Abstract
Transcriptional regulation of cold-responsive genes is crucial for exogenous melatonin-mediated cold tolerance in plants. Nonetheless, how melatonin regulates cold-responsive genes is largely unknown. In this study, we found that exogenous melatonin improved cold tolerance in watermelon by regulating expression of microRNAs (miRNAs). We identified a set of miRNAs that were regulated by melatonin under unstressed or cold conditions. Importantly, mRNA-seq analysis revealed that melatonin-induced downregulation of some miRNAs, such as miR159-5p, miR858, miR8029-3p, and novel-m0048-3p correlated with the upregulation of target genes involved in signal transduction (CDPK, BHLH, WRKY, MYB, and DREB) and protection/detoxification (LEA and MDAR) under cold stress. These results suggest that miRNAs may be involved in melatonin-mediated cold tolerance in watermelon by negatively regulating the expression of target mRNAs.
Keywords: cold stress, high-throughput sequencing, melatonin, microRNA, watermelon
Introduction
Plants have to endure various abiotic stresses due to their sessile life-style. In particular, cold stress is one of the destructive environmental stresses that considerably reduce both yield and quality of fruits and vegetables in tropics and subtropics (Rivero et al., 2001, 2002). Transitory as well as constant cold stress causes damage to cell membranes, which disrupts the balance between water uptake and transpiration, leading to dehydration in shoots. Cold stress decreases the photosynthesis rate by affecting stomatal movement (Foyer et al., 2002), and induces accumulation of reactive oxygen species (ROS) by disrupting electron transport system in both mitochondria and chloroplasts (Suzuki and Mittler, 2006). ROS at high concentration can damage membranes through lipid peroxidation, break DNA strand, and inactivate various vital enzymes (Cheng and Song, 2006).
To survive cold stress, plants have evolved a variety of stress response mechanisms that minimize damage, ensure proper cellular homeostasis, and enable plants to function under stressful condition. Molecular sensors distributed in different cellular compartments can sense any decrease in growth temperatures, and thereby generate secondary signals, such as Ca2+, ROS, and inositol 1,4,5-trisphosphate (InsP), and activate different transcriptional regulators, such as basic helix-loop-helix (BHLH), INDUCER OF CBF EXPRESSION (ICE) 1, C-repeat-binding factor (CBF), WRKY, and MYB, via the activation of phosphoprotein kinases, such as calcium-dependent protein kinases (CDPKs) and multiple mitogen-activated protein kinases (Xiong et al., 2002; Mahajan and Tuteja, 2005). Eventually, late embryogenesis abundant (LEA) proteins, chaperones, detoxification enzymes, pathogenesis-related proteins, mRNA/protein-binding proteins, proteinase inhibitors, transporters, lipid-transfer proteins, and enzymes required for osmoprotectant biosynthesis are induced to maintain normal physiological processes including photosynthesis (Janská et al., 2010).
Melatonin (N-acetyl-5-methoxytryptamine), a low molecular-weight molecule with an indole ring in its structure, is ubiquitous in living organisms (Hardeland et al., 2011; Tan et al., 2012). In vascular plants, melatonin was first identified in 1995 (Dubbels et al., 1995; Hattori et al., 1995) and numerous subsequent studies have established its important roles in plant growth, development, and defense against various abiotic and biotic stresses, such as salinity, drought, cold, excess copper, and pathogens (Zhang et al., 2014; Arnao and Hernández–Ruiz, 2015). Previously, the primary role of melatonin in stress mitigation was considered as a broad spectrum antioxidant that directly scavenges ROS and/or modulates cellular antioxidant system (Wang P. et al., 2012; Zhang and Zhang, 2014; Manchester et al., 2015). Recent studies have revealed that melatonin could also activate defense systems by regulating the expression of stress-responsive genes involved in signal transduction. For example, exogenous melatonin upregulates the expression of cold-responsive genes such as C2H2-type zinc finger transcription factor (ZAT) 10, ZAT12, CBFs, cold-responsive gene (COR) 15, and calmodulin-binding transcription activator (CAMTA) 1 under cold stress (Bajwa et al., 2014). Furthermore, Shi and Chan (2014) reported that the AtZAT6-activated CBF pathway might be essential for melatonin-mediated response to freezing stress in Arabidopsis. However, the mechanism by which melatonin regulates the expression of stress-responsive genes to activate defense networks is unclear.
MicroRNAs (miRNAs) with a length of 19–25 nucleotides (nt), are highly conserved, endogenous, single-stranded non-coding RNA molecules. An increasing number of studies have demonstrated that miRNAs are important regulators in plant development and stress responses. Generally, miRNAs regulate the response to biotic and abiotic stresses by binding to reverse complementary sequences, resulting in the cleavage or translational inhibition of target mRNAs (Khraiwesh et al., 2012; Sunkar et al., 2012). For instance, miR398 is downregulated to release its suppression of CSD1 and CSD2 in plant responses to oxidative stress (Sunkar et al., 2006). The increased sensitivity of TamiR159-overexpressing rice lines to heat stress suggests that the downregulation of TamiR159, which targets TaGAMYB1 and TaGAMYB2 in wheat, is involved in a heat stress-related signaling pathway, and therefore contributes to heat stress tolerance (Wang Y. et al., 2012). Transcriptional control of the expression of cold-responsive genes is well known, but miRNAs have recently been added to the suite of cold-responsive gene regulatory networks. Various miRNAs that target stress-related genes are significantly up- or downregulated during cold stress in a range of plant species (Khraiwesh et al., 2012; Sunkar et al., 2012).
Watermelon (Citrullus lanatus L.), one of the most economically important crops in the world, is highly sensitive to low temperatures (Rivero et al., 2002). In this study, by assessing the watermelon plants in terms of leaf phenotype, the maximum quantum yield of PSII (Fv/Fm), and relative electrolyte leakage (REL) under cold stress, we found that exogenous melatonin could enhance watermelon tolerance to cold stress. To understand the molecular mechanisms of melatonin-mediated cold tolerance in watermelon, we carried out a high-throughput miRNA and mRNA sequencing. The results showed that melatonin up- or downregulated a set of miRNAs under normal temperature or cold stress conditions. Importantly, under the cold stress, the downregulation of some miRNAs, such as miR159-5p, miR858, miR8029-3p, and novel-m0048-3p, by melatonin may cause the upregulation of putative target genes involved in signal transduction (CDPK, BHLH, WRKY, MYB, and DREB) and protection/detoxification (LEA and MDAR), suggesting that some miRNAs may be involved in melatonin-mediated cold tolerance in watermelon.
Materials and Methods
Plant Materials, Growth Conditions, and Treatments
Watermelon (C. lanatus L., cv. Y134) seeds were surface sterilized with 5% sodium hypochlorite (NaOCl) solution for 5 min and then rinsed with running water and distilled water. The sterilized seeds were soaked in distilled water for 8 h and sown directly in pots filled with a mixture of peat/vermiculite (3/1, v/v). Plants were grown in growth chambers with the following environmental conditions: a constant relative humidity of 60–70%, a 12-h photoperiod, 25/18°C (day/night), and a photosynthetic photon flux density of 600 μmol m-2 s-1. The plants were watered daily and fertilized with Hoagland’s nutrition solution at 1-day interval.
Seedlings at the four-leaf stage were sprayed with 150 μM melatonin solution for 3 days, with distilled water used as the control. The melatonin (Sigma-Aldrich, St. Louis, MO, USA) solutions were prepared by dissolving the solute in ethanol followed by dilution with Milli-Q water [ethanol/water (v/v) = 1/10,000]. Each plant was sprayed with 20 mL of solution. Twelve hours after the third spray of melatonin, the plants were exposed to cold stress (i.e., 4°C) for 36 h with a 12-h photoperiod and photosynthetic photon flux density of 600 μmol m-2 s-1. Leaf samples were harvested at different time-points after imposition of cold stress, such as at 0, 3, 6, 12, 24, and 36 h to analyze Cla020078 (CBF1) and Cla020702 (MYB) transcript levels, at 6 h to sequence miRNAs and mRNAs, and at 36 h to analyze cold tolerance.
Analysis of Chlorophyll Fluorescence and REL
The maximum photochemical efficiency of PSII (Fv/Fm) was measured with Portable Chlorophyll Fluorometer (PAM2500; Heinz Walz, Effeltrich, Germany), after the whole plants were dark adapted for 30 min. Minimal fluorescence (Fo) was measured during the weak measuring pulses and maximal fluorescence (Fm) was measured by a 0.8-s pulse light at 4,000 μmol m-2 s-1. Fv/Fm was determined using the third leaf of watermelon plants counting from the bottom to up. The calculation of Fv/Fm was done by the following formula as described by van Kooten and Snel (1990). Fv/Fm = (Fm - Fo)/Fm.
REL in the leaves was determined as previously described elsewhere (Zhou and Leul, 1998).
Library Construction and Sequencing of Small RNA
Eight independent small RNA libraries from plant leaves for four treatments (Control, CK; Melatonin, MT; Cold; Melatonin+Cold, MT-C), with two biological replicates for each treatment, were sequenced. The sequences have been deposited into the NCBI Sequence Read Archive database (SRP078211, SRA438995). Library construction and sequencing of small RNA were performed using a service provider Gene Denovo Co. (Guangzhou, China). Total RNA was extracted using miRNeasy Mini Kit (Cat#217004, QIAGEN GmBH, Germany) according to the manufacturer’s protocol. Total RNA integrity was measured on an Agilent 2100 Bioanalyzer system (Agilent) for quality control. A total of 16–35 nt RNA fragments were excised, purified from a PAGE gel, and ligated with 5′ and 3′ adaptors using T4 RNA ligase. Reverse transcription followed by PCR was used to create cDNA constructs based on the small RNA ligated with 3′ and 5′ adapters. Subsequently, the amplified cDNA constructs were purified from agarose gel, in preparation for sequencing analysis using the Illumina Genome Analyzer (Illumina, CA, USA) according to the manufacturer’s instructions.
Identification and Differential Expression Analysis of Known and Novel miRNAs
The raw sequences were firstly processed by Illumina’s Genome Analyzer Pipeline software to filter out the adapter sequences, low quality as well as low-copy sequences. Then, the extracted small RNA sequences with 15–26 nt in length were subjected to cucurbit species mRNAs, Rfam1 and Repbase2 to discard mRNA, rRNA, tRNA, snRNA, snoRNA and repeat sequences. Finally, the remaining unique sequences were analyzed by BLAST against miRBase (Release 21)3. Solexa sequences with identical or related (one mismatch) sequences from mature miRNAs were identified as known miRNAs (Meyers et al., 2008).
To identify potential novel miRNAs in watermelon, rest of the unmapped small RNA sequences were searched by BLAST against watermelon genome downloaded from cucumber genome database4. The mappable sequences were then folded into a secondary structure using UNAfold software5. Only the non-coding sequences which could form a perfect stem-loop structure and meet the criteria for miRNAs prediction (Meyers et al., 2008) were then considered to be a potential novel miRNA candidate.
We chose the miRNAs increased or decreased by more than twofold and P-value <0.01 in two treatments as the criterion for a melatonin or cold response. Then, the heat map of differentially expressed miRNAs expression profile was drawn with MultiExperiment Viewer version 4.0 and clustering analysis was performed using a hierarchical clustering method (Eisen et al., 1998).
Expression Analysis of Predicted Target Genes Based on High-Throughput Sequencing
Eight independent mRNA libraries from plant leaves for four treatments (Control, CK; Melatonin, MT; Cold; Melatonin+Cold, MT-C), with two biological replicates for each treatment, were sequenced. The sequences have been deposited into the NCBI Sequence Read Archive database (SRP078211, SRA438977). Library construction and sequencing of mRNA were performed using a service provider Gene Denovo Co. (Guangzhou, China). After removal of low quality sequences, the clean reads were mapped to the watermelon reference genome, allowing up to one mismatch. The differentially expressed genes were identified using the R package edgeR (Robinson et al., 2010). The expression level of each unigene was calculated and normalized to generate FPKM (fragments per kilobase of exon per million mapped fragments). The false discovery rate (FDR) was used to determine the threshold of the P-value in multiple tests. In this study, the FDR < 0.05 and fold change ≥1.5 were used as significance cut-offs of the gene expression differences.
Prediction of Putative Target Genes of Melatonin- or Cold-Responsive miRNAs
A plant miRNA target prediction server6, was used to predict putative miRNA target genes with default settings based on the library of Watermelon genome database, version 1. It reports all potential sequences complementary to an inquiring miRNA sequence with mismatches no more than a specified value for each mismatch type. The minimal weighed score ≤3.0 was applied in the prediction according to scoring schema of miRU by Zhang (2005). The other default settings as follows: length for complementarity scoring (hspsize), 20 bp; target accessibility-allowed maximum energy to unpair the target site (UPE), 25.0; flanking length around target site for target accessibility analysis, 17 bp in upstream/13 bp in downstream; range of central mismatch leading to translational inhibition, 9–11 nt (Brodersen et al., 2008).
RNA Extraction and Quantitative Real-time PCR Analysis
Two cold-sensitive genes Cla020078 (CBF1) and Cla020702 (MYB) were chose to perform quantitative real-time PCR (qRT-PCR). Total RNA was extracted using a RNA extraction kit (Tiangen, Beijing, China) according to the supplier’s instructions. DNA contamination was removed using a purifying column. One microgram of total RNA was reverse-transcribed using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan) following the supplier’s instructions. The gene-specific primers for qRT-PCR were designed based on their cDNA sequences, as follows: Cla020078 (F, AGCAGAGCCCTAACACAGGT; R, AATGGTCTTGAGTTGGG), Cla020702 (F, GATCCATTGACGGCACTAAC; R, TCGCTACAACGTCCTTCATC), and watermelon β-actin gene (F, CCATGTATGTTGCCATCCAG; R, GGATAGCATGGGGTAGAGCA) was used as an internal control (Kong et al., 2014). The qRT-PCR assays were performed using an iCycler Iq Multicolor PCR Detection System (Bio-Rad, Hercules, CA, USA). PCRs were performed using the SYBR Premix ExTaq II (2×) Kit (Takara, Tokyo, Japan). The PCR conditions consisted of denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. The quantification of mRNA levels was based on the method of Livak and Schmittgen (2001).
Statistical Analysis
The experiment was a completely randomized design with three replicates. Each replicate contained at least 10 plants. Analysis of variance was used to test for significance, and significant differences (P < 0.05) between treatments were determined using Tukey’s test.
Results
The Effects of Melatonin on Cold Stress Tolerance and Cold-Responsive Gene Expressions in Watermelon
Melatonin plays important regulatory roles in plant defense against various biotic and abiotic stresses (Zhang et al., 2014). In this study, we first analyzed the effects of exogenous melatonin on watermelon tolerance to cold stress. As shown in Figure 1, melatonin had a minimal effect on the leaf phenotypes, the maximum quantum yield of PSII (Fv/Fm), and REL at optimal growth temperatures. After 4°C treatment for 36 h, the blade edges of watermelons were wilted, and the Fv/Fm and REL were significantly reduced and increased, respectively. However, pretreatment with melatonin obviously alleviated cold-induced wilting of blade edges and the reduction and increase in leaf Fv/Fm and REL, respectively. For example, the Fv/Fm and REL decreased and increased by 15.45 and 19.82%, respectively in melatonin-pretreated plants after cold treatment, far less than the changes of 31.06 and 50.51% in control plants (Figure 1). Taken together, exogenous melatonin significantly enhanced watermelon tolerance to cold stress.
FIGURE 1.
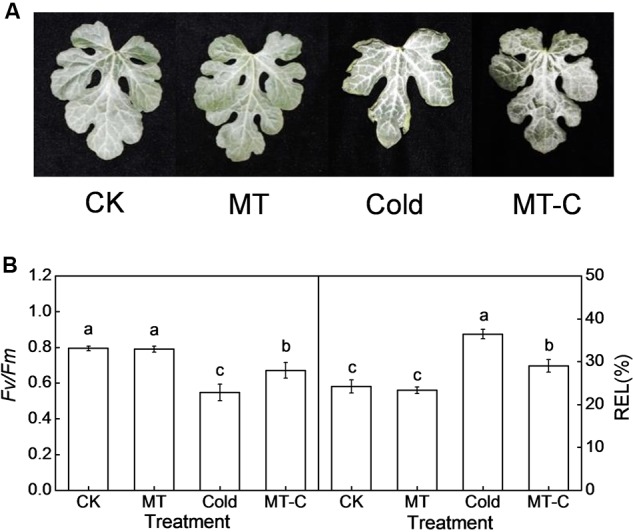
Changes in (A) the leaf phenotypes, (B) the maximum photochemical efficiency of PSII (Fv/Fm) and relative electrolyte leakage (REL) as influenced by melatonin or/and cold stress. Leaves of watermelon (Citrullus lanatus L.) seedlings at the four-leaf stage were pretreated with 150 μM melatonin (MT) for 3 days. Then, they were exposed to cold stress (i.e., 4°C) for 36 h. Data of Fv/Fm are the means of six replicates (±SD). Data of REL are the means of four replicates (±SD). Means denoted by the same letter did not significantly differ at P < 0.05 according to Tukey’s test. CK, Control; MT, Melatonin; Cold; MT-C, Melatonin+Cold.
To analyze the effects of melatonin on the expression of cold-responsive genes and to choose the appropriate time point for miRNA expression profiling, we examined the dynamic responses of Cla020078 (CBF1) and Cla020702 (MYB) to melatonin or/and cold treatment. Transcript levels of CBF1 and MYB were slightly induced by melatonin at optimal growth temperatures (Figure 2). After cold treatment (4°C), they were rapidly upregulated, reaching peak levels at 6 h, and subsequently declining to original levels at 36 h. Furthermore, pretreatment with melatonin improved the induction of CBF1 and MYB expression by cold stress. For instance, at 6 h after cold treatment, the expression levels of CBF1 and MYB in melatonin-pretreated plants were upregulated by 5.99- and 85.69-fold, respectively, far more than the increases of 3.18- and 43.38-fold observed in control plants.
FIGURE 2.
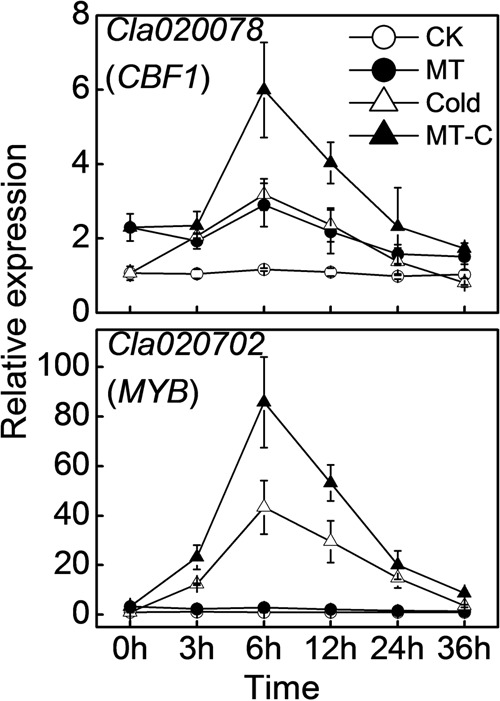
The dynamic response of the expression of Cla020078 (CBF1) and Cla020702 (MYB) to melatonin or/and cold stress. The plants were treated as described in Figure 1 and the leaf samples were harvested at indicated times (h) after cold treatment. Data are the means of four replicates (±SD). Means denoted by the same letter did not significantly differ at P < 0.05 according to Tukey’s test.
Analysis of MicroRNA Response to Melatonin or/and Cold Based on High-Throughput Sequencing
To examine whether miRNAs are involved in melatonin-mediated cold response in watermelon, we performed high-throughput sequencing analysis of miRNA in watermelon leaves treated with and without the melatonin and cold stress. A total of 9,704,684/10,831,539 (Control, CK-1/2), 10,124,858/12,383,237 (Melatonin, MT-1/2), 12,812,180/9,718,613 (Cold-1/2), and 9,806,063/11,489,173 (Melatonin+Cold, MT-C-1/2) raw reads were obtained (Supplementary Table S1). After the removal of rRNAs, tRNAs, snRNAs, and snoRNAs, a total of 5,327,079/5,914,071, 5,635,109/6,581,846, 6,219,825/5,085,935, and 5,409,138/6,272,887 sRNA sequences for CK-1/2, MT-1/2, Cold-1/2, and MT-C-1/2 were obtained, respectively. The majority of redundant reads were in the range of 19–26 nt, among which the most abundant sequences were 24 nt long in all libraries (Figure 3A). To identify miRNAs in watermelon, all sRNA sequences were compared to known plant miRNAs in miRBase Release 21. A total of 440 known unique miRNAs with high sequence similarity to known plant miRNAs were identified (Supplementary Table S2). By mapping all unique sRNA sequences to the watermelon genome and predicting the hairpin structures for their flanking sequences, 106 novel miRNAs candidates were identified. The expression level of each miRNA was normalized to generate TPM (tags per million) and the correlation of detected miRNA expression between two biological repeats for each treatment was analyzed. As shown in Figure 3B, there was a strong correlation (R = 0.99, P < 0.0001) between the two biological replicates for all four treatments, suggesting that the sequencing results are highly reliable.
FIGURE 3.
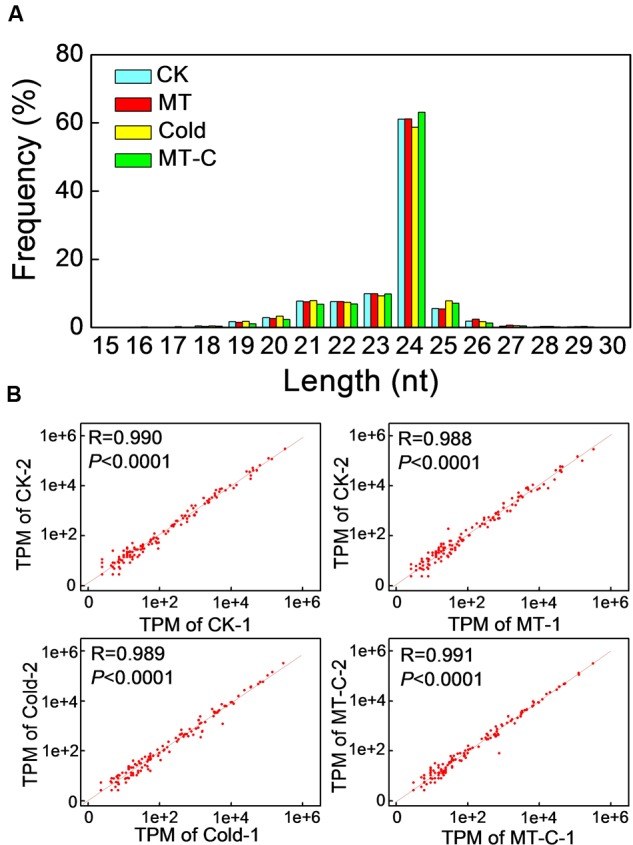
(A) Summary of the sequence lengths distribution and (B) the correlation between two biological repeats in melatonin or/and cold treatments. The plants were treated as described in Figure 1 and samples were harvested at 6 h after cold treatment.
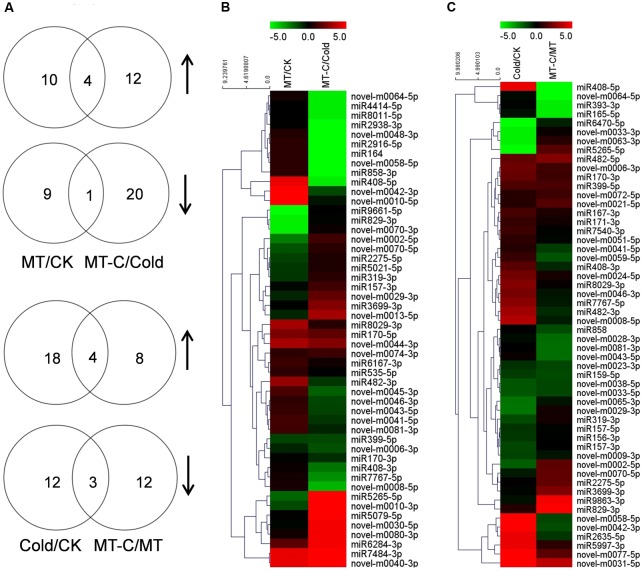
A total of 50 miRNAs were significantly up- or downregulated by melatonin under unstressed or cold conditions (Figure 4). Among them, four miRNAs such as miR170-5p, novel-m0074-3p, novel-m0044-3p, and novel-m0040-3p were upregulated, while one miRNA (miR399-5p) were up- and downregulated by melatonin pretreatment, under both unstressed and cold conditions. Ten and nine miRNAs were specifically up- and downregulated by melatonin at optimal growth temperatures, respectively. Twelve and 20 miRNAs were specifically up- and downregulated by melatonin after cold treatment for 6 h, respectively. A total of 54 miRNAs were significantly up- or downregulated by cold stress in control or melatonin-pretreated plants, respectively. Among them, four miRNAs (miR399-5p, miR170-3p, miR482-5p, and novel-m0031-5p) and three miRNAs (miR159-5p, novel-m0033-5p, and novel-m0038-5p) were up- and downregulated by cold stress, respectively, in both control and melatonin-pretreated plants. In total, 18 and 12 miRNAs were specifically up- and downregulated by cold in control plants, respectively. While 8 and 12 miRNAs were specifically up- and downregulated by cold in melatonin-pretreated plants, respectively.
FIGURE 4.
Analysis of differentially expressed miRNAs induced by melatonin or cold stress. (A) The Venn diagram illustrates number of up- (n-resize) and downregulated (s-resize) miRNAs in comparison of MT/CK, MT-C/Cold, Cold/CK, and MT-C/MT. Up- or downregulation of miRNAs was taken into account when a log2 fold change >1, and P-value <0.01. Heat map showing (B) melatonin or (C) cold induced differentially expressed miRNAs and Clustering analysis according to expression pattern. Complete hierarchical clustering carried out using Euclidean distance, color coding was done according to the scale given. Green indicates downregulation and red upregulation. CK, Control; MT, Melatonin; Cold; MT-C, Melatonin+Cold.
Biological Function Analysis of Differentially Expressed miRNAs via Prediction of Target mRNAs
To examine the biological functions of differentially expressed miRNAs, the miRNA sequences were searched against watermelon genomic sequences using the plant miRNA potential target finder7 to predict target mRNAs. miRNAs negatively regulate mRNAs by inducing their cleavage or repressing their translation (Bartel, 2004). In this study, a total of 505 mRNAs were predicted to be cleaved by miRNAs. Then, we performed RNA-seq analysis in watermelon leaves treated with and without the melatonin and cold stress using the high-throughput Illumina Solexa system. Reads from each sample were aligned to the C. lanatus reference genome. The expressions of predicted mRNAs in comparison of MT/CK, MT-C/Cold, Cold/CK, and MT-C/MT were analyzed. The expression patterns of only 49 detected mRNAs were opposite with respect to those of 21 corresponding miRNAs (Table 1). The negative correlation coefficients between miRNAs and their target mRNAs regulated by melatonin and cold were -0.74 (P < 0.001) and -0.48 (P < 0.001), respectively, suggesting that these 49 target mRNAs were likely cleaved by the corresponding miRNAs (Figure 5). Additionally, 34 miRNAs were searched against a total of 87 potential target mRNAs via translational inhibition (Supplementary Table S3). The functions of these mRNAs were annotated in the Cucurbit Genomics Database8.
Table 1.
Potential target mRNAs cleaved by differentially expressed miRNAs.
| miRNA | Target | Log2 fold change of miRNAs/targets | Description | |||
|---|---|---|---|---|---|---|
| MT/CK | MT-C/Cold | Cold/CK | MT-C/MT | |||
| miR159-5p | Cla001826 | -0.14/0.21 | -0.63/0.34 | -1.19∗/0.53 | -1.45∗/0.66∗ | Calcium-dependent protein kinase (CDPK) |
| miR170-3p | Cla006665 | -0.08/-0.04 | -1.09∗/0.31 | 2.02∗/-0.63∗ | 1.20∗/-0.28 | GRAS family transcription factor |
| miR171-3p | Cla006665 | 0.20/-0.04 | -0.31/0.31 | 1.04∗/-0.63∗ | 0.39/-0.28 | GRAS family transcription factor |
| miR399-5p | Cla008388 | -1.31∗/0.65∗ | -1.43∗/0.93∗ | 1.55∗/-3.83∗ | 1.43∗/-3.55∗ | Pentatricopeptide repeat protein 77 (PPRP77) |
| Cla005858 | /0.23 | /-0.18 | /-0.88∗ | /-1.29∗ | NEFA-interacting nuclear protein NIP30 | |
| miR482-5p | Cla014776 | -0.05/0.10 | 0.54/-0.85 | 2.01∗/-0.67 | 2.60∗/-1.61∗ | Os10g0422600 |
| Cla013089 | /0.43 | /-0.97 | /-3.27∗ | /-4.67∗ | Leucine-rich repeat family protein (LRR) | |
| Cla009419 | /0.12 | /-5.36 | /2.59∗ | /-8.06∗ | Unknown | |
| Cla016187 | /0.86 | /-1.31 | /-0.56 | /-2.57∗ | C2H2 zinc-finger transcription factor (C2H2-ZFTF) | |
| miR858 | Cla017745 | -0.08/-2.84 | -0.11/-0.23 | -0.04/1.29 | -1.02∗/3.90∗ | Late embryogenesis abundant (LEA) protein like |
| Cla017373 | /-0.67 | /0.30 | /1.50∗ | /2.47∗ | R2R3 MYB transcription factor | |
| miR2635-5p | Cla021237 | 7.03/-0.19 | -7.96/0.14 | 4.64∗/-0.70∗ | -1.04/-0.37 | Cryptic precocious CRP/Med12 |
| miR2916-5p | Cla001518 | 0.86/0.39 | -5.18∗/0.60∗ | 8.51/-1.31∗ | 0.00/-1.09∗ | DNA primase large subunit |
| miR5021-5p | Cla010003 | -1.08∗/0.64∗ | 0.59/-0.02 | -0.21/1.13∗ | 1.47/0.46 | 3-ketoacyl-CoA reductase 2 (KCR2) |
| Cla015503 | /1.53∗ | /-0.69 | /0.91 | /-1.32 | RING-H2 zinc finger protein (RING-H2 ZFP) | |
| Cla001461 | /1.33∗ | /0.25 | /0.02 | /-1.06 | Unknown | |
| Cla018974 | /1.09∗ | /0.36 | /-0.65 | /-1.38 | J023065D24 | |
| miR5997-3p | Cla009517 | 0.00/-0.12 | -0.88/0.33 | 4.64∗/-1.12∗ | 0.88/-0.67 | Vacuolar protein sorting-associated protein (VPS) 13 |
| Cla021487 | /-0.09 | /0.30 | /-0.73∗ | /-0.35 | Cysteine-rich repeat secretory protein (CRRS) 12 | |
| miR7540-3p | Cla015655 | 1.41/-0.15 | 0.17/1.01 | 1.19∗/-1.33∗ | -0.05/-0.18 | Fasciclin-like arabinogalactan protein (FLA) 2 |
| miR8029-3p | Cla018379 | 3.29∗/-0.17 | 0.80/0.70 | 2.60∗/-0.68∗ | 0.15/0.18 | Monodehydroascorbate reductase (MDAR) |
| Cla008020 | /0.01 | /0.49 | /-0.68∗ | /-0.21 | Cyclase/dehydrase | |
| Cla002580 | /-0.10 | /0.33 | /-0.80∗ | /-0.36 | Lysine-specific demethylase (LSD) 5A | |
| Cla012681 | /-0.29 | /0.36 | /-0.92∗ | /-0.27 | Pentatricopeptide repeat-containing protein (PPRP) | |
| Cla004188 | /-0.42 | /-0.04 | /-0.62∗ | /-0.24 | MYB transcription factor | |
| Cla004079 | /-1.03∗ | /-0.50 | /-3.11∗ | /-2.58 | Unknown | |
| Cla011614 | /-0.62∗ | /-0.18 | /-0.27 | /0.17 | Kinesin like protein | |
| Cla014313 | /-0.77∗ | /-0.23 | /-0.40 | /0.13 | Peroxidase (POD) | |
| novel-m0006-3p | Cla012226 | -0.70/-0.11 | -1.59∗/0.59∗ | 2.14∗/-0.25 | 1.26/0.46 | YABBY |
| hboxnovel-m0030-5p | Cla014681 | -0.11/0.88 | 4.84∗/-1.50∗ | -8.14/-2.35∗ | 0.13/-4.73∗ | K+ transporter |
| novel-m0045-3p | Cla003322 | 1.44/-0.47 | -1.89∗/0.71∗ | 0.97/-1.11∗ | -2.40/0.08 | Altered inheritance of mitochondria protein (AIMP) 32 |
| novel-m0048-3p | Cla014357 | 0.71/0.11 | -5.18∗/0.60∗ | 1.36/-2.46∗ | -7.90/-1.97∗ | Glycyl-tRNA synthetase (GARS) |
| Cla015456 | /-0.46 | /0.65∗ | /-1.87∗ | /-0.76 | Nitrate transporter (NRT) | |
| Cla022458 | /0.48 | /1.55∗ | /-1.03 | /0.05 | Dehydration-responsive element-binding protein (DREB) | |
| Cla017676 | /-0.24 | /1.22∗ | /-1.47 | /-0.01 | Kinesin-5 transcription factor | |
| Cla011198 | /0.00 | /0.74∗ | /0.89 | /1.63 | BHLH transcription factor | |
| Cla009841 | /0.18 | /0.73∗ | /-0.98 | /-0.43 | Ethylene-regulated nuclear protein ERT2-like | |
| Cla010176 | /-0.08 | /0.65∗ | /0.44 | /-0.29 | TCP transcription factor | |
| Cla006034 | /0.66 | /1.31∗ | /-0.15 | /0.50 | Ankyrin repeat-containing protein (Akr1p) | |
| Cla018866 | /-0.14 | /0.63∗ | /-0.49 | /0.27 | Nucleolar and coiled-body phosphoprotein (NOLC) 1 | |
| Cla016714 | /0.10 | /1.82∗ | /-2.37 | /-0.66 | Os02g0202300 | |
| Cla017355 | /0.05 | /0.71∗ | /-1.90∗ | /-1.23∗ | WRKY transcription factor | |
| Cla006039 | /-0.31 | /1.07∗ | /-2.55∗ | /-1.17 | Protein forked1 | |
| novel-m0058-5p | Cla015802 | 0.86/0.10 | -5.89∗/1.19∗ | 5.89∗/-3.96∗ | -1.36/-2.87∗ | Pentatricopeptide repeat-containing protein (PPRP) |
| Cla005115 | /0.15 | /0.80∗ | /-1.39∗ | /-0.74 | Chromatin remodeling complex subunit (SWI/SNF) | |
| novel-m0059-5p | Cla019857 | 1.98/-1.02 | -0.82 / 1.01 | 1.10/0.35 | -1.63∗/2.38∗ | TIR-NBS-LRR disease resistance protein |
| novel-m0070-5p | Cla005664 | -1.60∗/0.24 | 0.83/-0.35 | -0.50/0.00 | 1.93∗/-0.59∗ | Nitrate transporter (NRT) |
| Cla016054 | /0.96∗ | /0.22 | /-0.61 | /-1.35 | UDP-glucosyltransferase (GTase) 1 | |
| novel-m0077-5p | Cla004726 | 0.00/-0.13 | -0.24/0.46 | 5.58∗/-0.87∗ | 1.82/-0.28 | IQ-domain 10 |
| novel-m0081-3p | Cla004682 | 1.25∗/-0.46 | -0.69/0.17 | 0.06/0.14 | -2.17∗/0.77∗ | Serine/threonine-protein phosphatase (Ser/Thr-PPs) |
∗Indicates significant difference.
FIGURE 5.
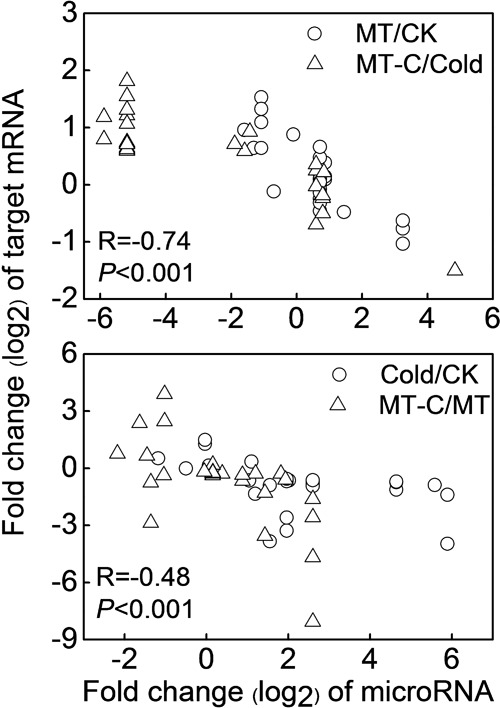
Correlation analysis between the expression of miRNAs and predicted targets by cleavage in comparison of MT/CK, MT-C/Cold, Cold/CK, and MT-C/MT.
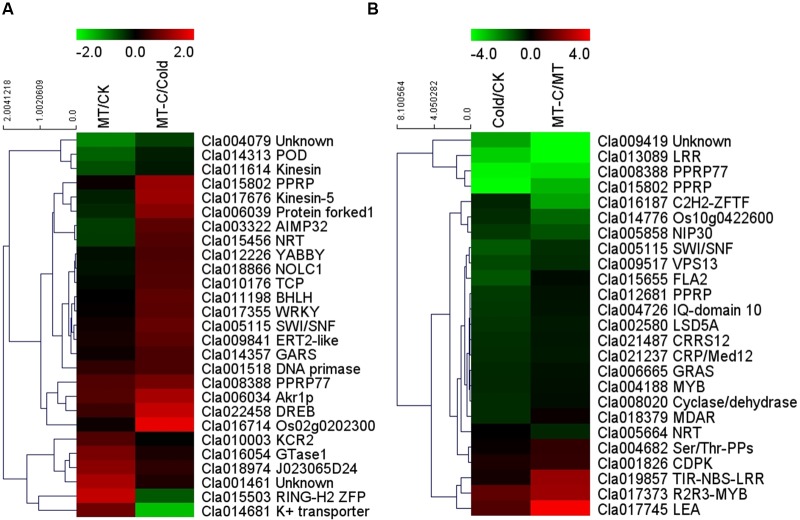
Some of the 27 potential target mRNAs cleaved by melatonin-responsive miRNAs were induced or repressed by melatonin at optimal growth temperatures, but not under cold stress (Figure 6A; Supplementary Table S4). Interestingly, most miRNAs including some cold-responsive genes, such as Cla011198 (BHLH), Cla017355 (WRKY), and Cla022458 (DREB, dehydration-responsive element-binding gene), were upregulated by melatonin during cold stress, but were not affected by melatonin under unstressed conditions. Most of 25 potential target mRNAs cleaved by cold-responsive miRNAs were repressed or unaffected by cold stress, except for Cla017745 (LEA) and Cla017373 (R2R3-MYB), those were upregulated (Figure 6B). However, melatonin pretreatment alleviated cold-induced repression of target mRNAs or improved cold-induced increases in target mRNAs. For instance, LEA and R2R3-MYB in melatonin-pretreated plants were upregulated by 5.53- and 14.91-fold under cold stress, far more than the changes of 2.82- and 2.44-fold in control plants, respectively. MYB, MDAR (monodehydroascorbate reductase), and PPRP (pentatricopeptide repeat protein) were significantly repressed by cold stress in control plants, but were minimally affected in melatonin-pretreated plants. Accordingly, a set of potential target mRNAs cleaved by corresponding miRNAs might be involved in the response of melatonin-pretreated plants to cold stress.
FIGURE 6.
Heat map showing (A) melatonin or (B) cold induced differential expression of the potential target mRNAs.
We also subjected the predicted target mRNAs to Gene Ontology classification based on their involvement in process in Cucurbit Genomics Database9 with watermelon 97103 v1 (Figure 7; Supplementary Figure S1). The target genes inhibited by miRNAs via cleavage and translational repression were involved in various biological processes, with 19 and 38 mRNAs involved in cellular process regulation, 18 and 31 mRNAs involved in responses to biotic or abiotic stress, and 5 and 10 mRNAs involved in signal transduction, respectively. Thus, a number of potential target mRNAs might be involved in watermelon responses to cold stress by direct or indirect ways.
FIGURE 7.
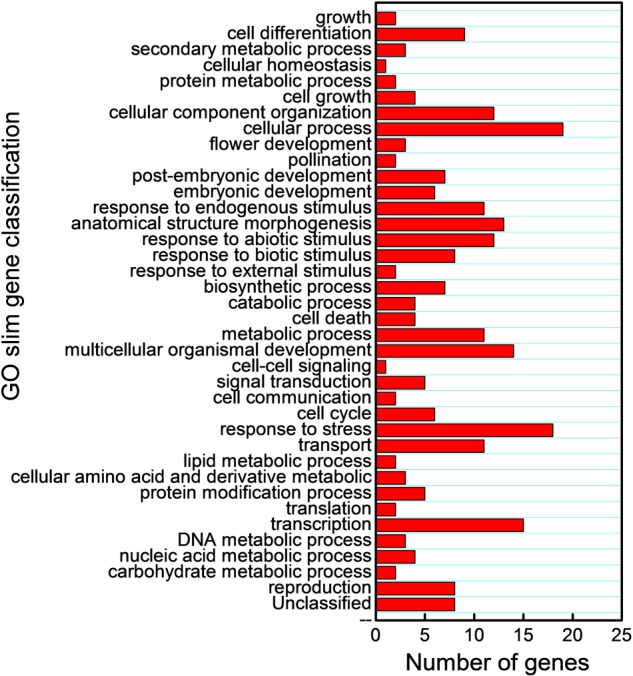
Gene Ontology classification of potential target genes cleaved by miRNAs based on their involvement in various biological processes.
Discussion
Melatonin Induces Cold Stress Tolerance and Cold-Responsive Gene Expressions in Watermelon
In recent years, melatonin has emerged as a focus of research in plant science. It functions in many aspects of plant growth and development, and regulates plant tolerance to abiotic stresses, such as salinity, drought, radiation, excess copper, and chemical stresses (Zhang et al., 2014). Recently, several studies have shown that exogenous melatonin with optimum concentrations enhances cold tolerance in Arabidopsis and Triticum aestivum L. (Bajwa et al., 2014; Shi and Chan, 2014; Turk et al., 2014), since the initial observation that exogenous melatonin attenuates cold-induced apoptosis in carrot suspension cells (Lei et al., 2004). In the current study, cold stress caused wilted blade edges, decreased maximum photochemical efficiency of PSII (Fv/Fm), and increased REL, which indicates membrane damage (Figure 1). Notably, in agreement with previous studies, the adverse effects of cold were significantly alleviated by the application of melatonin. Moreover, melatonin treatment up-regulated two cold-responsive genes Cla020078 (CBF1) and Cla020702 (MYB) in watermelon, which is in line with previous studies in Arabidopsis (Bajwa et al., 2014; Shi and Chan, 2014), suggesting that melatonin activates signaling pathways during cold stress via regulation of cold-responsive genes (Figure 2).
Melatonin Affects the Expression of a Number of MicroRNAs under Optimal Growth Conditions or Cold Stress
Transcriptional control of the expression of cold-responsive genes is well known (Chinnusamy et al., 2010), but miRNAs, which could regulate stress responses by controlling the expression of cognate target genes, have recently been added to the suite of cold-responsive gene regulatory networks. Comparative profiles of miRNA expression among various plant species (Arabidopsis, Brachypodium, and Populus) during cold stress revealed similarities and differences in miRNA regulation (Liu et al., 2008; Lu et al., 2008; Zhang et al., 2009). In this study, 440 known miRNAs and 106 novel miRNAs were identified using Solexa high-throughput sequencing (Supplementary Table S2). Based on an analysis of differentially expressed miRNAs, we identified a set of miRNAs including 16 known miRNAs and 21 novel miRNAs that were significantly up- or downregulated by cold stress, which exhibited similarities and differences with other species (Figure 4; Khraiwesh et al., 2012; Sunkar et al., 2012). The discrepancies in cold-responsive miRNA expression might be attributed to the differential responses of unrelated plant species and variation in experimental conditions and methods. Importantly, pretreatment with melatonin affected the expression of a set of miRNAs under unstressed or cold conditions, and changed the expression of cold-responsive miRNAs. These results suggest that a number of miRNAs are involved in melatonin-mediated enhancement in cold tolerance in watermelon. To the best of our knowledge, this is the first report on the interaction between melatonin and miRNAs in regulation of plant tolerance to stress, although a few studies have reported such interactions in animal cell lines (Lee et al., 2011; Sohn et al., 2015).
MicroRNAs May Be Involved in Melatonin-Mediated Cold Tolerance by Negatively Regulating Target mRNAs
To examine the functions of melatonin- or cold-responsive miRNAs, we predicted their target mRNAs. We detected 49 mRNAs with opposite expression patterns with respect to their corresponding miRNAs, and inferred that these mRNAs are likely cleaved by the corresponding miRNAs (Table 1). Among the 49 potential target mRNAs, several mRNAs were closely associated with plant defense against cold stress by activating signaling transduction or encoding functional proteins. These mRNAs include Cla001826, Cla011198, Cla022458, Cla004188/Cla017373, Cla017355, Cla017745, and Cla018379 which encode CDPK, BHLH transcription factor, DREB protein, MYB transcription factor, WRKY transcription factor, LEA protein, and MDAR, respectively. As Ca2+ sensors, CDPKs those can directly bind Ca2+, are major players in coupling cold stress signals to specific protein phosphorylation cascades, leading to the activation of downstream cold-responsive genes, such as ICE1, a MYC-type BHLH transcription factor (Janská et al., 2010). ICE1 is activated by low temperatures and triggers CBF/DREB expression, which promotes the accumulation of cold-responsive gene products such as LEA, antioxidant enzymes, and molecular chaperones (Janská et al., 2010). MYB and WRKY transcription factor gene families have been suggested to play important roles in the regulation of transcriptional reprogramming associated with plant responses to cold stress (Shinozaki et al., 2003; Zhu et al., 2005; Chen et al., 2012). Overexpression of some MYB or WRKY genes enhances plant tolerance to cold stress and activates the expression of a set of cold-responsive genes such as LEA, DREB2A, and genes encoding antioxidant enzymes (Dai et al., 2007; Niu et al., 2012; Yang et al., 2012). LEA is important for membrane stabilization and the protection of proteins from denaturation when the cytoplasm becomes dehydrated (Janská et al., 2010). MDAR is an important antioxidant enzyme that is critical to alleviate cold-induced oxidative damage (Chen and Paul, 2002). Importantly, these defense-related genes were significantly induced by melatonin, exhibiting contrasting expression patterns with respect to the corresponding miRNAs, including miR159-5p, miR858, miR8029-3p, and novel-m0048-3p (Figures 5 and 6). Accordingly, it could be inferred that some cold resistance pathways in melatonin-pretreated plants were promoted by a number of downregulated miRNAs under cold stress.
Some other potential target genes might also be involved in watermelon responses to cold stress by direct or indirect ways. For example, low temperatures can inhibit the uptake and transport of nitrate (Warren, 2009; Leffler et al., 2013; Dechatiwongse et al., 2014). The significantly upregulated expression of Cla015456 (NRT) by melatonin during cold stress suggests that the active transport of nitrate could be helpful in the acclimation of plants under adverse environmental conditions (Zhang et al., 2005). Additionally, the differentially expressed miRNAs were predicted to target a set of mRNAs via translational repression (Supplementary Table S3). Some of these mRNAs, such as MYB, MYC, and CYP450, are important in transcriptional regulation and protection in cold responses, respectively. However, whether these predicted target mRNAs are regulated by corresponding miRNAs via translational inhibition requires further studies.
Conclusion
We demonstrated that exogenous melatonin induced watermelon tolerance to cold stress, and this induction was associated with the upregulation of cold-responsive genes. Moreover, we provided evidence for the alteration of the expression profile of a set of miRNAs regulated by melatonin with putative cold-responsive gene targets. In particular, the downregulation of miR159-5p, miR858, miR8029-3p, and novel-m0048-3p by melatonin was coincident with the upregulation of putative target genes involved in signal transduction (CDPK, BHLH, WRKY, MYB, and DREB) and protection/detoxification (LEA and MDAR) under low-temperature conditions (Figure 8). To our knowledge, these results provide the first evidence for a potential regulatory role of miRNAs in melatonin-mediated cold tolerance in plants.
FIGURE 8.
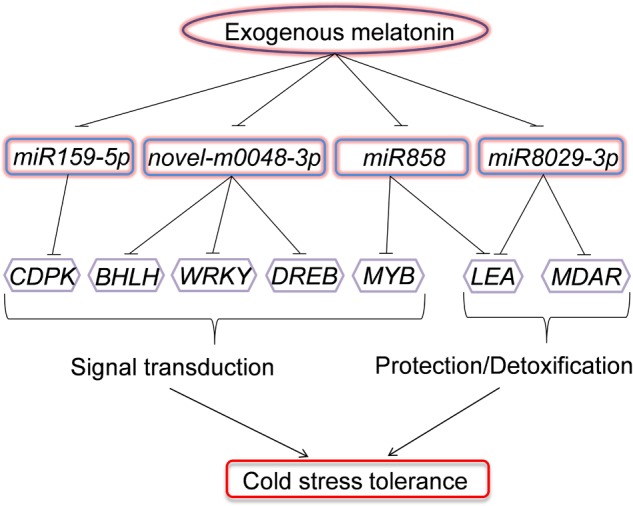
Model depicting the regulatory pathways of miRNAs in melatonin-mediated cold stress tolerance in watermelon. Under cold stress, application of exogenous melatonin could downregulate the expression of miR159-5p, miR858, miR8029-3p, and novel-m0048-3p. The downregulation of miRNAs results in the upregulation of their target genes involved in signal transduction (CDPK, BHLH, WRKY, MYB, and DREB) or protection/detoxification (LEA and MDAR) and enhance cold stress resistance.
Author Contributions
HL and XZ designed research; HL, YD, and JC performed research; HL, JH, HC, QL, CW, JM, YZ, JY, and XZ analyzed data; HL, YD, and XZ wrote and revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (No. 2016M592843), the Shaanxi Provincial Science and Technology Research and Development Project Fund, China (No. 2015NY091), the Modern Agro-industry Technology Research System of China (No. CARS-26-18), and the Fundamental Research Funds of Northwest A&F University (No. 2452015023).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01231
FIGURE S1 | Gene Ontology classification of potential target genes of miRNAs with translational repression based on their involvement in process.
TABLE S1 | Reads abundance of various classification of small RNAs in control plants (CK) and plants treated by melatonin (MT), Cold, and melatonin+cold (MT-C).
TABLE S2 | Identified known and novel miRNAs in watermelon.
TABLE S3 | Predicted target mRNAs of differentially expressed miRNAs with translational inhibition.
TABLE S4 | The FPKM value of melatonin- or cold-responsive potential target mRNAs in each treatment.
References
- Arnao M. B., Hernández–Ruiz J. (2015). Functions of melatonin in plants: a review. J. Pineal Res. 59 133–150. 10.1111/jpi.12253 [DOI] [PubMed] [Google Scholar]
- Bajwa V. S., Shukla M. R., Sherif S. M., Murch S. J., Saxena P. K. (2014). Role of melatonin in alleviating cold stress in Arabidopsis thaliana. J. Pineal Res. 56 238–245. 10.1111/jpi.12115 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190. 10.1126/science.1159151 [DOI] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819 120–128. 10.1016/j.bbagrm.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Chen W. P., Paul H. L. (2002). “Attenuation of reactive oxygen production during chilling in ABA-treated maize cultured cells,” in Plant Cold Hardiness, eds Li P. H., Palva E. T. (New York, NY: Springer; ), 223–233. 10.1007/978-1-4615-0711-6 [DOI] [Google Scholar]
- Cheng H. Y., Song S. Q. (2006). Species and organ diversity in the effect of hydrogen peroxide on superoxide dismutase activity in vitro. J. Integr. Plant Biol. 48 672–678. 10.1111/j.1744-7909.2006.00266.x [DOI] [Google Scholar]
- Chinnusamy V., Zhu J. K., Sunkar R. (2010). “Gene regulation during cold stress acclimation in plants,” in Plant Stress Tolerance: Methods and Protocols, ed. Sunkar R. (New York, NY: Springer; ), 39–55. 10.1007/978-1-60761-702-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Xu Y., Ma Q., Xu W., Wang T., Xue Y., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143 1739–1751. 10.1104/pp.106.094532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechatiwongse P., Srisamai S., Maitland G., Hellgardt K. (2014). Effects of light and temperature on the photoautotrophic growth and photoinhibition of nitrogen-fixing cyanobacterium Cyanothece sp. ATCC 51142. Algal Res. 5 103–111. 10.1016/j.algal.2014.06.004 [DOI] [Google Scholar]
- Dubbels R., Reiter R. J., Klenke E., Goebel A., Schnakenberg E., Ehlers C., et al. (1995). Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography mass spectrometry. J. Pineal Res. 18 28–31. 10.1111/j.1600-079X.1995.tb00136.x [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95 14863–14868. 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Vanacker H., Gomez L. D., Harbinson J. (2002). Regulation of photosynthesis and antioxidant metabolism in maize leaves at optimal and chilling temperatures: review. Plant Physiol. Biochem. 40 659–668. 10.1016/S0981-9428(02)01425-0 [DOI] [Google Scholar]
- Hardeland R., Cardinali D. P., Srinivasan V., Spence D. W., Brown G. M., Pandi-Perumal S. R. (2011). Melatonin–A pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 93 350–384. 10.1016/j.pneurobio.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Hattori A., Migitaka H., Iigo M., Itoh M., Yamamoto K., Ohtani-Kaneko R., et al. (1995). Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 35 627–634. [PubMed] [Google Scholar]
- Janská A., Maršík P., Zelenková S., Ovesna J. (2010). Cold stress and acclimation–what is important for metabolic adjustment? Plant Biol. 12 395–405. 10.1111/j.1438-8677.2009.00299.x [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Yuan J., Gao L., Zhao S., Jiang W., Huang Y., et al. (2014). Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE 9:e90612 10.1371/journal.pone.0090612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. E., Kim S. J., Youn J. P., Hwang S. Y., Park C. S., Park Y. S. (2011). MicroRNA and gene expression analysis of melatonin-exposed human breast cancer cell lines indicating involvement of the anticancer effect. J. Pineal Res. 51 345–352. 10.1111/j.1600-079X.2011.00896.x [DOI] [PubMed] [Google Scholar]
- Leffler A. J., James J. J., Monaco T. A. (2013). Temperature and functional traits influence differences in nitrogen uptake capacity between native and invasive grasses. Oecologia 171 51–60. 10.1007/s00442-012-2399-4 [DOI] [PubMed] [Google Scholar]
- Lei X. Y., Zhu R. Y., Zhang G. Y., Dai Y. R. (2004). Attenuation of cold-induced apoptosis by exogenous melatonin in carrot suspension cells: the possible involvement of polyamines. J. Pineal Res. 36 126–131. 10.1046/j.1600-079X.2003.00106.x [DOI] [PubMed] [Google Scholar]
- Liu H. H., Tian X., Li Y. J., Wu C. A., Zheng C. C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14 836–843. 10.1261/rna.895308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S. F., Sun Y. H., Chiang V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55 131–151. 10.1111/j.1365-313X.2008.03497.x [DOI] [PubMed] [Google Scholar]
- Mahajan S., Tuteja N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444 139–158. 10.1016/j.abb.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Manchester L. C., Coto–Montes A., Boga J. A., Andersen L. P. H., Zhou Z., Galano A., et al. (2015). Melatonin: an ancient molecule that makes oxygen metabolically tolerable. J. Pineal Res. 59 403–419. 10.1111/jpi.12267 [DOI] [PubMed] [Google Scholar]
- Meyers B. C., Axtell M. J., Bartel B., Bartel D. P., Baulcombe D., Bowman J. L., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20 3186–3190. 10.1105/tpc.108.064311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C. F., Wei W., Zhou Q. Y., Tian A. G., Hao Y. J., Zhang W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35 1156–1170. 10.1111/j.1365-3040.2012.02480.x [DOI] [PubMed] [Google Scholar]
- Rivero R. M., Ruiz J. M., García P. C., López -Lefebre L. R., Sánchez E., Romero L. (2001). Resistance to cold and heat stress: accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci. 160 315–321. 10.1016/S0168-9452(00)00395-2 [DOI] [PubMed] [Google Scholar]
- Rivero R. M., Ruiz J. M., García P. C., López-Lefebre L. R., Sánchez E., Romero L. (2002). Response of oxidative metabolism in watermelon plants subjected to cold stress. Funct. Plant Biol. 29 643–648. 10.1071/PP01013 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Chan Z. (2014). The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6-activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J. Pineal Res. 57 185–191. 10.1111/jpi.12155 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6 410–417. 10.1016/S1369-5266(03)00092-X [DOI] [PubMed] [Google Scholar]
- Sohn E. J., Won G., Lee J., Lee S., Kim S. H. (2015). Upregulation of miRNA3195 and miRNA374b mediates the anti-angiogenic properties of melatonin in hypoxic PC-3 prostate cancer cells. J. Cancer Educ. 6 19–28. 10.7150/jca.9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2051–2065. 10.1105/tpc.106.041673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Li Y. F., Jagadeeswaran G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17 196–203. 10.1016/j.tplants.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Mittler R. (2006). Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol. Plant. 126 45–51. 10.1111/j.0031-9317.2005.00582.x [DOI] [Google Scholar]
- Tan D. X., Hardeland R., Manchester L. C., Korkmaz A., Ma S., Rosales-Corral S., et al. (2012). Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. J. Exp. Bot. 63 577–597. 10.1093/jxb/err256 [DOI] [PubMed] [Google Scholar]
- Turk H., Erdal S., Genisel M., Atici O., Demir Y., Yanmis D. (2014). The regulatory effect of melatonin on physiological, biochemical and molecular parameters in cold-stressed wheat seedlings. Plant Growth Regul. 74 139–152. 10.1007/s10725-014-9905-0 [DOI] [Google Scholar]
- van Kooten O., Snel J. F. H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25 147–150. 10.1007/BF00033156 [DOI] [PubMed] [Google Scholar]
- Wang P., Yin L., Liang D., Li C., Ma F., Yue Z. (2012). Delayed senescence of apple leaves by exogenous melatonin treatment: toward regulating the ascorbate-glutathione cycle. J. Pineal Res. 53 11–20. 10.1111/j.1600-079X.2011.00966.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun F., Cao H., Peng H., Ni Z., Sun Q., et al. (2012). TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 7:e48445 10.1371/journal.pone.0048445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C. R. (2009). Why does temperature affect relative uptake rates of nitrate, ammonium and glycine: a test with Eucalyptus pauciflora. Soil Biol. Biochem. 41 778–784. 10.1016/j.soilbio.2009.01.012 [DOI] [Google Scholar]
- Xiong L., Schumaker K. S., Zhu J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl. 1), S165–S183. 10.1105/tpc.000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Dai X., Zhang W. H. (2012). A R2R3-type MYB gene, OsMYB2 is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63 2541–2556. 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. K., Lang P., Dane F., Ebel R. C., Singh N. K., Locy R. D., et al. (2005). Cold acclimation induced genes of trifoliate orange (Poncirus trifoliata). Plant Cell Rep. 23 764–769. 10.1007/s00299-004-0883-y [DOI] [PubMed] [Google Scholar]
- Zhang H. M., Zhang Y. (2014). Melatonin: a well-documented antioxidant with conditional pro-oxidant actions. J. Pineal Res. 57 131–146. 10.1111/jpi.12162 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu Y., Huan Q., Chong K. (2009). Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10:449 10.1186/1471-2164-10-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Sun Q., Zhang H., Cao Y., Weeda S., Ren S., et al. (2014). Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 66 647–656. 10.1093/jxb/eru336 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2005). miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 33 W701–W704. 10.1093/nar/gki383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Leul M. (1998). Uniconazole-induced alleviation of freezing injury in relation to changes in hormonal balance, enzyme activities and lipid peroxidation in winter rape. Plant Growth Regul. 26 41–47. 10.1023/A:1006004921265 [DOI] [Google Scholar]
- Zhu J. H., Verslues P. E., Zheng X. W., Lee B. H., Zhan X. Q., Manabe Y., et al. (2005). HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. U.S.A. 102 9966–9971. 10.1073/pnas.1009100107 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | Gene Ontology classification of potential target genes of miRNAs with translational repression based on their involvement in process.
TABLE S1 | Reads abundance of various classification of small RNAs in control plants (CK) and plants treated by melatonin (MT), Cold, and melatonin+cold (MT-C).
TABLE S2 | Identified known and novel miRNAs in watermelon.
TABLE S3 | Predicted target mRNAs of differentially expressed miRNAs with translational inhibition.
TABLE S4 | The FPKM value of melatonin- or cold-responsive potential target mRNAs in each treatment.