Abstract
Background:
Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) gene is an inhibitor of apoptosis that expresses in human embryonic tissues but it is absent in most healthy adult tissues. The copy number of BIRC5 has been indicated to be highly increased in tumor tissues; however, its association with the age of onset in breast cancer is not well understood.
Methods:
Forty tumor tissues of breast cancer were obtained from Tumor Bank of Cancer Institute, Imam Khomeini Hospital, Tehran, Iran. BIRC5 gene copy number variation (CNV) was evaluated using Multiplex Ligation-dependent Probe Amplification (MLPA) and then compared with the age of onset for each patient.
Results:
BIRC5 amplification was seen in 17.5% of cases. Also, a significant association was observed between BIRC5 gene amplification and individuals under 40 years of age (P=0.04).
Conclusion:
BIRC5 gene has the potential to be a marker for the detection and prognosis of cancer at an early age.
Keywords: BIRC5, amplification, Breast cancer, Amplification
INTRODUCTION
Breast cancer is an important health problem worldwide, and it is the second most common cancer among populations in both developed and developing countries. In Iran, breast cancer comprises 21.4% of female cancers[1].
There are several factors that can affect breast cancer prognosis. One of these factors is gene copy number variation (CNV)[2,3]. CNV changes have been introduced as an influential feature in human phenotypes and clinical diseases[4,5]. Genome copy number alterations are important factors in tumor development[6]. Since the result of cancer treatment depends on the early diagnosis of the disease, CNV determination in genes involved in cancer may be an effective step in the early detection of breast cancer[7]. Accumulating evidence suggests that breast cancer in patients under 40 is more aggressive and associated with poor outcome than in their older counterparts[8]. As there is an increasing outbreak of the disease in Iran, which is usually detected at advanced stages, the early detection of breast cancer would be helpful in reducing the mortality rate and improving patients’ prognosis[9,10].
Genomic changes, including increase in the expression levels or copy number of those genes which are involved in mitosis, cell cycle progression, embryogenesis, DNA reproduction, cell division, and proliferation, have been shown that are effective in development and progression of breast cancer[11-13].
Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) gene, also known as survivin, is a family member of the inhibitors of apoptosis. There are two copies of the aforementioned gene in the entire normal diploid cells that are located on 17q25 chromosome. The BIRC5 gene product is supposed to play a role in the prevention of apoptosis. It is also known that this gene participates in cell cycle progression and assists the cells to go through the cell cycle checkpoints. This gene has an important function both in tumorigenesis and tumor progression. In a study on pancreatic cancer, BIRC5 gene amplification has been reported[14]. BIRC5 gene amplification was also observed in lung cancer using multiplex ligation-dependent probe amplification (MLPA) technique[15]. In this regard, BIRC5 may have the potential to be considered as a therapeutic target in cancer.
With regard to the fact that BIRC5 is expressed at the embryonic stage but its expression is absent in normal adult tissues, changes in the copy number of this gene can possibly be effective in development and progression of cancer by preventing apoptosis in cells. Hence, this study was conducted to determine the association between BIRC5 genomic copy number variation and the age of onset in breast cancer.
MATERIALS AND METHODS
In this study, 40 tumor tissues of breast cancer and 6 normal breast tissues, as multiplex ligation-dependent probe amplification references, were randomly selected and obtained from National Tumor Bank of Iran, Cancer Institute, Imam Khomeini Hospital Complex, Tehran, Iran. Clinicopathological information such as stage, grade, and tumor size of each patient was also obtained from Tumor Bank for further analysis.
DNA extraction
DNA was extracted from breast tissue samples using the QIAamp DNA mini kit (Qiagen, USA) according to the manufacturer’s instruction. The quality and integrity of DNAs were evaluated by agarose gel electrophoresis. The concentration of high-quality extracted DNA was standardized by using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, USA).
Multiplex ligation-dependent probe amplification (MLPA)
The SALSA MLPA P078-C1 Breast Tumor probe kit (MRC, Holland) was used to determine BIRC5 gene amplification status. In each PCR reaction, three normal DNA samples and one no-template control (NTC) containing TE solution (0.1 mM EDTA+10 mM Tris-Hcl, pH 8.2) were included. All denaturation, hybridization, ligation, and PCR reactions were performed by Peqlab thermocycler (Germany). PCR products were then separated on an ABI3130 capillary sequencer (Applied Biosystems, USA).
Multiplex ligation-dependent probe amplification analysis
Analysis of BIRC5 gene copy number was carried out using GeneMarker ver 1.6 (softgenetics, USA). As BIRC5 has more than one probe in the provided kit, the mean of all the probe peaks of this gene was calculated. If the mean value was below 0.7, the respective gene was defined as lost, while values between 0.7–1.3 and >1.3 were assigned as normal and amplified, respectively[16,17].
Statistical Analysis
The statistical analyses were carried out by SPSS software package (PASW Statistics for Windows, Version 18.0. Chicago, USA). Significant index was checked by a factor of 95%, and P<0.05 was considered to indicate statistical significance.
RESULTS
In this study, DNA copy number changes in 40 tumor tissues and 6 normal breast tissues were evaluated by using MLPA. Of the total 40 tumor samples, 17.5% (7 cases) showed BIRC5 amplification. Examples of a breast cancer patient with no genomic copy number changes and a patient with BIRC5 gene amplification are depicted in Figure 1A and 1B, respectively.
Fig. 1.
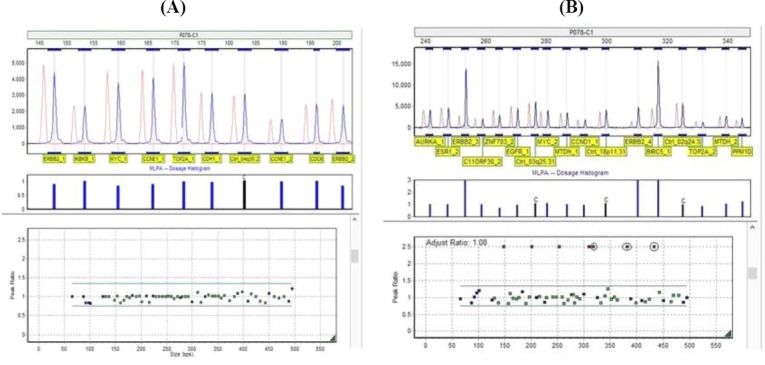
Multiplex ligation-dependent probe amplification (MLPA) analysis of two samples using genemarker software. (A) A breast cancer patient with no genomic copy number changes showing a tumor sample with no changes in BIRC5 genomic copy number. As it is evident, the height of the peaks of the sample (blue) is equal to that of the standard size (orange). (B) A breast cancer patient with BIRC5 gene amplification showing a tumor sample of a breast cancer patient with BIRC5 gene amplification. BIRC5 gene probes are marked with a circle.
The age range of 40 samples included in this study was between 29 to 77 years, and the average was 50.27 years. Among these patients who showed the BIRC5 gene amplification, 71% (5 cases) were under 40 years.
Analysis of the BIRC5 gene amplification and the comparison of changes with the age of onset of the disease showed a significant association between the breast cancer incidence in women under 40 years and an increase in BIRC5 copy number variation (P=0.04). Also, no correlation was found between the clinicopathological characteristics of the patients and BIRC5 gene amplification status (Table 1).
Table 1.
Distribution of BIRC5 CNVs in association with clinicopathological characteristics
| Clinicopathological characteristics | BIRC5 amplification (Number=7) (%) | P value | No BIRC5 amplification (Number=33) (%) | P value |
|---|---|---|---|---|
| Stage | ||||
| I | 0 (0) | 0.6 | 2 (5) | 0.6 |
| II | 5 (12.5) | 18 (45) | ||
| III | 2 (5) | 13 (32.5) | ||
| IV | 0 (0) | 0 (0) | ||
| Grade | ||||
| I | 2 (5) | 0.2 | 12 (30) | 0.5 |
| II | 4 (10) | 13(32.5) | ||
| III | 1 (2.5) | 8 (20) | ||
| Age | ||||
| ≤40 | 5 (12.5) | 0.04 | 6 (15) | 0.3 |
| >40 | 2 (5) | 27 (67) | ||
| Tumor size (cm) | ||||
| ≤2 | 0 (0) | 0.4 | 6 (15) | 0.6 |
| 2<T≤5 | 5 (12.5) | 23 (57) | ||
| >5 | 2 (5) | 4 (10) |
DISCUSSION
Although cancer is not prevalent in individuals under the age of 40, the incidence of the disease at a younger age may increase the risk of recurrence[18]. Breast cancer in young women is more acute with a poorer prognosis and overall survival in comparison with elder women diagnosed with the disease[11]. Hence, finding those factors that may be related to early onset of breast cancer can lead to faster diagnosis and longer period of disease free survival. BIRC5 is expressed in human embryonic tissues, while it is not detected in normal adult tissues[19]. The overexpression of this gene in tumor tissues has been already reported [20].
In the present study, the genomic copy number variation of BIRC5, which is supposed to be involved in breast cancer progression, was examined, and its association with the early-onset breast cancer was also evaluated. Also,. 17.5% of the patients showed an increase in BIRC5 gene copy number, and half of the patients under 40 years had amplified BIRC5 gene. Increase in the BIRC5 gene copy number in patients who were younger than 40 years showed a significant relationship between the two indices (P=0.04).
Using MLPA technique, Kornegoor et al.[21] have studied the genomic copy number alterations for several oncogenes including BIRC5 in males with breast cancer. They found that among 110 studied samples, 40% of the patients showed an increase in the number of genomic copies of BIRC5 gene. The differences between our findings and Kornegoor et al.[21] results may be due to alterations in patients’ sex, race, and the size of sample population. Moelans et al.[22] also used MLPA technique to compare genomic BIRC5 gene copy number changes in ductal carcinoma in situ and invasive ductal carcinoma, in which 20% of the patients with ductal carcinoma in situ type breast cancer and 12% of the patients with invasive ductal carcinoma type showed amplified BIRC5 gene copy number.
Using reverse transcription-PCR technique, Span et al.[23] measured mRNA level of this gene in 275 breast cancer patients. They observed a significant relationship between younger patients and BIRC5 mRNA level in tumor samples. Colak et al.[11] also detected genomic changes in BIRC5 gene among patients under 45 years by studying the whole genome mRNA expression in cancer, from non-invasive breast cancer to invasive breast cancer.
Sought to examine the relationship between the early onset of cancer and increase in the number of BIRC5 gene copies in tumors, Alaggio et al.[24] examined genomic copy number of the gene BIRC5 using FISH technique in malignant peripheral nerve sheath tumors. They concluded that the number of BIRC5 gene copies in 35% of the patients in the first two decades of their life was increased.
Baykara et al.[15] analyzed the copy number of 22 genes, including BIRC5, in 82 tumor tissues of lung cancer. Similar to our results they did not find any possible association with clinical parameters.
By examining the number of gene copies involved in cancer development and treatment, it is possible to determine the cancer progression and the onset age with stronger prognostic. With prospective studies, larger sample sizes, and confirmation of the predictive role between BIRC5 gene amplification and the early onset of breast cancer, it is possible to use BIRC5 gene as a marker for cancer detection and the onset age prediction, as well as the likelihood of recurrence of the disease. Moreover, the results of this study indicate that increase in the expression of BIRC5 gene may not only be due to variation in transcription factors but also may occur along with the increase in the gene copy number. It is also worth mentioning that analyzing gene expression at the mRNA level by real-time PCR or at the protein level by Western blot could help to confirm these results.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCE
- 1.Babu GR, Samari G, Cohen SP, Mahapatra T, Wahbe RM, Mermash S, Galal OM. Breast cancer screening among females in Iran and recommendations for improved practice:a review. Asian Pacific journal of cancer prevention. 2011;12(7):1647–1655. [PubMed] [Google Scholar]
- 2.Choy KW, Setlur SR, Lee C, Lau TK. The impact of human copy number variation on a new era of genetic testing. An international journal of obstetrics and gynaecology. 2010;117(4):391–398. doi: 10.1111/j.1471-0528.2009.02470.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nature genetics. 2007;39(7 Suppl):S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annual review of genomics and human genetics. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozlowski P, Jasinska AJ, Kwiatkowski DJ. New applications and developments in the use of multiplex ligation-dependent probe amplification. Electrophoresis. 2008;9(23):4627–4636. doi: 10.1002/elps.200800126. [DOI] [PubMed] [Google Scholar]
- 6.Cava C, Zoppis I, Mauri G, Ripamonti M, Gallivanone F, Salvatore C, Gilardi MC, Castiglioni I. Combination of gene expression and genome copy number alteration has a prognostic value for breast cancer. Conference proceedings:Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2013;2013:608–611. doi: 10.1109/EMBC.2013.6609573. [DOI] [PubMed] [Google Scholar]
- 7.Anders CK, Hsu DS, Broadwater G, Acharya CR, Foekens JA, Zhang Y, Wang Y, Marcom PK, Marks JR, Febbo PG, Nevins JR, Potti A, Blackwell KL. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. Journal of clinical oncology. 2008;6(20):3324–3330. doi: 10.1200/JCO.2007.14.2471. [DOI] [PubMed] [Google Scholar]
- 8.Gnerlich JL, Deshpande AD, Jeffe DB, Sweet A, White N, Margenthaler JA. Elevated breast cancer mortality in women younger than age 40 years compared with older women is attributed to poorer survival in early-stage disease. Journal of the American College of surgeons. 2009;08(3):341–347. doi: 10.1016/j.jamcollsurg.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghiasvand R, Adami HO, Harirchi I, Akrami R, Zendehdel K. Higher incidence of premenopausal breast cancer in less developed countries;myth or truth? BMC cancer. 2014;14:343. doi: 10.1186/1471-2407-14-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmore JG, Armstrong K, Lehman CD, Fletcher SW. Screening for breast cancer. The journal of the American medical association. 2005;93:1245–1256. doi: 10.1001/jama.293.10.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colak D, Nofal A, Albakheet A, Nirmal M, Jeprel H, Eldali A, Al-Tweigeri T, Tulbah A, Ajarim D, Malik OA, Inan MS, Kaya N, Park BH, Bin Amer SM. Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women. PloS one. 2013;8(5):e63204. doi: 10.1371/journal.pone.0063204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Molecular cancer. 2011;10(1):15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC. Gene expression profiles of human breast cancer progression. Proceedings of the national academy of sciences of the United States of America. 2003;100(10):5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahlamaki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, Karhu R, Kallioniemi A. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes, chromosomes and cancer. 2002;35(4):353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- 15.Baykara O, Bakir B, Buyru N, Kaynak K, Dalay N. Amplification of chromosome 8 genes in lung cancer. Journal of cancer. 2015;6(3):270–275. doi: 10.7150/jca.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moelans CB, de Weger RA, Monsuur HN, Vijzelaar R, van Diest PJ. Molecular profiling of invasive breast cancer by multiplex ligation-dependent probe amplification-based copy number analysis of tumor suppressor and oncogenes. Modern pathology. 2010;3(7):1029–1039. doi: 10.1038/modpathol.2010.84. [DOI] [PubMed] [Google Scholar]
- 17.Bunyan DJ, Eccles DM, Sillibourne J, Wilkins E, Thomas NS, Shea-Simonds J, Duncan PJ, Curtis CE, Robinson DO, Harvey JF, Cross NC. Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. British journal of cancer. 2004;91(6):1155–1159. doi: 10.1038/sj.bjc.6602121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel CA, Domchek SM. Breast cancer in young women. Breast cancer research. 2010;12(5):212. doi: 10.1186/bcr2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu JS, Shew JY, Huang CS. Immunohistochemical analysis of survivin expression in primary breast cancers. Journal of the formosan medical association. 2004;103(12):925–931. [PubMed] [Google Scholar]
- 20.Boidot R, Vegran F, Jacob D, Chevrier S, Gangneux N, Taboureau J, Oudin C, Rainville V, Mercier L, Lizard-Nacol S. The expression of BIRC5 is correlated with loss of specific chromosomal regions in breast carcinomas. Genes, chromosomes and cancer. 2008;47(4):299–308. doi: 10.1002/gcc.20533. [DOI] [PubMed] [Google Scholar]
- 21.Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MC, de Bruin PC, Oudejans JJ, Marchionni L, van Diest PJ. Oncogene amplification in male breast cancer:analysis by multiplex ligation-dependent probe amplification. Breast cancer research and treatment. 2012;135(1):49–58. doi: 10.1007/s10549-012-2051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moelans CB, de Wegers RA, Monsuurs HN, Maess AH, van Diest PJ. Molecular differences between ductal carcinoma in situ and adjacent invasive breast carcinoma:a multiplex ligation-dependent probe amplification study. Analytical cellular pathology (Amsterdam) Journal. 2011;34(3):475–482. doi: 10.1007/s13402-011-0043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Span PN, Tjan-Heijnen VC, Heuvel JJ, de Kok JB, Foekens JA, Sweep FC. Do the survivin (BIRC5) splice variants modulate or add to the prognostic value of total survivin in breast cancer? Clinical chemistry. 2006;52(9):1693–1700. doi: 10.1373/clinchem.2006.071613. [DOI] [PubMed] [Google Scholar]
- 24.Alaggio R, Turrini R, Boldrin D, Merlo A, Gambini C, Ferrari A, Dall’igna P, Coffin CM, Martines A, Bonaldi L, De Salvo GL, Zanovello P, Rosato A. Survivin expression and prognostic significance in pediatric malignant peripheral nerve sheath tumors (MPNST) PloS one. 2013;8(11):e80456. doi: 10.1371/journal.pone.0080456. [DOI] [PMC free article] [PubMed] [Google Scholar]


