Highlights
-
•
Viruses are sensed by host pattern recognition receptors during infection.
-
•
Viruses must inhibit the innate antiviral immune response for full replication.
-
•
Viruses can sequester innate immune proteins or target them for degradation.
-
•
RNA viruses encode proteases that cleave host innate immune proteins.
Abstract
Upon infection, both DNA and RNA viruses can be sensed by pattern recognition receptors (PRRs) in the cytoplasm or the nucleus to activate antiviral innate immunity. Sensing of viral products leads to the activation of a signaling cascade that ultimately results in transcriptional activation of type I and III interferons, as well as other antiviral genes that together mediate viral clearance and inhibit viral spread. Therefore, in order for viruses to replicate and spread efficiently, they must inhibit the host signaling pathways that induce the innate antiviral immune response. In this review, we will highlight recent advances in the understanding of the mechanisms by which viruses evade PRR detection, intermediate signaling molecule activation, transcription factor activation, and the actions of antiviral proteins.
Current Opinion in Microbiology 2016, 32:113–119
This review comes from a themed issue on Host-microbe interactions: viruses
Edited by Jonathan C Kagan
For a complete overview see the Issue and the Editorial
Available online 8th June 2016
http://dx.doi.org/10.1016/j.mib.2016.05.015
1369-5274/© 2016 Elsevier Ltd. All rights reserved.
Introduction
Upon virus infection, viral pathogen-associated molecular patterns (PAMPs) are sensed by host pattern recognition receptors (PRRs). PAMPs are unique features present in viruses that are not present in the host cell and therefore allow cells to distinguish self versus non-self to activate an immune response to infection. There are several types of PRRs that sense viral infection, including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and DNA sensors, which recognize both viral nucleic acids and proteins. Activation of these PRRs leads to signaling through adaptor proteins, such as MAVS and STING. These adaptor proteins then activate kinases and transcription factors that induce the expression of type I and III interferons (IFNs), as well as antiviral proteins (PRRs are reviewed in [1]). The antiviral innate immune signaling pathways highlighted in this review are shown in Figure 1, Figure 2, Figure 3, Figure 4 .
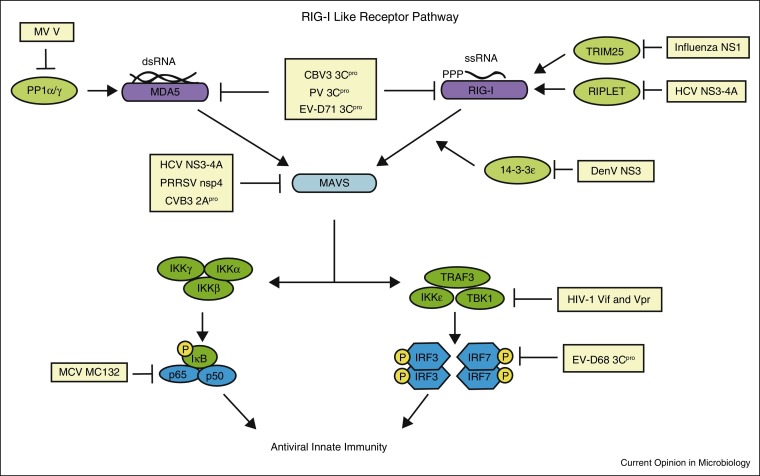
Figure 1.
Evasion of RIG-I-like receptor signaling by viruses. The RIG-I-like receptors RIG-I and MDA5 are activated by viral dsRNA in the cell cytoplasm. PP1α/γ dephosphorylates MDA5 to allow subsequent signaling. TRIM25 and RIPLET are E3 ubiquitin ligases that ubiquitinate RIG-I for its full activation. The 14-3-3ɛ protein mediates RIG-I translocation to the membrane to interact with MAVS. MAVS is the adaptor protein for both RIG-I and MDA5 and recruits downstream signaling molecules to mediate signaling to the transcription factors IRF3/7 and NFκB. Several aspects of this signaling pathway are inhibited by viruses, as shown here. Abbreviations: coxsackievirus B3 (CVB3), dengue virus (DenV), enterovirus 68 (EV-D68), enterovirus 71 (EV-D71), hepatitis C virus (HCV), human immunodeficiency virus (HIV), molluscum contagiosum virus (MCV), poliovirus (PV), porcine reproductive and respiratory syndrome virus (PRRSV), and West Nile virus (WNV).
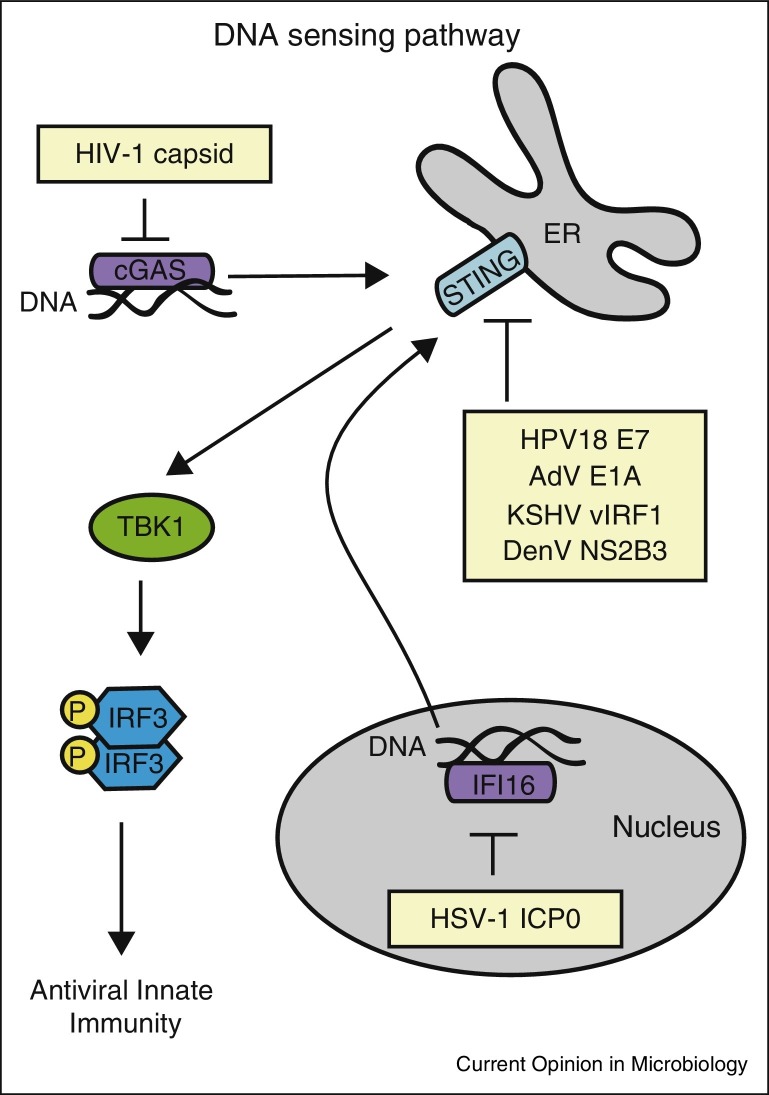
Figure 2.
Evasion of DNA sensors by viruses. Viral DNA is sensed by cGAS in the cytoplasm and IFI16 in the nucleus. cGAS and IFI16 signal to a common adaptor protein, STING, which recruits TBK1 to activate IRF3, which induces expression of type I IFN and other antiviral genes. IFI16, cGAS, and STING have all been inhibited by viruses, as shown here. Abbreviations: adenovirus (AdV), herpes simplex virus-1 (HSV-1), human immunodeficiency virus (HIV), human papillomavirus 18 (HPV18), and Kaposi's sarcoma associated herpes virus (KSHV).
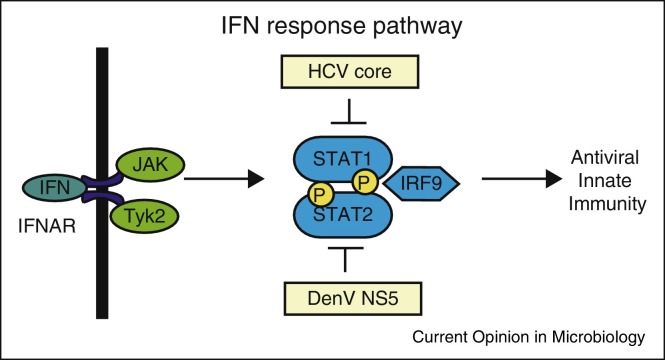
Figure 3.
Viral evasion of the IFN response pathway. Once induced, IFNs are secreted from the infected cell and signal in an autocrine and paracrine manner through the IFN receptor complex to activate the JAK/STAT pathway. This signaling leads to the activation of the ISGF3 complex, which consists of STAT1, STAT2, and IRF9, that translocates to the nucleus to induce ISGs with broad antiviral functions. Both STAT1 and STAT2 are inhibited by viruses, as shown here. Abbreviations: dengue virus (DenV), hepatitis C virus (HCV), and interferon α receptor complex (IFNAR).
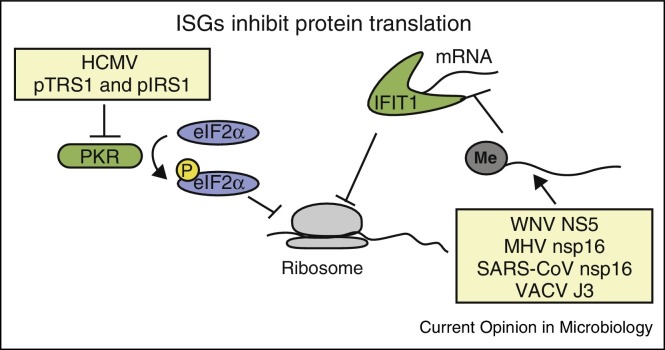
Figure 4.
Evasion of ISGs by viruses. Both PKR and IFIT1 are ISGs that limit mRNA translation in virally infected cells. PKR phosphorylates the eukaryotic initiation factor eIF2α to inhibit translation. Several viruses, including human cytomegalovirus (HCMV) shown here, inhibit this function of PKR. As IFIT1 binds to uncapped RNAs to prevent their translation, several viruses encode 2′-O methyltransferases that cap viral RNAs to prevent inhibition of their translation during infection. Abbreviations: murine hepatitis virus (MHV), severe acute respiratory syndrome coronavirus (SARS-CoV), vaccinia virus (VACV), and West Nile virus (WNV).
As full induction of antiviral innate immunity potently limits viral replication, viruses have evolved several general strategies to evade this innate immunity. A broad overview of many of the strategies to evade PRR signaling have been previously reviewed in detail [2, 3, 4•, 5•]. In this review, we describe the recent advances in viral evasion of the host antiviral innate immune response, including mechanisms for viral evasion of PRR detection, intermediate signaling molecule activation, transcription factor activation, and the actions of antiviral proteins.
Evasion or targeting of PRRs
Viruses have developed several ways to evade detection by PRRs. Many RNA viruses replicate in the cytoplasm where they are sensed by the cytoplasmic PRRs, MDA5 and RIG-I (Figure 1). In contrast, most DNA viruses replicate within the nucleus and can be sensed in the nucleus or in the cytoplasm by IFI16 or cGAS, respectively (Figure 2). Thus to avoid detection by the host innate immune system at their sites of replication, viruses have evolved several evasion strategies. For example, the positive-sense single-stranded (ss) RNA virus, dengue virus (DenV), replicates in the cytoplasm where its dsRNA would be expected to be detected by the cytoplasmic RNA sensor proteins RIG-I and MDA5 to induce type I IFN. However, new work has revealed that DenV induces membrane modifications that sequester the DenV RNA away from RIG-I and MDA5, resulting in poor induction of type I IFN [6]. It has been known for some time that hepatitis C virus (HCV), which like DenV is in the Flaviviridae family of viruses, similarly induces membrane rearrangements to house its replication machinery [7, 8]. Interestingly, it has now been suggested that similar to DenV, HCV-induced membrane rearrangement prevents recognition of HCV RNA by RIG-I [9••]. Indeed, a study by Neufeldt et al., showed that HCV co-opts host nuclear pore complex proteins (NPC) to the membranous web to regulate protein transport into the replication complex. As viral proteins have cryptic nuclear localization signals they can relocalize to these sites of replication; however, as RIG-I does not, it is excluded from the sites of replication. Intriguingly, addition of a nuclear localization signal to RIG-I, allows it to sense HCV RNA and activate IFN induction, suggesting that the NPC at the membranous web acts as a regulator to determine the proteins that can access the membranous web [9••]. This is the first demonstration that sequestered viral replication within rearranged cytoplasmic membranes actively prevents PRR sensing of viral RNA and the subsequent induction of IFN.
The PRRs RIG-I and MDA5 are frequently inhibited by viruses to prevent activation of IFN. These proteins are activated by specific PAMPs, with RIG-I recognizing ssRNA that contains a 5′ triphosphate as well as short double-stranded (ds) RNA molecules, and MDA5 sensing longer dsRNA molecules (reviewed in [3]). Enteroviruses, including poliovirus (PV), coxsackievirus B3 (CVB3), and enterovirus 71 (EV-D71), are positive-sense ssRNA viruses sensed in the cytoplasm by MDA5 and RIG-I. These enteroviruses encode two proteases, 2Apro and 3Cpro, required for viral polyprotein processing. However, 2Apro and 3Cpro have also been shown to cleave MDA5 and RIG-I, respectively [10]. This demonstrates that enteroviruses have converged on common strategies to evade multiple PRRs during infection further supporting the fact that PRR sensing is critical for limiting viral replication and spread.
Both RIG-I and MDA5 require a coordinated set of events to go from their inactive to active state (reviewed in [11]). One of the steps to activate RIG-I and MDA5 includes removal of inhibitory phosphorylation marks by the protein phosphatases PP1α and PP1γ [12]. The negative-sense ssRNA viruses, measles virus (MV) and Nipah virus, both inhibit MDA5 activation through the actions of their V protein [13, 14]. The V protein, which acts as an IFN antagonist, binds PP1α and PP1γ to prevent the dephosphorylation of MDA5 specifically during infection [13]. MV also utilizes a second strategy to prevent PP1α/γ dephosphorylation of MDA5. This strategy involves activation of a DC-SIGN signaling pathway that activates Raf1 kinase for activation of the PP1 inhibitor 1, which blocks PP1α/γ action [14].
Full activation of RIG-I requires the actions of a set of proteins, including TRIM25, RIPLET, and 14-3-3ɛ (reviewed in [11]). All of these proteins are targeted by viruses to prevent their activation. Both TRIM25 and RIPLET are E3 ubiquitin ligases that ubiquitinate RIG-I with K63-linked ubiquitin chains for its full activation. However, the negative-sense ssRNA virus, influenza virus, which is sensed by RIG-I [15, 16], evades the actions of both TRIM25 and Riplet [17, 18]. The viral NS1 protein binds to both TRIM25 and Riplet in a species-specific manner [18]. This prevents the activation of RIG-I during infection leading to a decreased induction of IFN [17, 18]. Recently, it was shown that the HCV NS3-4A protease complex, which has long been known to cleave MAVS and TRIF (see below) [19, 20, 21, 22, 23], also cleaves RIPLET to inhibit induction of IFN [24]. The 14-3-3ɛ protein, which binds to RIG-I to mediate translocation of RIG-I from the cytoplasm to interact with MAVS at intracellular membranes [25], is also inhibited by viruses. The 14-3-3ɛ protein binds proteins like RIG-I that contain phosphorylated serine or threonine at an Rxx(pS/pT)xP motif. Interestingly, the NS3 proteases of both DenV and West Nile virus (WNV) bind to 14-3-3ɛ via a phosphomimetic RxEP motif, suggesting that NS3 competitively inhibits RIG-I binding to 14-3-3ɛ, thus blocking translocation to MAVS to prevent induction of antiviral innate immunity [26••].
Viruses also evade sensing by PRRs by encoding proteins that protect the viral nucleic acids from sensors. The cytoplasmic PRR cGAS senses viral DNA in the cytoplasm (reviewed in [27]). During HIV-1 and HIV-2 infection, the viral complementary DNA (cDNA) within the virion is sensed by cGAS after infection [28]. However, the HIV-1 but not HIV-2 cDNA is protected within the viral capsid until it is transported into the nucleus for replication. The mechanism behind this protection is due to affinity of the HIV-1, but not HIV-2, capsid with the host protein cycophilin A (CypA) which stabilizes the viral capsid to prevent exposure of the viral cDNA to cGAS in the cytoplasm [28]. In addition to cGAS, viruses target IFI16, which senses DNA viruses that replicate in the nucleus. In particular, the herpes simplex virus-1 (HSV-1) immediate early protein ICP0 has E3 ubiquitin ligase activity that ubiquitinates IFI16, resulting in its degradation by the ubiquitin proteasome and loss of IFN induction [27, 29].
Targeting of adaptor proteins and their kinases
In addition to using viral proteases to cleave PRRs, as described above, viruses also utilize their proteases to target the downstream signaling molecules in antiviral innate immune pathways. In particular, the NS3-4A protease of HCV blocks antiviral signaling by cleaving at least three innate immune signaling molecules. The HCV NS3-4A protease prevents activation of the transcription factor IRF3 and induction of IFN by cleaving the signaling adaptor protein MAVS [19, 20, 21, 23]. The NS3-4A protease can also cleave TRIF, an adaptor protein for TLR3, a protein that senses viral dsRNA in the endosome [22]. Finally, as described above, NS3-4A also cleaves RIPLET [24]. As the HCV NS3-4A protease cleaves two molecules in the RIG-I signaling pathway (both RIPLET and MAVS), this suggests that either the virus is ensuring that the RIG-I pathway is inhibited or that RIPLET may have additional functions within innate immunity besides activating RIG-I.
Since MAVS activation coordinates IFN-induction by both RIG-I and MDA5, it is not surprising that viruses often target MAVS or proteins that regulate its function. In addition to HCV, the positive-sense RNA viruses, porcine reproductive and respiratory syndrome virus (PRRSV) and EV-D71, use the nsp4 cysteine protease [30] and 2Apro, respectively, to cleave MAVS during infection [31]. The DenV protease NS2B3 cleaves the mitofusins, MFN1 and MFN2, known to be positive (MFN1) or negative (MFN2) regulators of MAVS function [32, 33, 34, 35, 36]. Therefore, as their cleavage in DenV-infected cells results in increased virus replication, it suggests that cleavage of MFN1 (vs MFN2) is required to prevent the antiviral response in DenV-infected cells.
While MAVS is the adaptor for RNA virus sensing, STING is the adaptor for DNA virus sensing via the PRRs cGAS and IFI16 (reviewed in [5•]). Interestingly, Lau et al. determined that both the adenovirus E1A and human papilloma virus 18 (HPV18) E7 proteins bind to STING to prevent induction of type I IFN upon DNA transfection [37••]. Additionally, the Kaposi's sarcoma-associated herpes virus (KSHV) protein vIRF1 binds to STING and prevents its interactions with TBK1 and IRF3 to block IFN induction [38]. Importantly, this inhibition of IFN induction by KSHV was found to important for reactivation of KSHV from viral latency [38].
Intriguingly, several RNA viruses have mechanisms to block the function of STING, even though it is a known adaptor for DNA virus sensing (reviewed in [2]). The HCV NS4B protein, the DenV NS2B3 protease, and the yellow fever virus NS4B protein all block STING downstream signaling to IFN ([39] and reviewed in [40]). While the mechanism of how STING senses RNA viruses remains unclear, the fact that multiple RNA viruses have strategies to antagonize its function suggests that it must play a role in IFN induction during RNA virus infection (reviewed in [40]). Flaviviridae virus infection may damage mitochondria, leading to the releases of mitochondrial DNA that primes the innate immune response [41••]. Indeed, HCV infection induces mitophagy, and this results in decreased IFN induction suggesting that this induction of mitophagy is a viral mechanism to protect from mitochondrial DNA induction of type I IFN [42].
Viruses also target the kinases IKKɛ and TBK1, which transduce signals from MAVS or STING to activate antiviral transcription factors. IKKɛ is inhibited by the nucleoprotein of arenaviruses, including lymphocyte choriomeningitis virus and Lassa fever virus [43]. TBK1 is inhibited by both Vpr and Vif during HIV-1 infection of dendritic cells and macrophages to prevent its autophosphorylation and activation [44]. Further, both TBK1 and IKKɛ are inhibited by the ebola virus Vp35 protein to prevent their interactions with the transcription factors IRF3 and IRF7 [45]. Since these kinases can be activated by multiple PRR pathways, inhibition of the kinases broadly inhibits the antiviral innate immune response.
Targeting transcription factors
Viruses also directly inhibit transcription factors that act in the IFN induction and response pathways to prevent transcriptional activation of IFNs and interferon-stimulated genes (ISGs) during virus infection. To evade IFN induction, enterovirus 68 (EV-D68) 3Cpro cleaves IRF7 during infection [46]. The human poxvirus, molluscum contagiosum virus (MCV), a DNA virus, also evades IFN induction by using its MC132 protein to recruit the Elongin B/Elongin C/Cullin-5 ubiquitin ligase complex to ubiquitinate and degrade the p65 subunit of NFκB to prevent its activation [47]. To antagonize the transcriptional induction of the IFN response pathway, several viruses directly act on the STAT proteins. Both STAT1 and STAT2 are phosphorylated following IFN signaling thereby promoting their interaction with IRF9 to form the ISGF3 complex that transcriptionally activates ISGs (Figure 3). In particular, the DenV NS5 protein targets STAT2 for degradation, resulting in the ubiquitination and degradation of STAT2 [48]. Additionally, the HCV core protein dysregulates STAT1 signaling by increasing the levels of non-phosphorylated STAT1 in the cell [49]. Antagonism of transcription factors by viruses efficiently blocks IFN signaling and ISG induction.
Evasion of ISGs
Not surprisingly, viruses have evolved ways to inhibit the antiviral actions of ISGs that are induced by the IFN response pathway. These ISGs have broad mechanisms to confer antiviral activity [50]. In this section, we will focus on how viruses evade the antiviral actions of the ISGs IFIT1 and PKR (Figure 4). The IFIT proteins bind to uncapped RNA to prevent their translation. While many viruses have uncapped RNA and use internal ribosome entry sites for their translation (e.g. HCV), some viruses have evolved ways to cap their RNA to evade IFIT1 recognition. For example, Lassa fever virus and influenza virus snatch caps from host mRNAs. Additionally, many viruses encode proteins that can perform these capping functions (reviewed in [51]). In particular, the WNV NS5 protein contains 2′-Omethyltransferase (2′O-MT) activity to generate a cap 1 structure. This particular cap structure is not sensed by IFIT1 during infection therefore this allows the virus to evade restriction by IFIT1 [52]. Coronaviruses, positive-sense ssRNA viruses, also encode a 2′O-MT protein, nsp16 [53]. Similar to the MT activity of WNV NS5A, the MT activity of nsp16 is required for evasion of IFIT sensing during both murine hepatitis virus and severe acute respiratory syndrome coronavirus infection [52, 54]. Vaccinia virus, a DNA virus that replicates exclusively in the cytoplasm, also has a 2′O-MT and disruption of its activity results in increased susceptibility of vaccinia virus to IFIT protein restriction [52]. Taken together, many viruses evade the actions of IFIT1, demonstrating that IFIT1 has the capacity for potent restriction of viral replication.
The antiviral effector ISG PKR is one of the most common proteins targeted and inactivated by viruses (reviewed in [55]). Activation of this ISG by dsRNA results in PKR autophosphorylation, dimerization, and phosphorylation of eIF2α leading to decreased protein synthesis due to translational inhibition. This inhibition of translation affects both host and viral mRNAs, which ultimately decreases viral replication. A recent example of inhibition of PKR function was described during infection with human cytomegalovirus virus (HCMV), a DNA virus of the herpesvirus family [56]. This virus encodes two proteins, pTRS1 and pIRS1, that antagonize PKR to prevent its autophosphorylation and subsequent phosphorylation of eIF2α. Importantly, deletion of the viral pTRS1 and pIRS1 proteins leads to decreased expression of viral early and late proteins, resulting in decreased viral replication [56]. This suggests that these proteins are critical for HCMV to prevent the antiviral activity of PKR for its replication.
Conclusions
Evasion of the host antiviral innate immune response is critical for virus replication and spread. Viruses have several strategies to evade IFN induction and signaling to avoid the antiviral mechanisms of the host innate immune system. In fact, some viruses utilize multiple strategies to evade antiviral innate immune signaling, as is seen with HCV. This virus evades RIG-I detection of its replicating RNA in the membranous web by co-opting the NPC to regulate protein trafficking to the these sites of replication [9••]. It also cleaves MAVS, TRIF, and RIPLET to prevent downstream signaling to IRF3 and NFκB [19, 20, 21, 22, 23, 24]. Further, it induces mitophagy to limit IFN induction and it also inhibits the transcription factor STAT1 to prevent ISG induction [42, 49]. Taken together, there is a strong need for viruses to evade IFN induction and signaling to prevent activation of host innate immune system to allow for viral replication.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We would like to thank Christine Vazquez, Nandan Gokhale and Michael McFadden for helpful discussion and reading of the manuscript. This work was supported by the National Institutes of Health (K22 AI100935 — SMH, 5P30 AI064518 — SMH, and T32-CA009111 — DCB), the Duke University Center for AIDS Research (CFAR), and a Duke School of Medicine Whitehead Scholarship (SMH).
References
- 1.Sparrer K.M., Gack M.U. Intracellular detection of viral nucleic acids. Curr Opin Microbiol. 2015;26:1–9. doi: 10.1016/j.mib.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Z., Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe. 2016;19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Chiang J.J., Davis M.E., Gack M.U. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev. 2014;25:491–505. doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review describing the RLR signaling pathways and mechanisms that viruses use to evade these pathways.
- 5•.Orzalli M.H., Knipe D.M. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review describing the DNA sensing signaling pathways and mechanisms that viruses use to evade these pathways.
- 6.Uchida L., Espada-Murao L.A., Takamatsu Y., Okamoto K., Hayasaka D., Yu F., Nabeshima T., Buerano C.C., Morita K. The dengue virus conceals double-stranded RNA in the intracellular membrane to escape from an interferon response. Sci Rep. 2014;4:7395. doi: 10.1038/srep07395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egger D., Wolk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H.E., Bienz K., Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Neufeldt C.J., Joyce M.A., Van Buuren N., Levin A., Kirkegaard K., Gale M., Jr., Tyrrell D.L., Wozniak R.W. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12:e1005428. doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the first studies to show that viral-induced membrane modification for replication compartments excludes PRRs from these sites of viral replication.
- 10.Feng Q., Langereis M.A., Lork M., Nguyen M., Hato S.V., Lanke K., Emdad L., Bhoopathi P., Fisher P.B., Lloyd R.E. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gack M.U. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol. 2014;88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wies E., Wang M.K., Maharaj N.P., Chen K., Zhou S., Finberg R.W., Gack M.U. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis M.E., Wang M.K., Rennick L.J., Full F., Gableske S., Mesman A.W., Gringhuis S.I., Geijtenbeek T.B., Duprex W.P., Gack M.U. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe. 2014;16:19–30. doi: 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesman A.W., Zijlstra-Willems E.M., Kaptein T.M., de Swart R.L., Davis M.E., Ludlow M., Duprex W.P., Gack M.U., Gringhuis S.I., Geijtenbeek T.B. Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. Cell Host Microbe. 2014;16:31–42. doi: 10.1016/j.chom.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 16.Opitz B., Rejaibi A., Dauber B., Eckhard J., Vinzing M., Schmeck B., Hippenstiel S., Suttorp N., Wolff T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 17.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E., Farzan M., Inoue S., Jung J.U., Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajsbaum R., Albrecht R.A., Wang M.K., Maharaj N.P., Versteeg G.A., Nistal-Villan E., Garcia-Sastre A., Gack M.U. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 20.Loo Y.M., Owen D.M., Li K., Erickson A.K., Johnson C.L., Fish P.M., Carney D.S., Wang T., Ishida H., Yoneyama M. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foy E., Li K., Wang C., Sumpter R., Jr., Ikeda M., Lemon S.M., Gale M., Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 22.Li K., Foy E., Ferreon J.C., Nakamura M., Ferreon A.C., Ikeda M., Ray S.C., Gale M., Jr., Lemon S.M. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshiumi H., Miyashita M., Matsumoto M., Seya T. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H.M., Loo Y.M., Horner S.M., Zornetzer G.A., Katze M.G., Gale M., Jr. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Chan Y.K., Gack M.U. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol. 2016;17:523–530. doi: 10.1038/ni.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]; A study describing how the DenV protease acts as a phosphomimetic to bind to 14-3-3ɛ to prevent binding to RIG-I and translocation of RIG-I to MAVS.
- 27.Orzalli M.H., Broekema N.M., Diner B.A., Hancks D.C., Elde N.C., Cristea I.M., Knipe D.M. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci U S A. 2015;112:E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahaye X., Satoh T., Gentili M., Cerboni S., Conrad C., Hurbain I., El Marjou A., Lacabaratz C., Lelievre J.D., Manel N. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Orzalli M.H., DeLuca N.A., Knipe D.M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J., Xu S., Wang J., Luo R., Wang D., Xiao S., Fang L., Chen H., Jiang Y. Porcine reproductive and respiratory syndrome virus 3C protease cleaves the mitochondrial antiviral signalling complex to antagonize IFN-beta expression. J Gen Virol. 2015;96:3049–3058. doi: 10.1099/jgv.0.000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L.C., Chen S.O., Chang S.P., Lee Y.P., Yu C.K., Chen C.L., Tseng P.C., Hsieh C.Y., Chen S.H., Lin C.F. Enterovirus 71 proteins 2A and 3D antagonize the antiviral activity of gamma interferon via signaling attenuation. J Virol. 2015;89:7028–7037. doi: 10.1128/JVI.00205-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu C.Y., Liang J.J., Li J.K., Lee Y.L., Chang B.L., Su C.I., Huang W.J., Lai M.M., Lin Y.L. Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PLoS Pathog. 2015;11:e1005350. doi: 10.1371/journal.ppat.1005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasukawa K., Oshiumi H., Takeda M., Ishihara N., Yanagi Y., Seya T., Kawabata S., Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 35.Castanier C., Garcin D., Vazquez A., Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onoguchi K., Onomoto K., Takamatsu S., Jogi M., Takemura A., Morimoto S., Julkunen I., Namiki H., Yoneyama M., Fujita T. Virus-infection or 5’ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Lau L., Gray E.E., Brunette R.L., Stetson D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]; A study that demonstrates that transformed cells transfected with DNA do not induce IFN during infection because endogenous viral oncogenes inhibit STING.
- 38.Ma Z., Jacobs S.R., West J.A., Stopford C., Zhang Z., Davis Z., Barber G.N., Glaunsinger B.A., Dittmer D.P., Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015;112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D., Maringer K., Bernal-Rubio D., Shabman R.S., Simon V. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maringer K., Fernandez-Sesma A. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev. 2014;25:669–679. doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]; A study describing how mitochondrial stress leads to release of mitochondrial DNA into the cytoplasm, which can activate IFN signaling to prime the innate immune response during infection.
- 42.Kim S.J., Syed G.H., Khan M., Chiu W.W., Sohail M.A., Gish R.G., Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A. 2014;111:6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pythoud C., Rodrigo W.W., Pasqual G., Rothenberger S., Martinez-Sobrido L., de la Torre J.C., Kunz S. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKepsilon. J Virol. 2012;86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harman A.N., Nasr N., Feetham A., Galoyan A., Alshehri A.A., Rambukwelle D., Botting R.A., Hiener B.M., Diefenbach E., Diefenbach R.J. HIV blocks interferon induction in human dendritic cells and macrophages by dysregulation of TBK1. J Virol. 2015;89:6575–6584. doi: 10.1128/JVI.00889-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prins K.C., Cardenas W.B., Basler C.F. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang Z., Liu L., Lei X., Zhou Z., He B., Wang J. 3C protease of enterovirus D68 inhibits cellular defense mediated by interferon regulatory factor 7. J Virol. 2015;90:1613–1621. doi: 10.1128/JVI.02395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brady G., Haas D.A., Farrell P.J., Pichlmair A., Bowie A.G. Poxvirus protein MC132 from molluscum contagiosum virus inhibits NF-B activation by targeting p65 for degradation. J Virol. 2015;89:8406–8415. doi: 10.1128/JVI.00799-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison J., Laurent-Rolle M., Maestre A.M., Rajsbaum R., Pisanelli G., Simon V., Mulder L.C., Fernandez-Sesma A., Garcia-Sastre A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stone A.E., Mitchell A., Brownell J., Miklin D.J., Golden-Mason L., Polyak S.J., Gale M.J., Jr., Rosen H.R. Hepatitis C virus core protein inhibits interferon production by a human plasmacytoid dendritic cell line and dysregulates interferon regulatory factor-7 and signal transducer and activator of transcription (STAT) 1 protein expression. PLoS One. 2014;9:e95627. doi: 10.1371/journal.pone.0095627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dauber B., Wolff T. Activation of the antiviral kinase PKR and viral countermeasures. Viruses. 2009;1:523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziehr B., Vincent H.A., Moorman N.J. Human cytomegalovirus pTRS1 and pIRS1 antagonize protein kinase R to facilitate virus replication. J Virol. 2016;90:3839–3848. doi: 10.1128/JVI.02714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]