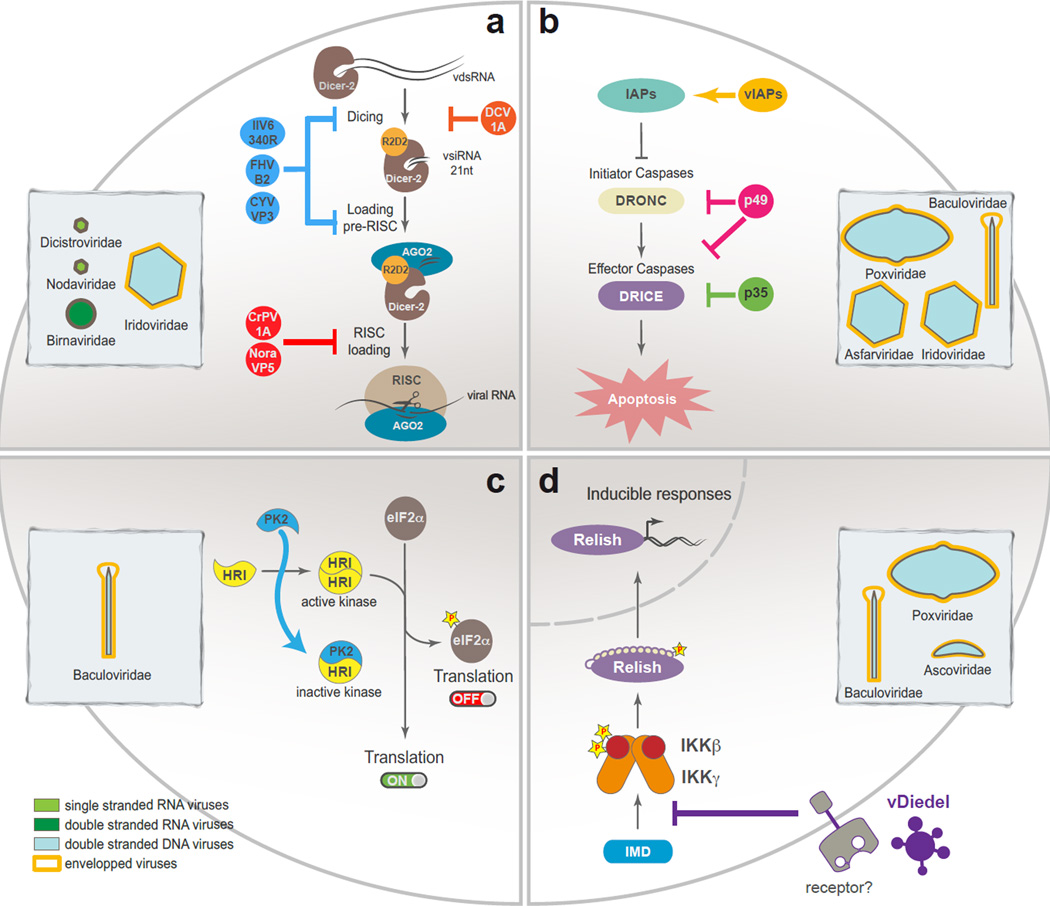
Figure 1. Insect antiviral pathways targeted by viruses.
(a) Multiple insect viruses encode virus suppressors of RNAi (VSRs) that can interfere with the siRNA pathway at different steps. Some VSRs can block activation by hiding dsRNA from the Dcr-2 while others can bind siRNAs or interfere with RISC directly. (b) Apoptosis triggered in infected cells can interfere with viral replication and is targeted by diverse virus-encoded mechanisms such as inhibitors of apoptosis proteins (IAPs) or caspase inhibitors of the p35 family (p35 and p49). (c) In insects, translation is inhibited in virus-infected cells by activation of HRI-like eIF2α kinases. Baculoviruses encode PK2, a homolog of eIF2α kinases, that can prevent phosphorylation of eIF2α and translation inhibition. (d) Regulation of the insect IMD/NF-κB pathway by the immunomodulatory cytokine Diedel is important to control both the antiviral response as well as homeostasis. Different insect viruses encode a co-opted version of this endogenous inhibitor of the IMD pathway. See text for more details.
Host antiviral pathways targeted by insect viruses
Viral suppressors drive the rapid evolution of the siRNA pathway
The siRNA pathway is activated by dsRNA commonly generated as a byproduct of viral replication. The nucleic acid sensor named Dcr-2, which contains helicase and RNase III domains, recognizes viral dsRNA. Dcr-2 processes the dsRNA into duplex siRNAs that associate with a specific argonaute protein known as AGO2. One of the strands of the duplex siRNA remains associated with AGO2 to form the RNA induced silencing complex (RISC). This multiple turnover nuclease can slice and degrade RNAs that possess complementary sequences. During viral infection, this mechanism is able to effectively silence viral RNAs and inhibit replication [13]. The importance of the siRNA pathway for the antiviral defense is underlined by the variety of viral suppressors of RNAi (VSRs) encoded by insect viruses, acting at different steps of the pathway (Figure 1a). Some VSRs directly bind long dsRNA and prevent processing by Dcr-2 (DCV-1A, FHV-B2, IIV6-340R, and VP3 from the birnaviruses Drosophila X virus and Culex Y virus) [7,39–41]. With the exception of DCV-1A, these suppressors bind siRNAs in vitro, suggesting they also inhibit the pathway postprocessing of long dsRNA. Other VSRs directly bind to AGO2 and inhibit target slicing (CrPV-1A and VP1 from Nora viruses) [42,43]. The importance of VSRs is highlighted by experiments on FHV demonstrating that the virus is efficiently controlled by the siRNA pathway upon deletion of the VSR B2 [44].
Nora viruses (NV) define a new family related to picornaviruses and have been identified in different drosophila species. They provide a good example of the co-evolution between VSRs and the host siRNA pathway machinery. Indeed, VP3 from NV isolated from D. immigrans can efficiently bind to and inhibit AGO2 from this species, but not from the related species D. melanogaster [43]. This species-specificity can be explained by the evolution of AGO2, which is among the fastest evolving genes in Drosophila [45]. Thus, the arms race between the virus and its host puts selective pressure onto genes encoding components of the siRNA pathway to escape targeting by VSRs. The rapid evolution of RNAi genes is also observed in mosquitoes, underlying the importance of this antiviral pathway at least in Dipteran insects [46].
Inhibitors of apoptosis in insect DNA viruses
Apoptosis can restrict viral replication and function as an antiviral response. The hallmark of apoptotic cell death is the activation of a cascade of cysteine proteases (caspases) involving initiator and effector enzymes. The activity of caspases is tightly regulated by cellular factors including the Inhibitors of Apoptosis Proteins (IAPs). Apoptosis can be rapidly activated during viral infection by different mechanism, such as induction of pro-apoptotic genes or degradation of labile IAPs. For example, in Drosophila cells infected with FHV activation of p53 leads to induction of the pro-apoptotic gene reaper that blocks the activity of IAPs [19]. In baculovirus infected Lepidopteran cells, inhibition of host translation leads to depletion of cellular IAPs, whose stability is tightly regulated [18]. Indeed, Lepidopteran and Drosophila IAPs contain N-terminal instability motifs that are targeted by different signaling pathways in response to virus infection [47].
Different insect DNA viruses belonging to the families of Baculo-like viruses (Baculovirus and Nudiviruses), Entomopoxviruses, Iridoviruses and Asfarviruses, encode inhibitors of apoptosis. These can be divided in two groups based on the mechanism of action: the viral IAPs and the p35-like suicide caspase inhibitors [48] (Figure 1b). In fact, IAPs were first described as viral proteins encoded by baculoviruses. Unlike their cellular counterparts, vIAPs lack an N-terminal instability motif [47]. As a result, they act by stabilizing cellular IAPs, thus preventing apoptosis in infected cells as recently shown for the prototypical baculovirus Op-IAP3 from Orgya pseudotsugata multiple nucleopolyhedrovirus [18]. The p35 suppressor from Autographa californica multiple nucleopolyhedrovirus functions as a substrate for effector caspases such as DRICE, while the related p49 suppressor from Spodoptera littoralis nucleopolyhedrovirus has the potential to block both initiator and effector caspases [48]. Deletion of the genes encoding suppressors of apoptosis results in increased cell death and attenuation of the infection.
Inhibition of translation as an antiviral mechanism in insects
Inhibition of translation is commonly observed in virus infected insect cells, suggesting that it could contribute to the control of viral replication. Indeed, control of translation initiation by eIF2α kinases is an important antiviral pathway in mammals. These enzymes phosphorylate the α subunit of translation initiation factor 2, which prevents translation initiation in eukaryotes. There are four eIF2α kinases in mammals, GCN2, HRI, PERK and PKR, of which PKR is the major antiviral player although GCN2 and PERK may also participate in specific cases. Insects lack PKR but most of them have orthologs for the other three eIF2α kinases [49]. Interestingly, HRI-like kinases seem to have independently developed an antiviral function in insects. Mammalian viruses often encode PKR inhibitors, which reinforce the role of this kinase in antiviral defense. Interestingly, a subset of Alphabaculoviruses encode a homolog of eIF2α kinases named PK2. This homolog interferes with dimerization and activation of eIF2α kinases(Figure 1c). During infection in vivo, it blocks HRI-like kinase-dependent phosphorylation of eIF2α and allows full viral replication. pk2 deficient baculoviruses induce high level of eIF2α phosphorylation and are attenuated [49].
Virokine suppressors of IMD controlled inducible response
The regulatory cytokine named Diedel was recently identified in Drosophila and shown to act as an inhibitor of the IMD pathway [50]. Diedel is essential to ensure fly survival during SINV infection by modulating the activation of the IMD pathway. Indeed, there are indications in different insects that the inducible response regulated by IMD/NF-κB pathway can contribute to antiviral defense [29–31]. How this pathway gets activated remains unclear at this stage, although in Culex mosquitoes it involves Dicer-2, the same dsRNA sensor as for the siRNA pathway [29].
Several insect viruses from the Ascovirus, Entomopoxvirus and Baculovirus families encode Diedel homologs [50] (Figure 1d). Furthermore, the Diedel homolog encoded by Spodoptera frugiperda ascovirus 1a can functionally substitute the cellular protein and inhibit the IMD pathway in Drosophila. Although the antiviral effector mechanism regulated by IMD remains unknown, the fact that several unrelated insect viruses encode Diedel homologs stresses the value of exploring further the contribution of this evolutionary conserved pathway.
Concluding remarks
In conclusion, there are multiple layers of insect antiviral defense that relies on conserved but also divergent pathways. Some mechanisms are insect-, tissue- or virus-specific, highlighting the importance to investigate virus-host interactions in the right context. For example, in the case of oral infections, viruses face tissue specific antiviral pathways in the gut and during systemic dissemination. Analysis of antiviral immunity in the gut deserves special attention in light of its importance to restrict dissemination of insect-borne viruses and the complexity imposed by the microbiota [8,24].
Full understanding of antiviral immunity is very challenging if the diversity of pathways approaches that of insect hosts. Nevertheless, these studies will be very instructive because they will reveal original antiviral strategies and weak spots in viruses. To meet this challenge, the community of insect immunologists can take advantage of new tools that are becoming available. For example, the genomic revolution is rapidly increasing the number of sequenced insect genomes and leading to the discovery of novel viruses [1– 3,15]. Thus, genomic data mining of divergent insect-viral pairs can help identify viral escape mechanisms and reveal novel antiviral pathways.
Highlights.
Insect antiviral immunity involves a diverse set of pathways
Unique caveats are associated with studies of insect antiviral pathways
An unprecedented diversity of viruses co-evolved with the insect immune system
Virus immune evasion mechanisms validate the importance of antiviral pathways
Acknowledgments
We thank Carine Meignin for discussions and preparation of the figure, Stéphanie Blandin and Nelson Martins for critical reading of the manuscript, and NIH (PO1 AI070167), ANR (ANR-13-BSV3-009), Infect-ERA (ANR-14-IFEC-0005), Investissements d’Avenir Programs (ANR-10-LABX-36 ANR-11-EQPX-0022), CNPq, Ciências sem fronteiras (PVE-400648/2013-0), CAPES and FAPEMIG for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 2. Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015:4. doi: 10.7554/eLife.05378. * Reference 2, together with 3 and 15, provide insight into the genetic diversity of insect viruses.
- 3.Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, Brouqui J-M, Bayne EH, Longdon B, Buck AH, et al. The Discovery, Distribution, and Evolution of Viruses Associated with Drosophila melanogaster. PLoS Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnettler E, Tykalová H, Watson M, Sharma M, Sterken MG, Obbard DJ, Lewis SH, McFarlane M, Bell-Sakyi L, Barry G, et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 2014;42:9436–9446. doi: 10.1093/nar/gku657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AWE, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayachandran B, Hussain M, Asgari S. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 2012;86:13729–13734. doi: 10.1128/JVI.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 8.Carissimo G, Pondeville E, McFarlane M, Dietrich I, Mitri C, Bischoff E, Antoniewski C, Bourgouin C, Failloux A-B, Kohl A, et al. Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E176–E185. doi: 10.1073/pnas.1412984112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chejanovsky N, Ophir R, Schwager MS, Slabezki Y, Grossman S, Cox-Foster D. Characterization of viral siRNA populations in honey bee colony collapse disorder. Virology. 2014;454–455:176–183. doi: 10.1016/j.virol.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Zografidis A, Van Nieuwerburgh F, Kolliopoulou A, Apostolou-Karampelis K, Head SR, Deforce D, Smagghe G, Swevers L. Viral Small-RNA Analysis of Bombyx mori Larval Midgut during Persistent and Pathogenic Cytoplasmic Polyhedrosis Virus Infection. J. Virol. 2015;89:11473–11486. doi: 10.1128/JVI.01695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFarlane M, Arias-Goeta C, Martin E, O’Hara Z, Lulla A, Mousson L, Rainey SM, Misbah S, Schnettler E, Donald CL, et al. Characterization of Aedes aegypti innate-immune pathways that limit Chikungunya virus replication. PLoS Negl. Trop. Dis. 2014;8:e2994. doi: 10.1371/journal.pntd.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gammon DB, Duraffour S, Rozelle DK, Hehnly H, Sharma R, Sparks ME, West CC, Chen Y, Moresco JJ, Andrei G, et al. A single vertebrate DNA virus protein disarms invertebrate immunity to RNA virus infection. eLife. 2014:3. doi: 10.7554/eLife.02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding S-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 14.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012;8:e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguiar ERGR, Olmo RP, Paro S, Ferreira FV, de Faria IJ, da S, Todjro YMH, Lobo FP, Kroon EG, Meignin C, Gatherer D, et al. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clem RJ. Viral IAPs, then and now. Semin. Cell Dev. Biol. 2015;39:72–79. doi: 10.1016/j.semcdb.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Settles EW, Friesen PD. Flock house virus induces apoptosis by depletion of Drosophila inhibitor-of-apoptosis protein DIAP1. J. Virol. 2008;82:1378–1388. doi: 10.1128/JVI.01941-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byers NM, Vandergaast RL, Friesen PD. Baculovirus Inhibitor-of-Apoptosis Op-IAP3 Blocks Apoptosis by Interaction with and Stabilization of a Host Insect Cellular IAP. J. Virol. 2016;90:533–544. doi: 10.1128/JVI.02320-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Behura SK, Clem RJ, Schneemann A, Becnel J, Severson DW, Zhou L. P53-mediated rapid induction of apoptosis conveys resistance to viral infection in Drosophila melanogaster. PLoS Pathog. 2013;9:e1003137. doi: 10.1371/journal.ppat.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamiable O, Arnold J, da Silva de Faria IJ, Proveti Olmo R, Bergami F, Meignin C, Hoffmann JA, Marques JT, Imler J-L. Analysis of the contribution of hemocytes and autophagy to Drosophila antiviral immunity. J. Virol. 2016 doi: 10.1128/JVI.00238-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nainu F, Tanaka Y, Shiratsuchi A, Nakanishi Y. Protection of Insects against Viral Infection by Apoptosis-Dependent Phagocytosis. J. Immunol. Baltim. Md 1950. 2015;195:5696–5706. doi: 10.4049/jimmunol.1500613. [DOI] [PubMed] [Google Scholar]
- 22.Kolliopoulou A, Van Nieuwerburgh F, Stravopodis DJ, Deforce D, Swevers L, Smagghe G. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PloS One. 2015;10:e0121447. doi: 10.1371/journal.pone.0121447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. Baltim. Md 1950. 2013;190:650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014;10:e1004230. doi: 10.1371/journal.ppat.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkling SH, Overheul GJ, van Mierlo JT, Arends D, Gilissen C, van Rij RP. The heat shock response restricts virus infection in Drosophila. Sci. Rep. 2015;5:12758. doi: 10.1038/srep12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paradkar PN, Trinidad L, Voysey R, Duchemin J-B, Walker PJ. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18915–18920. doi: 10.1073/pnas.1205231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paradkar PN, Duchemin J-B, Voysey R, Walker PJ. Dicer-2-dependent activation of Culex Vago occurs via the TRAF-Rel2 signaling pathway. PLoS Negl. Trop. Dis. 2014;8:e2823. doi: 10.1371/journal.pntd.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila. PloS One. 2009;4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 34. Cao C, Magwire MM, Bayer F, Jiggins FM. A Polymorphism in the Processing Body Component Ge-1 Controls Resistance to a Naturally Occurring Rhabdovirus in Drosophila. PLOS Pathog. 2016;12:e1005387. doi: 10.1371/journal.ppat.1005387. * References 34 and 35 identify polymorphisms in host genes encoding restriction factors that affect the outcome of infection in a virus-specific manner.
- 35. Martins NE, Faria VG, Nolte V, Schlötterer C, Teixeira L, Sucena É, Magalhães S. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5938–5943. doi: 10.1073/pnas.1400378111. * References 34 and 35 identify polymorphisms in host genes encoding restriction factors that affect the outcome of infection in a virus-specific manner.
- 36. Wilfert L, Long G, Leggett HC, Schmid-Hempel P, Butlin R, Martin SJM, Boots M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science. 2016;351:594–597. doi: 10.1126/science.aac9976. * Reference 36 reports a clear example of direct systemic viral infection in an insect and how this significantly impacted the global spread of an important bee pathogen.
- 37.Ferreira ÁG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L. The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog. 2014;10:e1004507. doi: 10.1371/journal.ppat.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh M-C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. Immunol. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 39.van Cleef KWR, van Mierlo JT, Miesen P, Overheul GJ, Fros JJ, Schuster S, Marklewitz M, Pijlman GP, Junglen S, van Rij RP. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014;42:8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronkhorst AW, van Cleef KWR, Venselaar H, van Rij RP. A dsRNA-binding protein of a complex invertebrate DNA virus suppresses the Drosophila RNAi response. Nucleic Acids Res. 2014;42:12237–12248. doi: 10.1093/nar/gku910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Rij RP, Saleh M-C, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010;17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Mierlo JT, Overheul GJ, Obadia B, van Cleef KWR, Webster CL, Saleh M-C, Obbard DJ, van Rij RP. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 2014;10:e1004256. doi: 10.1371/journal.ppat.1004256. ** Reference 43 provides an elegant demonstration of how the co-evolution between a host antiviral pathway and a viral suppressor can lead to species-specific interaction.
- 44.Han Y-H, Luo Y-J, Wu Q, Jovel J, Wang X-H, Aliyari R, Han C, Li W-X, Ding S-W. RNA-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: characterization of viral small interfering RNA populations in wild-type and mutant flies. J. Virol. 2011;85:13153–13163. doi: 10.1128/JVI.05518-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obbard DJ, Jiggins FM, Bradshaw NJ, Little TJ. Recent and recurrent selective sweeps of the antiviral RNAi gene Argonaute-2 in three species of Drosophila. Mol. Biol. Evol. 2011;28:1043–1056. doi: 10.1093/molbev/msq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernhardt SA, Simmons MP, Olson KE, Beaty BJ, Blair CD, Black WC. Rapid intraspecific evolution of miRNA and siRNA genes in the mosquito Aedes aegypti. PloS One. 2012;7:e44198. doi: 10.1371/journal.pone.0044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandergaast R, Mitchell JK, Byers NM, Friesen PD. Insect inhibitor-of-apoptosis (IAP) proteins are negatively regulated by signal-induced N-terminal degrons absent within viral IAP proteins. J. Virol. 2015;89:4481–4493. doi: 10.1128/JVI.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda M, Yamada H, Hamajima R, Kobayashi M. Baculovirus genes modulating intracellular innate antiviral immunity of lepidopteran insect cells. Virology. 2013;435:1–13. doi: 10.1016/j.virol.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 49. Li JJ, Cao C, Fixsen SM, Young JM, Ono C, Bando H, Elde NC, Katsuma S, Dever TE, Sicheri F. Baculovirus protein PK2 subverts eIF2α kinase function by mimicry of its kinase domain C-lobe. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4364–E4373. doi: 10.1073/pnas.1505481112. ** Reference 49 describes a refined mechanism of action by a viral suppressor targeting the control of translation initiation by eIF2α kinases.
- 50. Lamiable O, Kellenberger C, Kemp C, Troxler L, Pelte N, Boutros M, Marques JT, Daeffler L, Hoffmann JA, Roussel A, et al. Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2016;113:698–703. doi: 10.1073/pnas.1516122113. **Reference 50 reports an immunomodulatory cytokine encoded by Drosophila that is co-opted by different insect DNA viruses.