Abstract
PET studies allow in vivo imaging of the density of brain receptor species. The PET signal, however, is the sum of the fraction of radioligand that is specifically bound to the target receptor and the non-displaceable fraction (i.e. the non-specifically bound radioligand plus the free ligand in tissue). Therefore, measuring the non-displaceable fraction, which is generally assumed to be constant across the brain, is a necessary step to obtain regional estimates of the specific fractions.
The nondisplaceable binding can be directly measured if a reference region, i.e. a region devoid of any specific binding, is available. Many receptors are however widely expressed across the brain, and a true reference region is rarely available. In these cases, the nonspecific binding can be obtained after competitive pharmacological blockade, which is often contraindicated in humans.
In this work we introduce the genomic plot for estimating the nondisplaceable fraction using baseline scans only. The genomic plot is a transformation of the Lassen graphical method in which the brain maps of mRNA transcripts of the target receptor obtained from the Allen brain atlas are used as a surrogate measure of the specific binding. Thus, the genomic plot allows the calculation of the specific and nondisplaceable components of radioligand uptake without the need of pharmacological blockade.
We first assessed the statistical properties of the method with computer simulations. Then we sought ground-truth validation using human PET datasets of seven different neuroreceptor radioligands, where nonspecific fractions were either obtained separately using drug displacement or available from a true reference region. The population nondisplaceable fractions estimated by the genomic plot were very close to those measured by actual human blocking studies (mean relative difference between 2% and 7%). However, these estimates were valid only when mRNA expressions were predictive of protein levels (i.e. there were no significant post-transcriptional changes). This condition can be readily established a priori by assessing the correlation between PET and mRNA expression.
Keywords: Non-displaceable binding, PET, Lassen plot, Allen brain atlas, Pharmacological displacement, Pharmacological blockade, mRNA
Introduction
Positron emission tomography (PET) radioligands allow in vivo imaging of the distribution and availability of brain receptors. Among the criteria for a successful radioligand (Pike, 2009) a high level of specific binding is of prominent importance. The PET signal is a mixture of specific binding (i.e. the percentage of radioligand bound to the target receptor) and nondisplaceable uptake, which is the sum of nonspecific binding and free ligand in tissue (Innis et al., 2007). The ratio at equilibrium of the specifically bound radioligand to that of the nondisplaceable radioligand in tissue is the binding potential (BPND) which reflects the affinity of the radioligand for the target and more importantly the target availability in vivo (Innis et al., 2007). If the nondisplaceable component is known, then the specific component, and hence BPND, can be derived from the total activity.
The nondisplaceable binding can be directly measured if a reference region, i.e. a region truly devoid of the receptors under study, is available (Lammertsma et al., 1996; Lammertsma and Hume, 1996). The radioligand concentration in this region then becomes the reference for all regions, because the nondisplaceable binding is generally assumed to be constant in the brain (Lammertsma and Hume, 1996). Many receptors are however expressed across the whole human brain, and therefore a reference region is rarely available (Turkheimer et al., 2012). In these instances, measuring the nondisplaceable fraction requires an additional scan after administration of a competitive blocking agent (Lassen et al., 1995). This can be achieved without the complete blockade of all receptors, because the parameter of interest can be derived from a simple correlation using the variation of the total binding before and after administration of the blocking drug. This approach, generally described as the Lassen plot method (Cunningham et al., 2010; Lassen et al., 1995), has however two disadvantages: 1) blocking drugs may not be safe for human use at pharmacological doses or may not be available and 2) at least two separate PET scans are necessary for each subject.
Recently, Zanotti-Fregonara et al. (2015) introduced a method to estimate the specific and nondisplaceable components of a PET radioligand for metabotropic glutamate receptor 1 (mGluR1), based on the correlation of the regional density of the mGluR1 gene transcript with PET measurements of the expressed protein. This analysis is a variation of the Lassen plot, which measures the linear relationship of the tracer specific binding as a function of total distribution volume in brain. In Zanotti-Fregonara et al. (2015), the regional densities of the mGluR1 gene transcript in human brain were strongly and linearly correlated with regional densities (distribution volume, VT) of mGluR1 measured with PET. Furthermore, the extrapolated value of VT when mGluR1 gene transcript would equal 0 (i.e., in the absence of specific binding) was a reasonably accurate measure of tracer nondisplaceable volume of distribution (VND), based on pharmacological blockade in a monkey. This variation of the Lassen plot, which we call the “genomic plot”, has the significant advantage of estimating the specific and nondisplaceable components of radioligand uptake without the need for pharmacological blockade.
The purpose of this study was to assess the wider applicability of the genomic plot to estimate specific and nondisplaceable components of several PET targets, including six receptors and one enzyme. We first validated the statistical properties of the genomic plot with computer simulations to test its robustness against varying brain protein and mRNA patterns as well as varying mRNA–protein relations for different neurotransmitter systems. We then sought ground-truth validation using human PET datasets where estimates of the nondisplaceable fraction were available from blocking studies.
Material and methods
Theory
In a brain PET study, the radioligand activity in the tissue is typically the sum of a specific and a nondisplaceable component (Innis et al., 2007; Lassen et al., 1995; Mintun et al., 1984). Given n regional estimates, the total volume of distribution for the jth region (VT,j) is given by:
| (1) |
where VS,j is the regional specific distribution volumes for the jth region and VND represents the nondisplaceable distribution volume. The implicit assumption of Eq. (1) is that VND is constant across all brain regions, a common and generally valid assumption in brain neuroreceptor experiments (Lammertsma et al., 1996; Lammertsma and Hume, 1996; Lassen et al., 1995).
If a close relationship exists between mRNA expression and protein concentration, the transcriptome then reflects the in vivo distribution of the brain protein, and therefore it can be used as a proxy of the specific binding of the radioligand. This can be written as:
| (2) |
where mRNAj represents the vector of mRNA measurements for a given gene in the jth region and α is the scale factor between the transcript expression (either relative or absolute) and the tracer specific binding. Notably, the use of a constant value for α across the different brain regions is a strong assumption which holds only when a linear dependence exists between the mRNA expression and the correspondent protein density and when the occupancy by endogenous neurotransmitter is uniform across brain regions. When these conditions are met, by combining Eqs. (1) and (2), the distribution volume of the radioligand becomes linearly related to the mRNA expression of the target as
| (3) |
Following the convention introduced for the Lassen plot for PET occupancy studies (Lassen et al., 1995) and given the smaller noise of PET VT compared to mRNA data, we use VT as the independent variable, rewriting Eq. (3) as:
| (4) |
The linear regression of these two variables (x=mRNA and y=VT) generates a plot where the x-axis intercept is equal to the VND. We named this method the genomic version of the Lassen plot or simply “genomic plot”.
Simulated data
Monte-Carlo simulations were performed to assess the statistical robustness of the genomic plot (Eq. (4)) in a number of realistic stochastic conditions reflecting:
-
◾
Variability of the specific binding VS between brain regions (i.e. variability of VS between ROIs)
-
◾
Misspecification between mRNA and VS (i.e. variability of α between ROIs) indicating lack of linearity between gene expression and measured protein levels
-
◾
Variability of the tracer binding potential (approximated by VS/VND)
For the first condition, we simulated a spectrum of 5 cases corresponding to a VS variability ranging from 10% (homogenous VS distribution) to 50% (heterogeneous VS distribution). A lognormal distribution was used to avoid the generation of negative VS.
For the second condition, we simulated 11 different scenarios of α between-region variability, from 1% (no misspecification between mRNA and VS) to 100% (complete misspecification between mRNA and VS). The values were randomly generated by sampling a lognormal distribution with mean equal to 0.1 and standard deviation proportional to the variance of α. This range of values for α was chosen based on the results reported by Rizzo and colleagues (Rizzo et al., 2014), who showed that PET VT estimates (ml/cm3) are approximately one order of magnitude lower than the linearized mRNA expression (unit less) of the correspondent target proteins. The analysis included 3 brain PET tracers ([11C]WAY100635, [11C]CUMI101 and [11C]DPN) with the Allen Human Brain atlas as source for the genomic information (http://human.brain-map.org/).
For the last condition, three different scenarios were tested: a low-binding case (mean VS = 0.5 VND), an intermediate-binding case (mean VS = VND) and a high-binding case (mean VS = 2 VND). Mean VS values were used as reference to generate the regional specific bindings, and the between-region variability was defined accordingly to the first condition. These representative cases were chosen to cover the typical range of binding potential for a PET tracer (Guo et al., 2009).
VND was assumed constant for all simulations (VND = 2 ml/cm3), as used previously (Cunningham et al., 2010). Twelve ROIs were simulated for both PET and mRNA data, which is a typical number of regions when brain PET scan is matched with mRNA measures (Rizzo et al., 2014). For all conditions, 1000 noisy simulations were generated by adding Gaussian distributed noise (zero mean and 5% coefficient of variation, CV) to the total distribution volumes, independently for each simulated ROI. This procedure, as well as the noise level, was defined according to the literature (Cunningham et al., 2010). In total, 165,000 simulations (5 cases of VS variability × 11 cases of genomic misspecification × 3 levels of binding × 1000 simulations) were computed. A summary of the settings used for the simulations is reported in Table 1.
Table 1.
Simulation variables.
| Nondisplaceable binding (VND) | |
| Mean (ml/cm3) | 2 |
| Type of distribution | Constant |
| Specific binding (Vs) | |
| Mean (ml/cm3) | 1 (low-binding case), 2 (intermediate-binding case) and 4 (high-binding case) |
| CV (σ/mean) | 10%, 20%, 30%, 40% and 50% |
| Type of distribution | Lognormal |
| Scale factor between mRNA and Vs (α) | |
| Mean (ml/cm3) | 0.1 |
| CV (σ/mean) | 1%, 2%, 3%, 4%, 5%, 10%, 20%, 30%, 40%, 50% and 100% |
| Type of distribution | Lognormal |
| Other variables | |
| Number of ROIs per simulated dataset | 12 |
| Number of simulations per scenario | 1000 |
CV: coefficient of variation; σ: standard deviation.
For each simulated scenario, VND was estimated using the genomic plot (Eq. (4)), and the results compared with the correspondent simulated values. Percentage mean bias (%bias) and root mean square error (%RMSE) were used as indexes of performance:
| (5) |
| (6) |
where N is number of simulations (N = 1000), is the simulated nondisplaceable distribution volume and VND,i is the ith estimated nondisplaceable distribution volume. The squared Pearson's correlation coefficients (R2) between simulated mRNA and simulated VT were also analysed to assess the impact of the different tested conditions on the relationship between these two quantities.
In vivo positron emission tomography data
To assess the applicability of the method to clinical data, we applied the genomic plot to the following brain PET tracers: 1)[11C]WAY100635, targeting the serotonin 5-HT1A receptor (Pike et al., 1996); 2)[11C]Ro15-4513 targeting the GABA α5 receptor (Lingford-Hughes et al., 2002); 3)[11C]LY2795050 targeting the kappa opioid receptor (Zheng et al., 2013); 4)[18F]FIMX, targeting the metabotropic glutamate receptor 1 (Xu et al., 2013); 5)[11C]NOP-1A targeting the nociceptin/orphanin FQ peptide receptor (Pike et al., 2011); 6)[11C](R)rolipram, targeting the phosphodiesterase 4 enzyme (Fujita et al., 2005); 7)[11C]Raclopride, targeting the dopamine D2 receptors (Ehrin et al., 1985).
The choice of these radioligands was driven by the availability of in vivo VND measurements usable as the gold standard to validate the estimates obtained with the genomic plot (Table 2). Both in house data and blocking studies from literature were considered:
[11C]WAY100635: Fifteen male healthy subjects (35.7 ± 10.5 years old) underwent a 95-min dynamic PET study in an ECAT EXACT3D (Siemens/CTI, Knoxville, TN, USA) scanner after a bolus injection of 301 ± 12 MBq of [11C]WAY100635. The data were analysed as previously reported (Bose et al., 2011). The data were acquired at Imanet PET centre, London (UK).
[11C]Ro15-4513: Data from previously reported study (Stokes et al., 2014) of four healthy male subjects (41.5 ± 4.4 years old) scanned twice were considered. All subjects underwent 90-min dynamic scan on an ECAT HR + 962 scanner (CTI/Siemens, Knoxville, TN, USA) camera after injection of ~450 MBq of [11C]Ro15-4513. The data were acquired at the Imanet PET centre, London (UK).
[11C]LY2795050: Sixteen healthy volunteers (24 to 56 years old; 8 males/8 females) underwent a 90-min dynamic PET scan on a High Resolution Research Tomograph (Siemens Medical Solutions, Knoxville, TN, USA) after intravenous administration of tracer over 1 min by an automatic pump (injected dose: 334 ± 149 MBq). Full details on PET procedures, arterial data extraction and processing are reported in (Naganawa et al., 2014). PET data were acquired at the Yale PET centre, New Haven, Connecticut (USA).
[18F]FIMX: Twelve healthy controls (28 ± 10 years old; 4 males/8 females) underwent a 120-min dynamic PET scan after intravenous administration of 189 ± 3.4 MBq of tracer. Full details on PET procedures, arterial data extraction and processing are reported in (Zanotti-Fregonara et al., 2015). PET data were acquired at the NIH PET centre, Bethesda, Maryland (USA).
[11C]NOP-1A: Data from previously reported study (Lohith et al., 2014) of eleven healthy subjects (from 22 to 42 years old; 8 males/3 females) were considered. All the subjects underwent 120-min dynamic scan on an Advance tomograph (GE Medical Systems, Waukesha, WI) after bolus injection of 713 ± 79 MBq of [11C]NOP-1A. Full details on PET protocol and data and processing are reported in Lohith et al. (2014) and Tonietto et al. (2015). PET data were acquired at the NIH PET centre, Bethesda, Maryland (USA).
[11C](R)rolipram: Twelve healthy controls (28 ± 11 years old; 10 males/2 females) were included in the analysis (Zanotti-Fregonara et al., 2011). All PET images were acquired using an Advance tomograph (GE Medical Systems, Waukesha, WI, USA) after a bolus injection of 695 ± 152 MBq of [11C](R)-rolipram The data were analysed as previously reported in (Rizzo et al., 2013). PET data were acquired at the NIH PET centre, Bethesda, Maryland (USA).
[11C]Raclopride: Ten healthy controls (46.7 ± 14.1 years old; 8 males/2 females) were acquired using a ECAT EXACT HR++ tomograph (CTI/Siemens 966; Siemens, Knoxville, TN) after a bolus injection of 180–186 MBq (Pavese et al., 2006). PET data were acquired at the Imanet PET centre, London (UK).
Table 2.
Performance of the genomic plot in the measured PET datasets.
| Tracer | Target | mRNA vs. VT correlation (R2) | N. ROIs | Estimated VND (ml/cm3) | Expected VND (ml/cm3) | Ref(s) |
|---|---|---|---|---|---|---|
| [11C]WAY100635 | Serotonin 5-HT1A receptor | 0.91 | 12a | 0.30 ± 0.02 | 0.28 ± 0.43† | Cunningham et al. (2010) |
| [11C]Ro15-4513 | GABA α5 receptor | 0.80 | 10b | 2.14 ± 0.24 | 2.13 ± 0.80† | Myers et al. (2015) |
| [11C]LY2795050 | Kappa opioid receptor | 0.25 | 11c | 0.26 ± 3.43 | 1.69 ± 0.13† | Naganawa et al. (2014) |
| [18F]FIMX | Metabotropic glutamate receptor 1 | 0.99 | 11d | 0.47 ± 0.02 () | 0.58 ± 0.03‡ () | Zanotti-Fregonara et al. (2015) |
| [11C]NOP-1A | Nociceptin/orphanin FQ peptide receptor | <0.01 | 9e | n.a. | 7.08 ± 0.47!! | Kimura et al. (2011) |
| [11C](R)rolipram | Phosphodiesterase 4 enzyme | <0.01 | 12a | n.a. | 0.60 ± 0.12‡ () | (Unpublished data from the NIH) |
VT: total distribution volume; VND: nondisplaceable distribution volume fp: free plasma fraction; n.a.: not available.
Region segmentations:
Frontal lobe, Parietal lobe, Temporal lobe, Occipital lobe, Cingulate gyrus, Insula, Striatum, Globus pallidum, Basal forebrain, Amygdala, Thalamus, Cerebellum.
Frontal lobe, Parietal lobe, Temporal lobe, Occipital lobe, Cingulate gyrus, Hippocampus, Insula, Striatum, Thalamus, Cerebellum.
Frontal lobe, Temporal lobe, Occipital lobe, Cingulate gyrus, Hippocampus, Insula, Striatum, Globus pallidum, Amygdala, Thalamus, Cerebellum.
Frontal lobe, Parietal lobe, Temporal lobe, Occipital lobe, Cingulate gyrus, Hippocampus, Insula, Striatum, Amygdala, Thalamus, Cerebellum.
Frontal lobe, Parietal lobe, Temporal lobe, Occipital lobe, Cingulate gyrus, Striatum, Thalamus, Cerebellum, Brainstem.
Type of blocking
Blocking study in humans;
Blocking study in monkeys corrected for fp interspecies differences;
Blocking study in monkeys NOT corrected for fp interspecies differences.
For all these studies, ethical approval was granted independently by the internal ethical committees of the different institutes. Regional VT estimateswere quantifiedwith a nonlinearweighted least square estimator applied to the 2-tissue compartmental model. For [11C]Raclopride, the outcome parameter was BPND, obtained with a simplified reference tissue model using the cerebellum as the reference region. For each tracer, regional population VT (or BPND for [11C]Raclopride) estimates were obtained by averaging results across subjects prior to genomic plot analysis (Supplementarymaterial –Tables 1–7). Notably all the datasets were acquired independently (experimental conditions change from study to study) and processed according to the goals of the clinical studies. For example, the brain segmentation was inconsistent between datasets, because for each case the region contouring was defined to best match the tracer tissue distribution. These characteristics represented the best conditions to test the flexibility, robustness and general applicability of the genomic plot.
mRNA data and Allen Human Brain atlas
The mRNA transcription maps were downloaded from the Allen Human Brain atlas (ABA) (http://www.brain-map.org) (Hawrylycz et al., 2012). This dataset is derived from six healthy donors (42.5 ± 13.4 years old, 5 male/1 female) and contains more than 29,000 gene expression profiles sampled throughout the brain. On average, 617 ± 242 samples were collected for each donor and the brain structures are sampled proportionally to their volume. For two out of the six brains, samples were collected from both hemispheres. For the remaining four, tissue samples for microarray analysis were limited to the left hemisphere.
Full details about the procedures for the tissue collection and processing, the microarray experimental design and execution, and the data quality control up to the integration of the data into the online resource are reported in the supplementary data of Hawrylycz et al. (2012).
The expression profiles of the genes downloaded from ABA were: HTR1A for [11C]WAY100635, GABRA5 for [11C]Ro15-4513, OPRK1 for [11C]LY2795050, GRM1 for [18F]FIMX, OPRL1 for [11C]NOP-1A, PDE4A, PDE4B, PDE4C and PDE4D for [11C](R)rolipram, and DRD2 for [11C]Raclopride. For all genes and all probes, mRNA expression was downloaded at the highest spatial resolution possible (i.e., each value represented a physical tissue sample) in log2 expression intensity. A summary of the data used for the analysis is available in the supplementary material.
mRNA data analysis
The individual mRNA expressions, originally mapped in the native brain MRI space, were spatially normalised to the standard stereotaxic space (MNI/ICBM152) using the transformation matrices available from the ABA data portal. Additionally, the anatomical classifications of the sample labels were moved on the image space to obtain brain segmentation consistent with the sample/structure/coarse levels of resolution, as defined in ABA. Based on this match, the mRNA samples belonging to the same ROIs were averaged across different donors. The ROIs considered were: frontal, parietal, temporal and occipital lobes, cingulate gyrus, hippocampus, insula, striatum, globus pallidum, amygdala, thalamus, cerebellum and brainstem. These regions were chosen to guarantee an adequate brain coverage and a sufficiently large number of samples per region (>10 per region and per donor).
Most of the gene expressions included in the ABA database are measured with two or more probes. The presence of multiple probes for the same gene, whose expression profiles may be inconsistent, does not allow the use of the mean mRNA expression as representative of the true mean transcript expressions. Thus, it is fundamental for any type of application to select only the probe that best describes the real transcript profile and discard the remaining ones. According to Hawrylycz et al. (2012), for each gene we selected a unique probe, representative of the gene expression. Among the available probes, the one with the least skewed distribution across donors was chosen (Fig. 1).
Fig. 1.
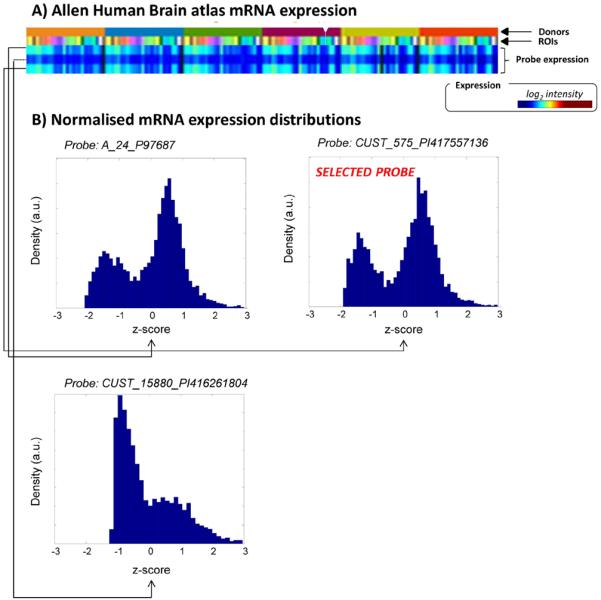
Example of probe selection for the 5TH1A receptor. A) Probe expression profiles as derived from the Allen Human Brain atlas (http://human.brain-map.org/) searching for HTR1A gene(setting: course level, log2 intensity, 6 donors). B) Normalised mRNA expression distributions (z-score) for the 3 probes of the HTR1A gene. For each probe the data from all the donors are reported. The probe of choice was CUST_575_PI417557136. The selection was done by comparing the normalised distributions and by selecting the one with the maximum correspondence to the highest z-score value (for further information please refer to MENGA's software manual — www.fair.dei.unipd.it/software).
The processing of the mRNA data was performed with MENGA (Multimodal Environment for Neuroimaging and Genomic Analysis) (Rizzo et al., 2016). This software package allows the integration of the mRNA transcript maps from ABA with any neuroimaging modality and calculates the gene vs. image cross-correlation statistics. MENGA software and manual are available at www.fair.dei.unipd.it/software. All analyses, including genomic plot implementation, were performed using Matlab® 2012b (The MathWorks, Inc., Natick, MA, USA) on a Windows 7 computer.
Genomic plot analysis
First, mRNA data were converted from log2 intensity into linear scale. This preliminary step is necessary to maintain a linear relationship between mRNA measures and gene transcript intensity. The values were then linearly regressed with the population VT values from the corresponding PET tracer to determine whether the relative gene transcript was proportional to VT. The squared Pearson's correlation coefficient (R2) was used to quantify the linear correlation. The VND values estimated by the genomic plot were then compared with the measured VND values obtained from the blocking studies available in the literature from human or nonhuman primates. For human data, the estimated VND values were directly compared to the measured ones. For primate data, interspecies differences were accounted for by correcting VND measurements for the tracer plasma free fractions of the two species ( for humans and for monkeys, respectively). This resulted in:
| (7) |
where is the nondisplaceable distribution volume measured in blocking studies in nonhuman primate and is its equivalent in humans. Eq. (7) implicitly assumes that the free fraction of ligand in the nondisplaceable tissue compartment (fND) is unchanged between species. The procedure was implemented consistently with the approach presented by Zanotti-Fregonara et al. (2015).
[11C]Raclopride is a particular case. Since this tracer targets dopamine D2 receptors, a true reference region (i.e. the cerebellum) is available. Therefore, [11C]Raclopride can be quantified with BPND, without any peripheral blood measurement. If the assumption used for VT (i.e. mRNA predicts the protein density in vivo) holds for [11C]Raclopride, then the intercept of the regression line should cross the origin of the axes.
Results
Simulated data
Fig. 2 shows some representative genomic plots applied to different cases of simulated data with VND = 2 ml/cm3 and BPND = 1 (intermediate-binding case). When the genomic misspecification (i.e. the misspecification between mRNA and VS) is minimal (CV α = 1%, Fig. 2A and C), mRNA vs. VT correlation is very high (R2 > 0.99) and the bias is <1%. On the contrary, when misspecification is maximal (CV α = 100%), the correlation between transcript and VT decreases (R2 = 0.40 and R2 = 0.48, Fig. 2B and D respectively) and this affects the precision and accuracy of VND quantification. Notably, when the VS variability between regions is maximal (CVVS = 50%, Fig. 2C and D), the performance is better than when the specific binding is uniform across regions (CVVS = 10%, Fig. 2A and B respectively).
Fig. 2.

Examples of simulated data. Representative genomic plots in different simulated conditions: 10% of VS variability and 1% of genomic misspecification (A); 10% of VS variability and 100% of genomic misspecification (B); 50% of VS variability and 1% of genomic misspecification (C); 50% of VS variability and 100% of genomic misspecification (D). In all the cases VND and mean VS are equal to 2 ml/cm3(corresponding to intermediate-binding simulated scenario).
These results are confirmed by the overall method performance across the different simulated scenarios (Fig. 3). The correlation matrix (Fig. 3A) follows a bimodal behaviour: the correlation values are high (R2 > 0.8) when the level of genomic misspecification is low (CV α < 20%); with higher values for higher VS variability. On the contrary, the lowest correlation values (R2 < 0.2) are found when genomic misspecification is high (CV α > 30%). Similar to the previous cases, the higher the VS variability between regions the better the mRNA vs. VT correlation. These results are independent from VND and from the simulated level of specific binding.
Fig. 3.
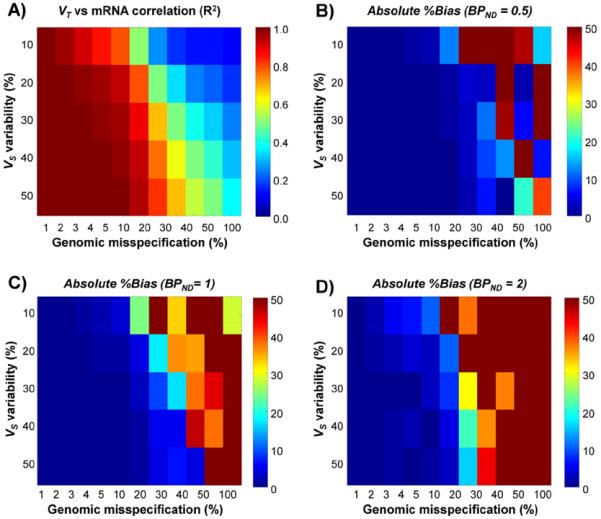
Performance of the genomic plot in simulated data. A) VT vs. mRNA mean correlation (R2) as function of VS variability and genomic misspecification. B-D) Absolute %bias as function of VS variability and genomic misspecification in low-binding scenario (BPND = 0.5), intermediate-binding scenario (BPND = 1) and high-binding scenario (BPND = 2) respectively.
In agreement with the correlation matrix results, %bias is also similarly dependent on genomic misspecification and VS variability (Fig. 3B–D). The best performance is found for the lowest misspecification (CV α = 1%) and the highest VS variability (CVVS = 50%), and the worst results are obtained in the opposite case (CV α = 100%, CVVS = 10%). Differently from the correlation analysis, the level of specific binding of the radioligand affects the overall performance: the lower the VS/VND ratio, the lower the bias. The %bias average (mean ± SD) is 70% ± 636% (median –2.4%) for high-binding case, 17% ± 54% (median –1.5%) for intermediate-binding case, and –13% ± 70% (median –0.6%) for low-binding case. Consistently, %RMSE medians range from 82% in the high-binding case to 10% in the low-binding case. The variability of %bias highlights the high sensitivity of genomic plot performances to the ability of mRNA to correctly predict the in vivo protein density: the reliability of the method is inversely proportional to the genomic misspecification.
This is important piece of information, as the correlation between mRNA and VT can be used to predict the performance, here intended in terms of bias, of the genomic plot. Defining 95% of confidence interval of the estimates, simulation results suggested that the genomic plot can yield an absolute %bias < 5% when R2 > 0.4 for low-binding scenario, R2 > 0.6 for intermediate-binding scenario and R2 > 0.85 for the high-binding scenario. Similarly, an absolute %bias < 10% could be obtained with a correlation R2 > 0.25 for low-binding scenario, R2 > 0.4 for intermediate-binding scenario and R2 > 0.75 for the high-binding scenario. Notably, these thresholds are valid only within the particular settings used for the simulations and need to be further validated by analysing actual measured data.
Performances in vivo measured datasets
Table 2 reports a summary of the genomic results in the measured PET datasets. A variable degree of correlation between mRNA and VT was found among the different tracers. The highest correlation was found for [18F]FIMX, [11C]WAY100635 and [11C]Ro15-4513 (R2 > 0.80) while a low correlation was obtained for [11C]LY2795050 (R2 = 0.25). For [11C]NOP-1A and [11C](R)rolipram, no correlation was found (R2 < 0.1). Notably, the different number and type of regions used for the comparison did not appear to play a role in the correlation between PET imaging and mRNA expression. For all tracers, 9 to 12 ROIs were analysed (Table 2).
Similarly to the results obtained in simulated data, the best consistency between estimated and measured VND values was obtained for the tracers in which genomic plot showed the highest correlation between PET and mRNA expression (Fig. 4). Specifically these were [11C]WAY100635 (estimated VND = 0.30 ml/cm3 – measured VND 0.28 ± 0.43 ml/cm3) and [11C]Ro15-4513 (estimated VND = 2.14 ml/cm3 – measured VND 2.13 ± 0.80 ml/cm3), followed by [18F]FIMX (estimated VND/fp = 138 ml/cm3 – measured VND/fp 170 ml/cm3).
Fig. 4.
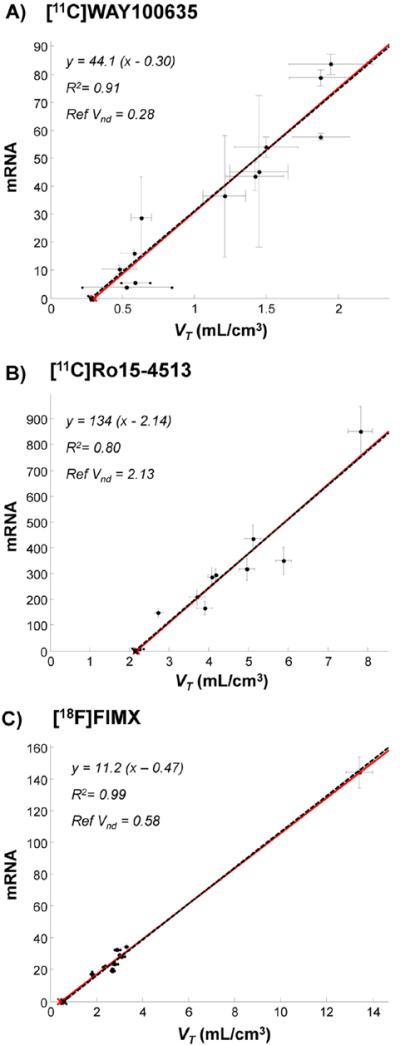
Genomic plot in PET measured data: successful cases. A) [11C]WAY100635 (15 subjects), B) [11C]Ro15-4513 (4 subjects) and C) [18F]FIMX (12 subjects). Data points (●) indicate regional population values. Both VT (x-axis) and mRNA (y-axis) standard errors are reported. Red lines refer to genomic plot regressions. Black dashed lines refer to the data regression with x-intercepts forced to the reference VND values, i.e. the VND values as derived from independent blocking studies. Please note that with [18F]FIMX the correlation is driven by a very high uptake region (i.e. cerebellum). When this is removed from the plot, the correlation is still significant (R2 = 0.65).
As predicted by the lack of any correlation between mRNA and PET, inaccurate results were obtained for [11C]LY2795050 (estimated VND = 0.26 ml/cm3 – measured VND 1.69 ± 0.13 ml/cm3), while for [11C]NOP-1A and [11C](R)rolipram non-physiological estimates were returned by the genomic graphical analysis (Fig. 5).
Fig. 5.

Genomic plot in PET measured data: unsuccessful cases. A) [11C]LY2795050 (16 subjects), B) [11C]NOP-1A (11 subjects) and C) [11C](R)rolipram (12 subjects). Data points (●) indicate regional population values. Both VT (x-axis) and mRNA (y-axis) standard errors are reported. Red lines refer to genomic plot regressions. Black dashed lines refer to the data regression with x-intercepts forced to the reference VND values, i.e. the VND values as derived from independent blocking studies.
Finally, as predicted from theoretical considerations, the regression line between mRNA data of D2 receptors and BPND values of [11C]Raclopride crossed the origin of the axes (Fig. 6).
Fig. 6.
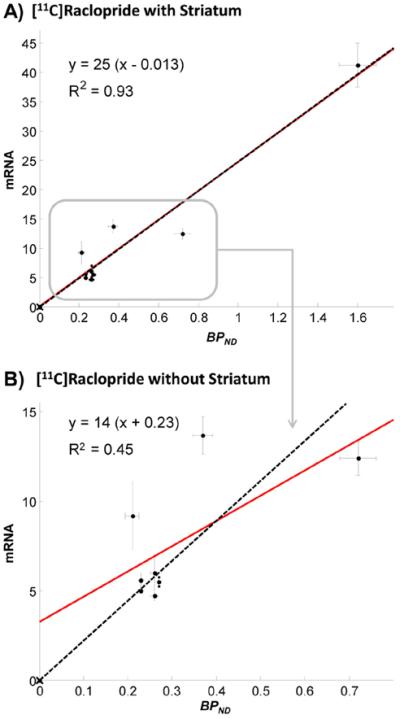
Genomic plot application to [11C]Raclopride brain PET data. The figure shows the regression between mRNA D2 receptor expressions and [11C]Raclopride BPND estimates as computed with the simplified reference tissue model, using the cerebellum as reference region (10 subjects). Analyses were performed in the following region of interest: frontal lobe, parietal lobe, temporal lobe, occipital lobe, cingulate gyrus, hippocampus, insula, striatum and thalamus (A). When the striatum is removed the correlation dropped and with it the performance of the method (B). Both BPND (x-axis) and mRNA (y-axis) standard errors are reported. Red lines refer to genomic plot regressions. Black dashed lines refer to the data regression with x-intercepts forced to the origin.
Impact of probe selection
Since for all genes multiple probes exists on mRNA microarrays we further investigated the application of the genomic plot to [11C]WAY100635, [11C]Ro15-4513 and [11C]LY2795050 (i.e. the tracers with human blocking study available) using secondary probes that were not originally selected as representative; the result is reported in Fig. 7. For each tracer most of the probes presented similar correlations with PET tracer activity and led to consistent VND estimates. Notably, these probes were also highly cross-correlated among each other (mean R2 = 0.92 for [11C]WAY100635, 0.95 for [11C]Ro15-4513 and 0.94 for [11C]LY2795050). Nevertheless, some probes did not correlate with the nominated probes nor with in vivo imaging (R2 < 0.01) and did not yield physiological VND estimates. These probes were CUST_15880_PI416261804 for [11C]WAY100635 and CUST_495_PI416408490 for [11C]LY2795050. Notably, both probes were characterized by a high number of low-expressed samples hence indicating that only probes with high affinity and abundant binding to the specific mRNA species should be selected for further processing.
Fig. 7.

Impact of probe selection on [11C]WAY100635 (A), [11C]Ro15-4513 (B) and [11C]LY2795050 (C) analysis. Reference VND estimates as obtained from blocking studies (red bars) are compared with those obtained from genomic plot applications (blue bars). For each tracer all the probes available from the Allen Human Brain database were tested. Dark blue bars refer to VND estimates as obtained from the probes selected a priori from the database. Light blue bars refer to VND estimates derived by the probes discarded from the initial selection. For each probe the VND standard error and the correlation with PET tracer uptake are also reported (squared Pearson's correlation coefficient, R2). Probes are sorted in descending order based on their correlation values with PET.
These results were confirmed when the analysis was extended to the tracers for which only nonhuman blocking studies were available. For both [11C]NOP-1A and [11C](R)rolipram, the use of secondary probes con-firmed a non-significant correlation between mRNA and PET VT (R2 < 0.1) and yielded negative VND estimates. Also for [18F]FIMX, the application of the genomic plot using secondary mRNA probes led to not physiological results. Notably these secondary probes were characterized by higher between-donor variability than the primary one. High correlation was found between primary and secondary probes for the DRD2 gene (R2 = 0.85 ± 0.10), resulting in no significant changes in the genomic plot performance.
Discussion
In this work, we presented an original approach for estimating the specific and nondisplaceable fractions of brain PET radioligands without using pharmacologically blocked studies.
The method derives the non-specific volume of distribution VND from the linear relationship between the relative regional density of the mRNA transcripts of the target receptor (considered as proxy of the tracer specific binding) and the regional volumes of distribution of the radioligand at baseline.
The proposed approach can be considered as a genomic variant of the Lassen plot (Lassen et al., 1995), and therefore was named genomic plot. It is worth noting that several versions of the Lassen plot have been previously introduced, where non-specific fractions were obtained either in the absence of a drug free scan (multiple drug levels but no baseline) (Cunningham et al., 2010), with partial blocking (Owen et al., 2014), or in the presence of varying affinities across subjects due to genetic polymorphisms (Guo et al., 2014). With the genomic plot, we have introduced the use of transcript densities as a proxy for protein density and the specific volume VS.
Method applicability
Because the Allen Human Brain atlas contains the transcript maps of 29,179 human brain proteins, the genomic plot potentially has potentially wide applicability. In practice, a number of conditions affect the performance of the method. One critical assumption is related to the ability of gene transcript to predict protein density. At least three factors may potentially explain the poor correlations reported in the literature between mRNA and protein concentrations, and these may not be mutually exclusive (Maier et al., 2009). First, post-transcriptional mechanisms (e.g. transcriptional and post-transcriptional splicing, translational modifications and protein complex formations) influence the degree to which mRNA expression translates into protein expression; second, proteins may differ substantially in their in vivo half-lives; third, technologies may not be perfectly accurate in measuring either mRNA or protein content. In addition, limitations in postmortem tissue availability and quality may further degrade the quality of genomic maps and their predictive level. In general, the strength of correlation between mRNA and in vivo protein density varies among genes depending on their function: it is stronger for genes related to cellular structure and lower or negligible for those related to cell development and the regulation of cellular function (Guo et al., 2008; Rizzo et al., 2014).
Other implicit assumptions for the applicability of the genomic plot regard the existence of a perfect correspondence between genomic and imaging samples a uniform occupancy of the endogenous neuro-transmitter across the regions. For the first condition we developed a method that scrupulously couples brain imaging with genomic data. By taking advantage of the detailed spatial information of the ABA database, we were able to generate a univocal correspondence between imaging and mRNA samples. This procedure is implemented in the MENGA software. The heterogeneity of endogenous binding, instead, is more difficult to control and may potentially limit the applicability of the genomic plot. It is important to highlight that the characteristics of the PET population must match with those of the mRNA donors. Since the presence of neurological or psychiatric diseases can unevenly affect the tracer uptake across brain regions, we recommend the use only with healthy controls.
Nevertheless, for all tested cases, both in simulations and measured data, we found that when mRNA expressions are well correlated to the PET signal at baseline the genomic plot yields accurate and precise estimates of the tracer non-specific fraction. On the contrary, when this condition is not met, the method was ineffective and its bias directly followed the misspecification between mRNA and PET. Thus, the correlation between mRNA and VT can be verified beforehand to predict whether the genomic plot will work for a given radioligand. Our simulations and analyses of clinical data found a mean bias lower than 10% when the correlation between mRNA and VT(R2 was greater than 0.75. This threshold can be relaxed with low or intermediate-binding tracers (BPND ≤ 1), i.e. when the VT carries more information on the VND than VS. Similarly, both simulations and clinical analyses showed that a higher correlation can be obtained more easily when the specific binding is variable between regions rather than uniformly distributed across the brain.
Method performances
The results obtained by the genomic plot on measured PET datasets in terms of accuracy matched quite well the a priori expectation for the different systems. VND quantification was unreliable for [11C]LY2795050, [11C]NOP-1A and [11C](R)rolipram. The first two are radioligands for the opioid system. For this system, well-known post-translational events control the production of mRNA variants and differentially modify proteins from each opioid receptor gene (Wei et al., 2004). By contrast, the target of [11C](R)rolipram is an intracellular enzyme, PDE4, which is part of the cAMP cascade. The bioactivity of this enzyme, and its affinity for [11C](R)rolipram, is modulated through phosphorylation (Fujita et al., 2005). In particular, phosphorylation is lost in non-living tissues and therefore the relationship between [11C](R)rolipram binding in vivo and mRNA expression post-mortem may not be preserved.
In contrast, serotoninergic receptors, with the exception of the 5-HT2C sub-type (Burns et al., 1997), are not known to undergo post-translational modifications, which explains the precision of the genomic plot for [11C]WAY100635. Also, the analysis of [11C]Ro15-4513 and [18F]FIMX yielded VND estimates in agreement with those obtained by occupancy studies in human and nonhuman primate respectively. Notably, it is unknown whether the mRNA expressions of GABA α5 and mGlu1 receptors can predict in vivo protein distribution (Berthele et al., 1999; Wisden et al., 1992), although this is likely, given the high correlation between mRNA and PET uptake (R2 > 0.9). Moreover, both GABA α5 and mGluR1 are widespread across brain and present higher expression values in some regions (hippocampus and forebrain for GABA α5, cerebellum for mGluR1). This uneven distribution of PET up-take represents the ideal condition for using the genomic plot.
Finally, the genomic plot yielded reliable results also for [11C]Raclopride data. [11C]Raclopride, a dopamine D2 radioligand, is commonly quantified using a reference region, usually the cerebellum. The outcome parameter, BPND, does not require blood sampling or correction for plasma free fraction, since both the input function and the free fraction are supposed to be equal across the different brain regions. As predicted from theoretical considerations, the intercept of the regression between mRNA and BPND values crossed the origin of the axes.
The validation of the method was limited by the blocking studies available in our centres and in the published literature. It should be noted that, compared to other studies in which only a single tracer was used for validation (e.g. Cunningham et al., 2010 or Turkheimer et al., 2012), the applicability of the methodology presented in this study was tested on multiple tracers directed to different brain systems. In particular, the use of nonhuman primate data can be justified under several aspects: 1) Nonhuman primate data have been used to validate the Lassen plot (e.g. Cunningham et al., 2010); 2) Nonhuman primates are often used when blocking studies are not feasible in humans; 3) The application of the genomic plot to nonhuman data allows assessing the method in suboptimal conditions.
Single subject applicability
Differently from the other Lassen plot methods, the genomic plot has been tested on population averages rather than on individual data. As a result, the method yields only population VND estimates instead of single subject results. There are several reasons behind this choice, mainly related to the mRNA data characteristics. First, genomic maps and PET images cannot be derived from the same subject but belong to different populations. Second, VND values are generally quite similar among subjects of the same species, and population values are often used as measure of tracer non-specific binding performance.
Indeed, when we tried to match single PET subjects to the population mRNA, we observed significant between-subject variability (Supplementary Material – Fig. 1) and percentage bias (mean ± SD = 10% ± 5%; range 1%:18%) (Supplementary Material – Fig. 2). For this particular analysis, we used [11C]WAY100635 because this tracer showed the best correlation with genomic expression and [11C]Raclopride, for which the individual reference region was used as reference for the non-displaceable binding of each subject. Based on these results, we recommend the use of the genomic plot only at the population level.
Methodological considerations
The genomic plot is based on a standard linear regression. Despite the simplicity of the methodology it is important to take into account the impact of axes convention or the type of estimator. Both VT and mRNA values are noisy, and when used as independent variables the noise can bias the final estimates. In our analyses, inverting the axes (mRNA on x-axis and VT on y-axis – corresponding to Eq. (3)) led to significantly higher bias and variability for all the tested scenarios (data not shown). This is explained by the fact that regional population VT values are generally less noisy than the correspondent mRNA data and therefore preferable as independent variables. Nevertheless, both the noise level and the variability of measurements can change depending on the radioligand and the system under study. In this case, alternative estimation methods, which take into account noise on both axes (Bekker, 1986), may be considered (e.g. likelihood estimation (Ogden, 2003) or orthogonal regression (Varga and Szabo, 2002)). These methods, similarly to weighted estimation approaches, necessitate assumptions about the noise distributions of the measurements. Realistic assumptions are difficult to make: both VT and mRNA are used as population averages in which it is necessary to distinguish the true inter-subject biological variability to random noise. Further investigations to solve this issue are necessary, although our analysis with the standard regression showed good performance, similarly to all the other versions of the Lassen plot (Cunningham et al., 2010; Lassen et al., 1995).
Probe selection
In the Allen Human Brain database, only 30% of the genes are univocally represented by a single probe. More than 50% are described by two probes, and 19% by three or more. For the most studied proteins the number of available probes can be greater than 30 (see for example monoamine oxidase A) (Zanotti-Fregonara et al., 2014). The availability of multiple probes is not necessarily an advantage as the different spec-ificity and sensitivity of the probes to the target can lead to inconsistent expression profiles across the brain for the same gene. Therefore, probe selection becomes a critical processing step as it heavily impacts on the final results. Notably, due to the between-probe discrepancy, probe averaging does not represent a viable strategy because the mean expression can be more inaccurate than the single probe profiles.
In this work we implemented a data-driven method for probe selection. To ensure that the method is generally applicable, no biological assumptions were used. The approach used between-donor consistency (e.g. correlation of probes expression across subjects) and abundancy of probe binding to select the most representative expression profile for a given gene. Notably the comparison between mRNA and imaging can be exploited in the opposite direction, i.e. by using the PET to test mRNA expression measurements. Large numbers of postmortem human studies use gene transcript density as a surrogate for protein levels. When a PET radioligand exists for a protein target, the genomic plot can validate that assumption and help selecting which of the multiple transcripts should be used as a surrogate measure.
Conclusion
The genomic variant of the Lassen plot, or genomic plot, allows the estimation of a PET radioligand nondisplaceable fraction in the absence of reference region and without requiring target-competition studies. The method has general applicability to any neuroreceptor PET tracer, because it relies on mRNA brain maps of the whole human genome. Nevertheless, its precision is dependent on the ability of mRNA expression to predict protein density. Therefore, the consistency between mRNA and PET should be verified a priori.
Supplementary Material
Acknowledgments
We thank the Allen Institute for the support with the human brain atlas.
Financial support This study was supported by the Programme Grant “Quantitative methodologies for Positron Emission Tomography” (UK Medical Research Council No. G1100809/1), by the Padova University grant “Neuroimaging Genetics: Models and Methods to Integrate Brain Phenotype and Genotype” (2013), and by the Intramural Research Programme of the National Institute of Mental Health (USA) project number ZIAMH002852 under clinical trials.gov identifier NCT02230592.
Footnotes
Disclosure The authors do not have any duality of interest.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2016.01.058.
References
- Bekker PA. Comment on identification in the linear errors in variables model. Econ.: J. Econ. Soc. 1986:215–217. [Google Scholar]
- Berthele A, Platzer S, Laurie DJ, Weis S, Sommer B, Zieglgänsberger W, Conrad B, Tölle TR. Expression of metabotropic glutamate receptor subtype mRNA (mGluR1-8) in human cerebellum. Neuroreport. 1999;10:3861–3867. doi: 10.1097/00001756-199912160-00026. [DOI] [PubMed] [Google Scholar]
- Bose SK, Mehta MA, Selvaraj S, Howes OD, Hinz R, Rabiner EA, Grasby PM, Turkheimer FE, Murthy V. Presynaptic 5-HT1A is related to 5-HTT receptor density in the human brain. Neuropsychopharmacology. 2011;36:2258–2265. doi: 10.1038/npp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Cunningham VJ, Rabiner EA, Slifstein M, Laruelle M, Gunn RN. Measuring drug occupancy in the absence of a reference region: the Lassen plot re-visited. J. Cereb. Blood Flow Metab. 2010;30:46–50. doi: 10.1038/jcbfm.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrin E, Farde L, de Paulis T, Eriksson L, Greitz T, Johnström P, Litton J-E, Nilsson JLG, Sedvall G, Stone-Elander S. Preparation of 11 C-labelled Raclopride, a new potent dopamine receptor antagonist: preliminary PET studies of cerebral dopamine receptors in the monkey. Int. J. Appl. Radiat.Isot. 1985;36:269–273. doi: 10.1016/0020-708x(85)90083-3. [DOI] [PubMed] [Google Scholar]
- Fujita M, Zoghbi SS, Crescenzo MS, Hong J, Musachio JL, Lu J-Q, Liow J-S, Seneca N, Tipre DN, Cropley VL. Quantification of brain phosphodiesterase 4 in rat with (R)-[11 C] rolipram-PET. NeuroImage. 2005;26:1201–1210. doi: 10.1016/j.neuroimage.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y, Chen X, Li L, Wu S, Chen Y, Jiang H, Tan L, Xie J, Zhu X, Liang S, Deng H. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008;40:426–436. doi: 10.1111/j.1745-7270.2008.00418.x. [DOI] [PubMed] [Google Scholar]
- Guo Q, Brady M, Gunn RN. A biomathematical modeling approach to central nervous system radioligand discovery and development. J. Nucl. Med. 2009;50:1715–1723. doi: 10.2967/jnumed.109.063800. [DOI] [PubMed] [Google Scholar]
- Guo Q, Owen DR, Rabiner EA, Turkheimer FE, Gunn RN. A graphical method to compare the in vivo binding potential of PET radioligands in the absence of a reference region: application to [ 11C] PBR28 and [ 18F] PBR111 for TSPO imaging. J. Cereb. Blood Flow Metab. 2014;34:1162–1168. doi: 10.1038/jcbfm.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein S, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J. Cereb. Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Fujita M, Hong J, Lohith TG, Gladding RL, Zoghbi SS, Tauscher JA, Goebl N, Rash KS, Chen Z. Brain and whole-body imaging in rhesus monkeys of 11C-NOP-1A, a promising PET radioligand for nociceptin/orphanin FQ peptide receptors. J. Nucl. Med. 2011;52:1638–1645. doi: 10.2967/jnumed.111.091181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. NeuroImage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- Lammertsma A, Bench C, Hume S, Osman S, Gunn K, Brooks D, Frackowiak R. Comparison of methods for analysis of clinical [11C] raclopride studies. J. Cereb. Blood Flow Metab. 1996;16:42–52. doi: 10.1097/00004647-199601000-00005. [DOI] [PubMed] [Google Scholar]
- Lassen N, Bartenstein P, Lammertsma A, Prevett M, Turton D, Luthra S, Osman S, Bloomfield P, Jones T, Patsalos P. Benzodiazepine receptor quantification in vivo in humans using [11C] flumazenil and PET: application of the steady-state principle. J. Cereb. Blood Flow Metab. 1995;15:152–165. doi: 10.1038/jcbfm.1995.17. [DOI] [PubMed] [Google Scholar]
- Lingford-Hughes A, Hume SP, Feeney A, Hirani E, Osman S, Cunningham VJ, Pike VW, Brooks DJ, Nutt DJ. Imaging the GABA-benzodiazepine receptor sub-type containing the α5-subunit in vivo with [11C] Ro15 4513 positron emission tomography. J. Cereb. Blood Flow Metab. 2002;22(7):878–889. doi: 10.1097/00004647-200207000-00013. [DOI] [PubMed] [Google Scholar]
- Lohith TG, Zoghbi SS, Morse CL, Araneta MDF, Barth VN, Goebl NA, Tauscher JT, Pike VW, Innis RB, Fujita M. Retest imaging of [11 C] NOP-1A binding to nociceptin/orphanin FQ peptide (NOP) receptors in the brain of healthy humans. NeuroImage. 2014;87:89–95. doi: 10.1016/j.neuroimage.2013.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Mintun MA, Raichle ME, Kilbourn MR, Wooten GF, Welch MJ. A quantitative model for the in vivo assessment of drug binding sites with positron emission tomography. Ann. Neurol. 1984;15:217–227. doi: 10.1002/ana.410150302. [DOI] [PubMed] [Google Scholar]
- Myers J, Comley RA, Gunn RN. XXVIIth International Symposium on Cerebral Blood Flow, Metabolism and Function & XIIth International Conference on Quantification on Brain Function with PET. Vancouver, Canada: 2015. Quantification of [11C]RO15-4513 specific binding and selectivity in vivo. [Google Scholar]
- Naganawa M, Zheng M-Q, Nabulsi N, Tomasi G, Henry S, Lin S-F, Ropchan J, Labaree D, Tauscher J, Neumeister A. Kinetic modeling of 11C-LY2795050, a novel antagonist radiotracer for PET imaging of the kappa opioid receptor in humans. J. Cereb. Blood Flow Metab. 2014;34:1818–1825. doi: 10.1038/jcbfm.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden RT. Estimation of kinetic parameters in graphical analysis of PET imaging data. Stat. Med. 2003;22:3557–3568. doi: 10.1002/sim.1562. [DOI] [PubMed] [Google Scholar]
- Owen DR, Guo Q, Kalk NJ, Colasanti A, Kalogiannopoulou D, Dimber R, Lewis YL, Libri V, Barletta J, Ramada-Magalhaes J. Determination of [(11) C] PBR28 binding potential in vivo: a first human TSPO blocking study. J. Cereb. Blood Flow Metab.: Off. J. Int. Soc. Cereb. Blood Flow Metab. 2014;34:1256. doi: 10.1038/jcbfm.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavese N, Gerhard A, Tai YF, Ho AK, Turkheimer F, Barker RA, Brooks DJ, Piccini P. Microglial activation correlates with severity in Huntington disease A clinical and PET study. Neurology. 2006;66(11):1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- Pike VW. PET radiotracers: crossing the blood-brain barrier and surviving metabolism. Trends Pharmacol. Sci. 2009;30:431–440. doi: 10.1016/j.tips.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike VW, McCarron JA, Lammertsma AA, Osman S, Hume SP, Sargent PA, Bench CJ, Cliffe IA, Fletcher A, Grasby PM. Exquisite delineation of 5-HT 1A receptors in human brain with PET and [carbonyl-11 C] WAY-100635. Eur. J. Pharmacol. 1996;301:R5–R7. doi: 10.1016/0014-2999(96)00079-9. [DOI] [PubMed] [Google Scholar]
- Pike VW, Rash KS, Chen Z, Pedregal C, Statnick MA, Kimura Y, Hong J, Zoghbi SS, Fujita M, Toledo MA. Synthesis and evaluation of radioligands for imaging brain nociceptin/orphanin FQ peptide (NOP) receptors with positron emission tomography. J. Med. Chem. 2011;54:2687–2700. doi: 10.1021/jm101487v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Veronese M, Zanotti-Fregonara P, Bertoldo A. Voxelwise quantification of [(11)C](R)-rolipram PET data: a comparison between model-based and data-driven methods. J. Cereb. Blood Flow Metab. 2013 doi: 10.1038/jcbfm.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Veronese M, Heckemann RA, Selvaraj S, Howes OD, Hammers A, Turkheimer FE, Bertoldo A. The predictive power of brain mRNA mappings for in vivo protein density: a positron emission tomography correlation study. J. Cereb. Blood Flow Metab. 2014 doi: 10.1038/jcbfm.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Veronese M, Expert P, Turkheimer F, Bertoldo A. MENGA: a new comprehensive tool for the integration of neuroimaging data and the Allen human brain transcriptome atlas. Plos One. 2016 doi: 10.1371/journal.pone.0148744. http://dx.doi.org/10.1371/journal.pone.0148744 (in press) [DOI] [PMC free article] [PubMed]
- Stokes PR, Myers JF, Kalk NJ, Watson BJ, Erritzoe D, Wilson SJ, Cunningham VJ, Barros DR, Hammers A, Turkheimer FE. Acute increases in synaptic GABA detectable in the living human brain: a [11 C] Ro15-4513 PET study. NeuroImage. 2014;99:158–165. doi: 10.1016/j.neuroimage.2014.05.035. [DOI] [PubMed] [Google Scholar]
- Tonietto M, Veronese M, Rizzo G, Zanotti-Fregonara P, Lohith TG, Fujita M, Zoghbi SS, Bertoldo A. Improved models for plasma radiometabolite correction and their impact on kinetic quantification in PET studies. J. Cereb. Blood Flow Metab. 2015 doi: 10.1038/jcbfm.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer FE, Selvaraj S, Hinz R, Murthy V, Bhagwagar Z, Grasby P, Howes O, Rosso L, Bose SK. Quantification of ligand PET studies using a reference region with a displaceable fraction: application to occupancy studies with [(11)C]-DASB as an example. J. Cereb. Blood Flow Metab. 2012;(32):70–80. doi: 10.1038/jcbfm.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Szabo Z. Modified regression model for the Logan plot. J. Cereb. Blood Flow Metab. 2002;22:240–244. doi: 10.1097/00004647-200202000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L-N, Law P-Y, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Front. Biosci. 2004;9:1665. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisden W, Laurie DJ, Monyer H, Seeburg PH. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. I. Telencephalon, diencephalon, mesencephalon. J. Neurosci. 1992;12:1040–1062. doi: 10.1523/JNEUROSCI.12-03-01040.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Zanotti-Fregonara P, Zoghbi SS, Gladding RL, Woock AE, Innis RB, Pike VW. Synthesis and evaluation in monkey of [18F] 4-Fluoro-N-methyl-N-(4-(6-(methylamino) pyrimidin-4-yl) thiazol-2-yl) benzamide ([18F] FIMX): a promising radioligand for PET imaging of brain metabotropic glutamate receptor 1 (mGluR1) J. Med. Chem. 2013;56:9146–9155. doi: 10.1021/jm4012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti-Fregonara P, Zoghbi SS, Liow J-S, Luong E, Boellaard R, Gladding RL, Pike VW, Innis RB, Fujita M. Kinetic analysis in human brain of [11 C](R)rolipram, a positron emission tomographic radioligand to image phosphodiesterase 4: a retest study and use of an image-derived input function. NeuroImage. 2011;54:1903–1909. doi: 10.1016/j.neuroimage.2010.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti-Fregonara P, Leroy C, Rizzo G, Roumenov D, Trichard C, Martinot J-L, Bottlaender M. Imaging of monoamine oxidase-A in the human brain with [11C] befloxatone: quantification strategies and correlation with mRNA transcription maps. Nucl. Med. Commun. 2014;35:1254–1261. doi: 10.1097/MNM.0000000000000196. [DOI] [PubMed] [Google Scholar]
- Zanotti-Fregonara P, Rong X, Zoghbi SS, Liow JS, Fujita M, Veronese M, Gladding RL, Rallis-Frutos D, Hong J, Pike VW, Innis RB. The positron emission tomographic radioligand 18F-FIMX images and quantifies metabotropic glutamate receptor 1 in proportion to the regional density of its gene transcript in human brain. J. Nucl. Med. 2015 doi: 10.2967/jnumed.115.162461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M-Q, Nabulsi N, Kim SJ, Tomasi G, Lin S.-f., Mitch C, Quimby S, Barth V, Rash K, Masters J. Synthesis and evaluation of 11C-LY2795050 as a κ-opioid receptor antagonist radiotracer for PET imaging. J. Nucl. Med. 2013;54:455–463. doi: 10.2967/jnumed.112.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


