Abstract
A recent study from our laboratory found that 18F-FIMX is an excellent PET radioligand for quantifying metabotropic glutamate receptor 1 (mGluR1) in monkey brain. This study evaluated the ability of 18F-FIMX to quantify mGluR1 in humans. A second goal was to use the relative density of mGluR1 gene transcripts in brain regions to estimate specific uptake and nondisplaceable uptake (VND) in each brain region.
Methods
After injection of 189 ± 3 MBq of 18F-FIMX, 12 healthy volunteers underwent a dynamic PET scan over 120 min. For 6 volunteers, images were acquired until 210 min. A metabolite-corrected arterial input function was measured from the radial artery. Four other subjects underwent whole-body scanning to estimate radiation exposure.
Results
18F-FIMX uptake into the human brain was high (SUV = 4–6 in the cerebellum), peaked at about 10 min, and washed out rapidly. An unconstrained 2-tissue-compartment model fitted the data well, and distribution volume (VT) (mL·cm−3) values ranged from 1.5 in the caudate to 11 in the cerebellum. A 120-min scan provided stable VT values in all regions except the cerebellum, for which an acquisition time of at least 170 min was necessary. VT values in brain regions correlated well with mGluR1 transcript density, and the correlation suggested that VND of 18F-FIMX was quite low (0.5 mL·cm−3). This measure of VND in humans was similar to that from a receptor blocking study in monkeys, after correcting for differences in plasma protein binding. Similar to other 18F-labeled ligands, the effective dose was about 23 µSv/MBq.
Conclusion
18F-FIMX can quantify mGluR1 in the human brain with a 120- to 170-min scan. Correlation of brain uptake with the relative density of mGluR1 transcript allows specific receptor binding of a radioligand to be quantified without injecting pharmacologic doses of a blocking agent.
Keywords: mGluR1, 18F-FIMX, PET, gene transcripts
APET radioligand for metabotropic glutamate receptor 1 (mGluR1) would be useful for exploring the potential role of this receptor in the pathophysiology of neuropsychiatric disorders and to facilitate the development of novel therapeutic drugs. However, design of PET radioligands for mGluR1 is challenging. Among the numerous demanding criteria that a prospective radioligand must fulfill (1), high affinity and selectivity are particularly difficult to achieve for mGluR1 imaging agents. In fact, mGluR1 belongs to the same family subgroup as mGluR5 and has a similar structure, DNA sequence, and function. Therefore, ligands engineered to bind to mGluR1 tend to cross-react with mGluR5. Indeed, several mGluR1 radioligands have been reported in the literature (2), but most were stopped at the preclinical level because of some unfavorable imaging characteristics.
To date, only 2 mGluR1 PET radioligands have been evaluated in humans. The first, the isothiazole derivative mGluR1 antagonist 11C-LY2428703, showed good characteristics both in vitro and in vivo in rats (3). It was, however, unsuitable for both monkey and human imaging because of low brain uptake, which was possibly caused by high binding to plasma proteins (4). The second, 11C-ITMM, was also successfully validated in rodents (5). Disadvantages in humans, however, included an overall low brain uptake and slow uptake and washout; as a result, 11C-ITMM is unlikely to quantify mGluR1 within the time constraints of the half-life (20 min) of 11C (6).
We recently developed a novel mGluR1 radioligand, 18F-FIMX, which has excellent imaging characteristics based on in vitro properties and in vivo imaging in monkey brain. 18F-FIMX has high affinity (half maximal inhibitory concentration, 5.1 nM) for human mGluR1, relatively low affinity for mGluR5 (8 µM (7)), suitable lipophilicity (LogD = 2.5), and a high ratio of specific (VS) to nondisplaceable uptake (VND) in monkey brain (8). Thus, one aim of this study was to evaluate the ability of 18F-FIMX to quantify mGluR1 in the human brain.
An important performance characteristic of any radioligand is its ratio of signal to noise—more specifically, its ratio of VS to VND. In the absence of any receptor-free region, measuring this ratio typically requires administering pharmacologic doses of a blocking agent that often does not exist for human use, especially for novel targets. For drug doses that do not completely block all receptors, the Lassen plot (9) uses a simple regression of VS to total distribution volume (VT) to estimate VND; this assumes that VND is the same in all brain regions, which is almost always the case.
This paper explored whether the relative regional density of mGluR1 gene transcript could be used as a substitute for VS, thereby allowing VND to be estimated without injecting any blocking drug. The rationale for this approach is that many gene transcripts are linearly related to the expression of the protein—that is, linearly proportional to the density of the receptor (10). For any given protein target, one must first confirm that the gene transcript (messenger RNA [mRNA]) is linearly proportional with protein density. Thus, we first explored whether mGluR1 gene transcript is proportional to receptor density measured with PET and then used this linear correlation to estimate the VND of 18F-FIMX. This allowed VS and the ratio of VS to VND to be calculated in each brain region. Furthermore, because the relative density of about 30,000 gene transcripts in regions of the human brain are now available in the Allen Brain Atlas (11), this method is potentially useful for numerous other PET imaging targets to estimate VS to VND of the associated radioligand. Thus, the 2 aims of this paper were to evaluate the ability of 18F-FIMX to quantify mGluR1 in the human brain and to explore whether the relative density of mGluR1 gene transcript could be used to estimate the ratio of VS to VND of the radioligand. In addition, because VND is typically similar across species, we validated the estimations in humans by calculating the VND of 18F-FIMX in a monkey after complete receptor blockade.
MATERIALS AND METHODS
Radioligand Preparation
18F-FIMX was produced from 18F-fluoride ion and an N-Boc–protected (phenyl)aryliodonium salt precursor as previously described (8). The radioligand was prepared according to our investigational new drug application (#119,521), available at http://pdsp.med.unc.edu/snidd/. The radioligand was obtained with high radiochemical purity (>99%) and specific activity at the time of injection of 61.8 ± 36.7 GBq/µmol. The mass dose was 0.09 ± 0.12 nmol/kg (range, 0.02–0.46 [n = 16]).
Subjects
Twelve healthy subjects (4 men, 8 women; age ± SD, 28 ± 10 y; weight ± SD, 73 ± 15 kg) underwent a brain scan, and an additional 4 subjects (2 men, 2 women; age ± SD, 31 ± 14 y; weight ± SD, 62 ± 8 kg) underwent a whole-body scan. All subjects were free of medical and neuropsychiatric diseases, as determined by medical history, physical examination, electrocardiogram, and laboratory blood and urine tests. Vital signs were monitored before ligand injection and then during and after completion of the scan. Urinalysis and blood lab tests were repeated within a few hours of completion of the PET scan. The study was approved by the Institutional Review Board, and all subjects signed a written informed consent form.
Plasma Measurements
After radioligand injection, arterial blood samples (1.5 mL each) were drawn from the radial artery at 15-s intervals until 120 s, followed by 3- to 5-mL samples at 3, 5, 10, 15, 20, 30, 45, 60, 75, 90, 105, and 120 min. For the 6 subjects who underwent a longer scan, additional samples were drawn at 135, 150, 165, 180, 195, and 210 min. The whole blood was first centrifuged to separate plasma. The concentration of parent radioligand in plasma was then measured by high-performance liquid chromatography in each blood sample (some blood samples around the peak of the input function, when the parent concentration is almost 100%, were interpolated) (12). All plasma input functions (and whole-blood curves) were well fitted by a triexponential function with gaussian weighting and used for kinetic modeling. The plasma free fraction (fP) was measured by ultrafiltration (13).
Image Acquisition and Analysis
Image acquisition and analysis are presented in the supplemental data (available at http://jnm.snmjournals.org).
Quantification of 18F-FIMX Binding
VT was calculated using both compartmental and noncompartmental models. Noncompartmental models included the Logan graphical analysis (14), Ichise’s multilinear analysis MA1 (15) and MA2 (15), Zhou’s relative-equilibrium Gjedde–Patlak bi-graphical analysis (RE-GP) (16), standard spectral analysis (SA) (17), and rank-shaping SA (RSSA) (18). Analyses were performed at the region and the voxel level (supplemental data). We also tested the possibility of obtaining measures of 18F-FIMX binding noninvasively, by replacing the arterial input function with a pseudoreference region. 18F-FIMX binding was thus calculated using a simplified reference tissue model (SRTM), an SUVR, and brain uptake itself (SUV) (supplemental data).
mGluR1 Transcript as Surrogate of Receptor Density
mRNA transcription maps were obtained from the 6 donors of the Allen Human Brain Atlas (http://human.brain-map.org/) (11). Three probes related to the expression of mGluR1 were available for each donor: A_23_P30976 (probe A), CUST_14602_PI416261804 (probe B), and CUST_320_PI416408490 (probe C). Regional mRNA values were obtained by averaging the individual samples of each region (i.e., the coarse resolution of the Atlas), expressed in log2 scale. Because mRNA was sampled from both the caudate and the putamen for all donors, values in the striatum were compared with the average of the caudate and putamen on PET. Similarly, mRNA values in the cingulum were compared with the average of anterior and posterior cingulum on the PET images. Both mRNA and VT values were preanalyzed with an autocorrelation analysis. As described by Rizzo et al. (10), the purpose of the autocorrelation analysis was to estimate data consistency with respect to intrasubject and intersubject variability. We first performed a Pearson analysis for each probe among the different donors, covering all possible combinations, to assess whether the values from different subjects were correlated and could thus be averaged to obtain 3 sequences of mRNA values corresponding to the 3 probes. We then performed the same autocorrelation among the 3 probes to select a representative one. The third autocorrelation was performed among the PET 2-tissue-compartment model VT values of the different subjects to assess whether these data could be averaged to obtain regionwise representative values of 18F-FIMX binding. Finally, the mRNA expression values of the representative probe were correlated to the averaged 2-tissue-compartment model/VT values of 18F-FIMX.
Estimation of VND of 18F-FIMX Using mGluR1 Transcript Density
We postulated that mRNA expression would be proportional to VS, mGluR1 would not be subject to significant posttranscriptional changes in vivo, and VND would be constant among the different brain regions. The mRNA values, expressed as log2 in the Allen Atlas, were first linearized and then linearly regressed against the 2-tissue-compartment model/VT values obtained with the 120-min scans (for the cerebellum, the average of the value at 210 min [n = 6] was used, because it is more stable). Following the Lassen plot method (9,19), the x-axis intercept is equal to VND. VND estimated in humans was compared with that measured in a monkey after pharmacologic blockade (supplemental data).
Whole-Body Imaging and Dosimetry
Whole-body imaging and dosimetry are presented in the supplemental data.
Statistical Analysis
Goodness-of-fit by nonlinear least-squares analysis was evaluated using the Akaike information criterion, model selection criterion, and F statistics (20). P values of less than 0.05 were considered significant. The precision of the estimates was expressed as a percentage and equaled the ratio of the SE divided by the value itself. The SE was obtained from the diagonal of the covariance matrix (21) using the generalized form of the error propagation equation (22). Smaller values indicate higher precision. VT values obtained using the different techniques, both at the regional and at the voxel level, were compared with the reference 2-tissue-compartment model VT. Comparisons were performed with repeated-measures ANOVA for VT values in the various regions, except the cerebellum, using SPSS (version 22 for Windows; SPSS Inc.). The cerebellum was excluded from the ANOVA analysis because it did not reach full stability at 120 min. Variability in binding values was defined as coefficient of variation = SD/mean × 100%. Group data are expressed as mean ± SD, except the kinetic microparameters, which are expressed as median values.
RESULTS
PET Brain Imaging with 18F-FIMX
Pharmacologic Effects
In the 12 subjects who underwent brain scanning and the 4 subjects who underwent whole-body scanning, no adverse events were reported either during or after the scans. In addition, no effects were noted on blood and urine tests, blood pressure, electrocardiogram, or respiratory rate.
Plasma Analysis
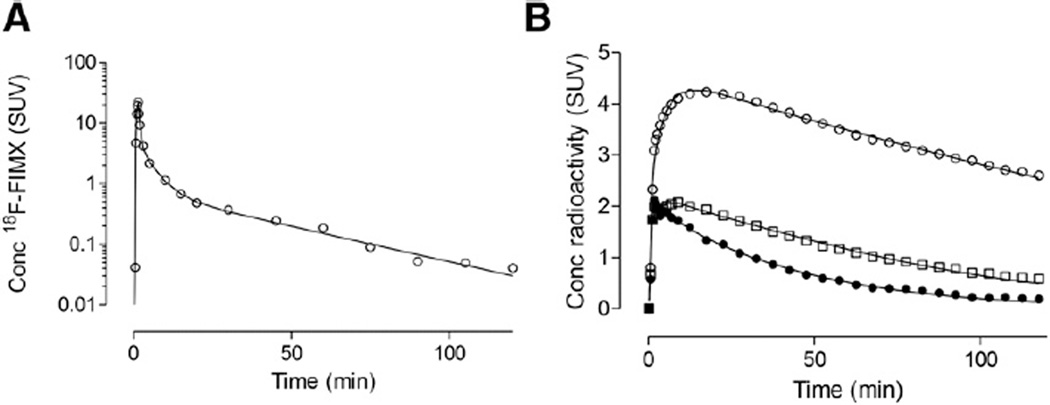
Unchanged 18F-FIMX in the arterial plasma peaked at 25 ± 4 SUV about 1.5 min after injection and rapidly declined thereafter (Fig. 1A). Parent radioligand represented 44% ± 7% of radioactivity in plasma at 10 min and 20% ± 6% at 120 min. For the subjects who underwent a longer scan, the parent fraction declined further to reach 11% ± 6% at 210 min. The parent concentration in some blood samples from those who underwent the longer scans (i.e., after 120 min) was higher than that of the preceding sample, suggesting that the parent measurement at later times may be inaccurate. At least 5 radiometabolites were observed (Supplemental Fig. 1), although all appeared less lipophilic than the parent. The fP was 0.34% ± 0.06% (mean ± SD from 12 subjects).
FIGURE 1.
(A) Metabolite-corrected arterial input function acquired over 120 min in a representative subject, fitted with a triexponential function. (B) Time–activity curves of brain regions of a subject fitted with 2-tissue-compartment model. Cerebellum (○) had highest uptake, thalamus (□) had intermediate uptake, and caudate nucleus (●) had lowest uptake.
Brain Images
Brain uptake peaked about 10 min after injection and then rapidly decreased (Fig. 1B). The cerebellum generally peaked later, at about 20 min. Nevertheless, the washout slope from the cerebellum was similar to that from the other regions. Consistent with the known distribution of mGluR1 in the mammalian brain (23,24), a high uptake was observed in the cerebellum (SUVpeak 4–6), whereas the remaining regions showed intermediate (thalamus) or low concentrations; SUVpeak was typically between 2 and 4.
Compartmental Analyses
The brain time–activity curves were better fit by an unconstrained 2-tissue-compartment model than by a 1-tissue-compartment model. The 2-tissue-compartment model had lower mean Akaike information criterion (370) and higher mean model selection criterion (5.1) scores than those for the 1-tissue-compartment model (Akaike information criterion = 431; model selection criterion = 3.2). The superiority of the 2-tissue-compartment model was confirmed by F statistics for all subjects. Moreover, spectral analysis showed at least 2 equilibrating components in each region for all subjects.
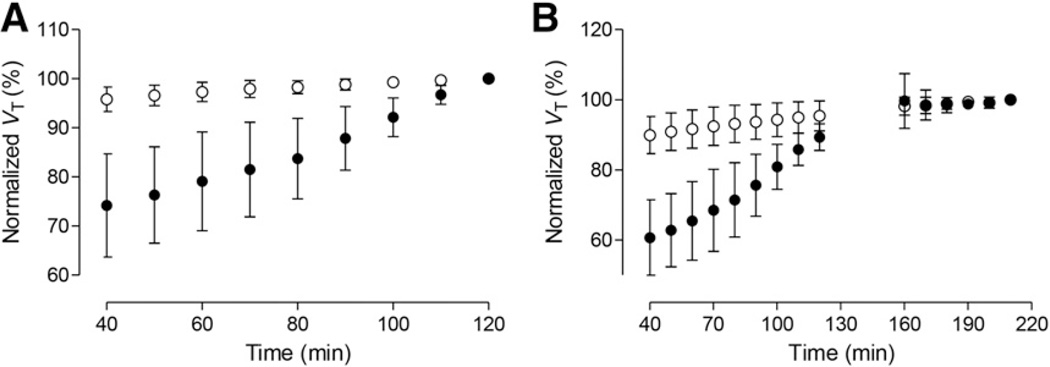
VT values were stable within the 120-min scan in all regions except the cerebellum (Fig. 2A). For noncerebellar regions, the VT value at 120 min was about 95% of the value at 210 min (Fig. 2B). In contrast, the cerebellum became relatively stable only later; its VT values increased slowly (by <5%) only from 170 to 210 min. Two subjects displayed some aberrant VT values after 120 min (the VT values were higher at 160–180 min than at 210 min; removing these 2 subjects from the time–stability analysis did not significantly affect the results).
FIGURE 2.
Time–stability analysis. VT results are normalized to 2-tissue-compartment model/VT values measured at 120 min (n = 12) (A) and 210 min (n = 6) (B). ● = cerebellum and ○ = average of other brain regions. Although good stability was reached by 120 min in low-uptake regions of brain, cerebellum still showed upward trend at 120 min and eventually almost stabilized by 170 min. Higher SD bars after 160 min denote presence of subjects with aberrant VT values.
Although the microparameters (K1 – k4) had only fair identifiability (Supplemental Table 1), the macroparameter VT was well identified: the SE for the cerebellum was 3.83% and that of the other brain regions was 0.81% on average. Regional VT values (mL·cm−3) had about a 7-fold range from 1.48 ± 0.17 in the caudate to 11.0 ± 1.9 in the cerebellum (Supplemental Table 2).
The free fraction fP for 18F-FIMX in plasma was low (0.34% ± 0.06%), and random errors in its measurement may have increased the coefficient of variation (mean/SD × 100%) of VT/fP (23%) compared with VT (13%) in 12 subjects.
Noncompartmental Analyses
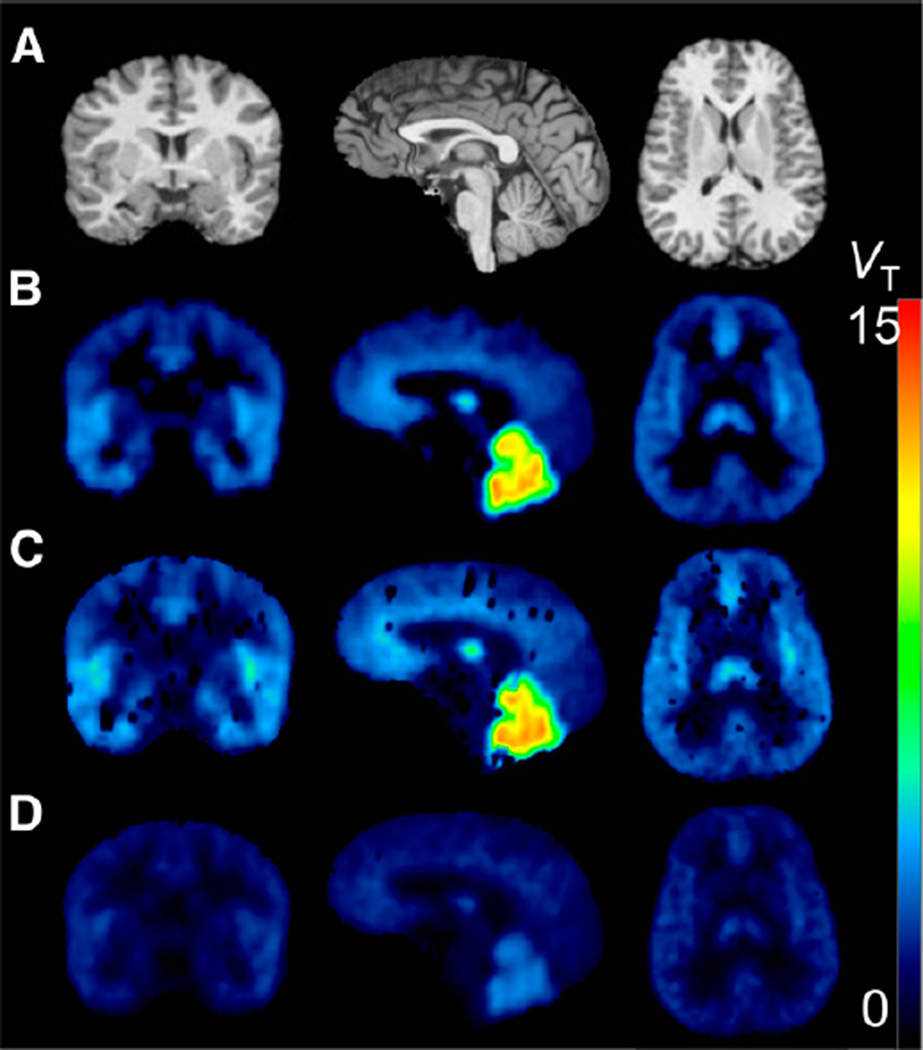
All alternative techniques were consistent with a 2-tissue-compartment model at the region level. At the voxel level, the 2 SA techniques yielded poor results (Fig. 3; supplemental data).
FIGURE 3.
Parametric images in a representative subject. (A) MRI. (B) Logan. (C) Standard SA. (D) RSSA. Images obtained with Ichise’s multi-linear analysis 1 (MA1) and Zhou’s RE-GP are visually indistinguishable from Logan images and are not shown. Parametric maps obtained with SA (C) contained many failed voxels randomly scattered across the whole brain. RSSA (D) yielded maps of good quality, but VT values were severely underestimated compared with 2-tissue-compartment model.
Bloodless Quantification
We found that either uptake in each brain region from 50 to 80 min (SUV50–80) or ratio in each brain region from 50 to 80 min (SUVR50–80) might be used as a bloodless substitute for VT (supplemental data).
Estimating VND Using mGluR1 Gene Transcript
To estimate VND from regional densities of mGluR1 gene transcripts, we sequentially performed 4 analyses: autocorrelation to determine which of the 3 published transcripts was appropriate to use, determination of whether relative gene transcript was proportional to VT, estimation of VND from this correlation, and measurement of VND in monkey brain after complete receptor blockade.
Autocorrelation
The values of each donor tightly correlated with those of any other donor for all 3 transcripts available in the Allen Brain Atlas (Pearson r value ranged from 0.765 to 0.963, all P < 0.0001). The mRNA expression values were then averaged among the subjects to obtain the final values for each probe. The autocorrelation between the probes showed a high correlation between probes A and B (r = 0.897, P = 0.0004), whereas the correlation of probe C to the other 2 was weaker, although still significant (r = 0.667, P = 0.035, and r = 0.688, P = 0.028, respectively). Because probe B showed the best correlation with the other 2 probes, it was chosen to perform cross-correlations with PET VT values. The between-subject autocorrelation for the 2-tissue-compartment model/VT showed high coefficients (r > 0.99), and the VT values of the 12 subjects were therefore averaged to obtain representative 18F-FIMX VT values.
Correlation of Gene Transcript and VT
The mRNA expression values were highly correlated to the mean VT values (r = 0.965, P < 0.0001). Notably, this high correlation was predominantly driven by the cerebellum, which is characterized by high mRNA expression and high PET binding. However, the correlation was highly significant even after the cerebellum was excluded from the analysis (r = 0.823, P = 0.0035). These results suggest that the relative density of gene transcripts can be used as a substitute for VS in the typical Lassen plot.
Estimating VND in Human Brain
Following the assumptions of the Lassen plot, the VND of 18F-FIMX was estimated as the x-intercept of the plot of mGluR1 transcript density versus VT across brain regions (Fig. 4). That is, when specific binding is absent (x-intercept), the value of VT equals VND. By this calculation, VND was only 0.5 mL·cm−3 (Fig. 4), suggesting that most of the total uptake in the brain was specific. For example, in the highest-density region (cerebellum), VT at 210 min was 13.4 mL·cm−3 and VS was thus 12.9 mL·cm−3 (13.4 − 0.5). In the lowest-density region (caudate), VT was about 1.5 mL·cm−3 and VS was thus 1 mL·cm−3 (1.5 − 0.5).
FIGURE 4.
Linear regression analysis between mRNA expression values measured by mRNA probe B for mGluR1 gene and average 2-tissue-compartment model/VT values of 18F-FIMX. VT of cerebellum, right-most data point, was calculated from 210 min of image acquisition. Linear regression shown in this graph, which includes cerebellum, has R2 of 0.992 and x-intercept of 0.5 mL·cm−3, which corresponds to VND of 18F-FIMX. When cerebellum is removed, R2 becomes 0.648 and x-intercept decreases to 0.3 mL·cm−3.
VND/fP was similar in human and nonhuman primates (~150 and 170 mL·cm−3, respectively) (supplemental data).
Whole-Body Biodistribution and Dosimetry
The effective dose was 23.4 µSv/MBq on average (supplemental data).
DISCUSSION
This study found that 18F-FIMX is an excellent PET radioligand for imaging and quantifying mGluR1 in the human brain. The distribution of its relatively high brain uptake reflected that of mGluR1. Uptake was quantified as VT with excellent identifiability using a 2-tissue-compartment model and several other noncompartmental methods. VT was stable after 120 min of scanning in all regions except the cerebellum (which required 170 min), and this time stability suggests that radiometabolites did not accumulate in the brain. We also report here a novel approach for estimating VND in the human brain using the relative density of the target gene transcript. This value of VND in humans was estimated by modifying the Lassen plot and confirmed by a receptor blocking study in monkey. Finally, whole-body scanning showed that radiation exposure from 18F-FIMX was similar to that from other 18F-labeled radioligands.
18F-FIMX appears to be superior to 11C-ITMM, the only other mGluR1 radioligand to provide a signal in the human brain (6). 11C-ITMM had an overall lower brain uptake (1–2.5 SUV) and a slower uptake and washout. In particular, the cerebellum was still in plateau 90 min after 11C-ITMM injection (6). Although time–stability and the identifiability of the parameters (especially VT and k4) were not reported in the original paper, it is possible that 11C-ITMM does not allow VT values to be reliably estimated within the feasible imaging time for 11C, whose half-life is 20 min. The high density of mGluR1 in the cerebellum may require a longer-lived radionuclide such as 18F; even then, as we found, 170 min were required to obtain time stable VT values.
Compartmental and Noncompartmental Analyses
18F-FIMX binding was better described by a 2- than a 1-tissue-compartment model, consistent with identification of 2 kinetically distinguishable pools of radioligand—namely, specifically bound and nondisplaceable. VT (mL·cm−3) values had a 7-fold range, from 1.5 in the caudate to 11 in the cerebellum (Supplemental Table 2). In contrast, the VT values of 11C-ITMM had only a 3-fold range (from 0.5 to 2.6 mL/cm3), perhaps reflecting an underestimation and lack of equilibrium in the cerebellum (6).
In theory, VT values should be corrected by the percentage of radioligand not bound to plasma proteins (VT/fP), because only this fraction is available for exchange with the tissues. However, fP measurement can be an important source of noise, especially when fP is very low. In this study, the fP value for 18F-FIMX was quite low (0.34% ± 0.06%), and imprecision in its measurement presumably caused a higher coefficient of variation for VT/fP (23%) than for VT (13.2%) (Supplemental Table 3). This higher variability would require larger sample sizes to detect statistically significant differences between groups.
All noncompartmental techniques provided good and substantially equivalent results at the region level (Supplemental Table 2). Quantification at the voxel level was, however, more challenging. The VT values obtained with Logan, MA1, and RE-GP were similar to those obtained by the 2-tissue-compartment model at the region level, with an average bias for all regions (excluding the cerebellum) ranging from −6.5% for Logan to −2.1% for RE-GP. Notably, MA1 is known to reduce noise-induced bias of parametric images (15) and, indeed, in the present study yielded a lower bias than Logan (−3.5% vs. −6.5%, P < 0.05).
Although SA/VT values also closely matched 2-tissue-compartment model/VT (−3% average bias, P > 0.05), SA parametric maps contained many outlying voxels randomly scattered across the whole brain, making them unsuitable for statistical parametric comparisons. This finding is not unusual when SA is applied at the voxel level (25). Finally, the maps obtained with RSSA severely underestimated actual VT values, and this underestimation was region-dependent, for example, greater in the cerebellum (−74%) than other regions (−33%).
Bloodless Methods
This study assessed 3 noninvasive (i.e., bloodless) methods of quantifying 18F-FIMX binding: a reference tissue model (i.e., SRTM), a ratio of brain radioactivity in 1 region compared with another region (i.e., SUVR), and merely the uptake in the region (i.e., SUV). When either the caudate or the cerebellum was used as the reference region, the binding potential relative to the nondisplaceable compartment of SRTM showed higher intersubject variability than VT (Supplemental Table 3), had poor time stability, and did not even converge in some regions with intermediate density of receptors. The poor performance of SRTM may be due to the fact that no true reference region (i.e., devoid of receptors) exists for mGluR1, and the regions used as reference have a VT value that is close to that of the other regions. For example, if our estimate of VND (0.5 mL·cm−3) is accurate, then two thirds of VT (1.5 mL·cm−3) in the caudate was actually VS (1.0 mL·cm−3). Whatever the cause of its poor performance, SRTM should not, in our opinion, be used to measure mGluR1 with 18F-FIMX.
In contrast to SRTM, SUVR50–80 (using the caudate as a reference region) showed the lowest variability of all techniques (9.9%). SUVR might thus be a practical alternative to full kinetic modeling. However, the use of SUVR in any particular disease will first require confirmation that the sizeable specific binding in caudate does not differ significantly between groups (e.g., patients and controls) or between conditions (e.g., baseline and receptor blocked).
The SUV50–80 was well correlated with VT (Supplemental Fig. 2). SUV50–80 is arguably the simplest bloodless method and does not require a reference region or demonstration that the reference region is equivalent between groups.
mGluR1 Gene Transcript to Estimate Specific and VND of 18F-FIMX
To estimate the specific and nondisplaceable components of 18F-FIMX, we used a version of the Lassen plot, which measures the linear relationship of specific binding as a function of total uptake in the brain. As an overview, the y-axis of the Lassen plot is VS or a variable proportional to VS, and the x-axis is VT or a variable proportional to VT. In its simplest form (i.e., VS vs. VT), when VS = 0, the x-intercept = VND. That is, when there is no VS (e.g., in a receptor-free region or after complete receptor blockade), the measured value of VT will equal VND. As mentioned, proportional variables can be substituted for VS or for VT. For example, Owen et al. (26) showed that VS can be substituted with 2 variables: VT baseline minus VT after partial blockade in numerous brain regions, as the resulting variable is proportional to VS and the percentage occupancy is the same in all regions when the radioligand occupies a tracer percentage (<5%–10%) of receptors; and for translocator protein radioligands, VT (high-affinity binders) minus VT (mixed-affinity binders) is also proportional to VS and similar across brain regions, as the genetic effect is uniform in brain. In this paper, we introduced the use of transcript density as a substitute for VS.
This technique has potentially broad applicability, as the Allen Brain Atlas has regional densities for about 30,000 human brain transcripts. However, this technique is applicable only if gene transcript density is linearly proportional to protein density—that is, when the target protein is not subject to significant posttranscriptional changes in vivo. Although this condition was met for 18F-FIMX, this may not always be the case (10). A more extensive validation of the robustness of our modified Lassen plot, in particular by taking human blocked studies as a reference, is under way. By linearly regressing the regional density of mGluR1 gene transcript and VT, we estimated VND (i.e., the x-intercept) to be only 0.5 mL·cm−3. Parametric VT images showed that no region had a consistent number of voxels whose value was about 0.5 mL·cm−3. Likely, all brain regions contain a considerable percentage of specific binding and none would be a pure reference region (according to the Allen Atlas, even white matter expresses a certain amount of mRNA transcript). The accuracy of our VND estimate was tested against the measured VND in a nonhuman primate, as the amount of nonspecific binding in the brain is considered to be constant across species (27). Our results showed that VND/fP was similar in human and nonhuman primates (about 150 and 170 mL·cm−3, respectively). Notably, when the cerebellum was removed from the analysis and the linear regression was performed only with the remaining regions, the x-intercept decreased from 0.5 to 0.3 mL·cm−3. This suggests a potential limitation of our modified Lassen plot: when the regional VT values of a radioligand are similar, the VND obtained can be sensitive to the noise of linear fitting. Therefore, the Lassen plot should preferably be used with radioligands with uneven distributions in the brain.
Dosimetry
According to the pathway we previously proposed for evaluating novel first-in-human 18F-labeled radioligands (28), we began the study by obtaining a whole-body scan with a low injected activity (74 MBq) in a healthy volunteer. The aim of this first scan was to verify that the radioligand did not abnormally accumulate in a radiosensitive organ, but it was widely distributed in the body, so that higher activities could be injected for brain imaging. We then proceeded to perform brain imaging by injecting higher activities (~200 MBq) and finally completed the dosimetry part of the study by imaging 3 more subjects with a whole-body scan. The 18F-FIMX effective dose (23.4 µSv/MBq) was similar to the average value of other 18F-labeled ligands (28). It could be argued that if a new radioligand shows a typical widespread distribution in the body, the final effective dose is likely to approach the typical average value (28). In this case, dosimetry studies might be avoided, and the average value might be used for radioprotection purposes in subsequent clinical studies.
CONCLUSION
18F-FIMX is a new PET radioligand with excellent properties to quantify mGluR1 in the human brain. 18F-FIMX binding was well quantified with both compartmental and noncompartmental methods using a 120-min scan. The high density of mGluR1 in the cerebellum was also accurately quantified, although at least 170 min was required. Regional VT values of 18F-FIMX were strongly correlated with mGluR1 transcript density, showing that 18F-FIMX uptake was consistent with the expected distribution of mGluR1. Finally, we introduced a modified Lassen plot that uses publicly available data on the regional densities of protein transcripts to estimate specific receptor binding of a radioligand, thereby avoiding the need for receptor blocking studies.
Supplementary Material
Acknowledgments
Ioline Henter provided excellent editorial assistance. Alicia Woock and Kimberly Jenko provided excellent laboratory assistance for the measurement of the arterial input functions.
The costs of publication of this article were defrayed in part by the payment of page charges. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734. This work was supported by the Intramural Research Program of the National Institute of Mental Health (project no. ZIAMH002852, under clinicaltrials.gov identifier NCT02230592), National Institutes of Health (IRP-NIMH-NIH).
Footnotes
DISCLOSURE
No other potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Pike VW. PET radiotracers: crossing the blood-brain barrier and surviving metabolism. Trends Pharmacol Sci. 2009;30:431–440. doi: 10.1016/j.tips.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, Huang Y. In vivo imaging of the metabotropic glutamate receptor 1 (mGluR1) with positron emission tomography: recent advance and perspective. Curr Med Chem. 2014;21:113–123. doi: 10.2174/09298673113209990217. [DOI] [PubMed] [Google Scholar]
- 3.Zanotti-Fregonara P, Barth VN, Liow JS, et al. Evaluation in vitro and in animals of a new 11C-labeled PET radioligand for metabotropic glutamate receptors 1 in brain. Eur J Nucl Med Mol Imaging. 2013;40:245–253. doi: 10.1007/s00259-012-2269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanotti-Fregonara P, Barth VN, Zoghbi SS, et al. 11C-LY2428703, a positron emission tomographic radioligand for the metabotropic glutamate receptor 1, is unsuitable for imaging in monkey and human brains. EJNMMI Res. 2013;3:47. doi: 10.1186/2191-219X-3-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujinaga M, Yamasaki T, Yui J, et al. Synthesis and evaluation of novel radioligands for positron emission tomography imaging of metabotropic glutamate receptor subtype 1 (mglur1) in rodent brain. J Med Chem. 2012;55:2342–2352. doi: 10.1021/jm201590g. [DOI] [PubMed] [Google Scholar]
- 6.Toyohara J, Sakata M, Oda K, et al. Initial human PET studies of metabotropic glutamate receptor type 1 ligand 11C-ITMM. J Nucl Med. 2013;54:1302–1307. doi: 10.2967/jnumed.113.119891. [DOI] [PubMed] [Google Scholar]
- 7.Satoh A, Nagatomi Y, Hirata Y, et al. Discovery and in vitro and in vivo profiles of 4-fluoro-N-[4-[6-(isopropylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl]-N-methylbenzam ide as novel class of an orally active metabotropic glutamate receptor 1 (mGluR1) antagonist. Bioorg Med Chem Lett. 2009;19:5464–5468. doi: 10.1016/j.bmcl.2009.07.097. [DOI] [PubMed] [Google Scholar]
- 8.Xu R, Zanotti-Fregonara P, Zoghbi SS, et al. Synthesis and evaluation in monkey of [18F]4-fluoro-N-methyl-N-(4-(6-(methylamino)pyrimidin-4-yl)thiazol-2-yl) benzami de ([18F]FIMX): a promising radioligand for PET imaging of brain metabotropic glutamate receptor 1 (mGluR1) J Med Chem. 2013;56:9146–9155. doi: 10.1021/jm4012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham VJ, Rabiner EA, Slifstein M, Laruelle M, Gunn RN. Measuring drug occupancy in the absence of a reference region: the Lassen plot re-visited. J Cereb Blood Flow Metab. 2010;30:46–50. doi: 10.1038/jcbfm.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzo G, Veronese M, Heckemann RA, et al. The predictive power of brain mRNA mappings for in vivo protein density: a positron emission tomography correlation study. J Cereb Blood Flow Metab. 2014;34:827–835. doi: 10.1038/jcbfm.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoghbi SS, Shetty HU, Ichise M, et al. PET imaging of the dopamine transporter with 18F-FECNT: a polar radiometabolite confounds brain radioligand measurements. J Nucl Med. 2006;47:520–527. [PubMed] [Google Scholar]
- 13.Gandelman MS, Baldwin RM, Zoghbi SS, Zea-Ponce Y, Innis RB. Evaluation of ultrafiltration for the free fraction determination of single photon emission computed tomography (SPECT) tracers: β-CIT, IBF, and iomazenil. J Pharm Sci. 1994;83:1014–1019. doi: 10.1002/jps.2600830718. [DOI] [PubMed] [Google Scholar]
- 14.Logan J, Fowler JS, Volkow ND, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(−) cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10:740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- 15.Ichise M, Toyama H, Innis RB, Carson RE. Strategies to improve neuroreceptor parameter estimation by linear regression analysis. J Cereb Blood Flow Metab. 2002;22:1271–1281. doi: 10.1097/01.WCB.0000038000.34930.4E. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Ye W, Brasic JR, Wong DF. Multi-graphical analysis of dynamic PET. Neuroimage. 2010;49:2947–2957. doi: 10.1016/j.neuroimage.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham VJ, Jones T. Spectral-analysis of dynamic PET studies. J Cereb Blood Flow Metab. 1993;13:15–23. doi: 10.1038/jcbfm.1993.5. [DOI] [PubMed] [Google Scholar]
- 18.Turkheimer FE, Hinz R, Gunn RN, Aston JAD, Gunn SR, Cunningham VJ. Rank-shaping regularization of exponential spectral analysis for application to functional parametric mapping. Phys Med Biol. 2003;48:3819–3841. doi: 10.1088/0031-9155/48/23/002. [DOI] [PubMed] [Google Scholar]
- 19.Lassen NA, Bartenstein PA, Lammertsma AA, et al. Benzodiazepine receptor quantification in vivo in humans using [11C]flumazenil and PET: application of the steady-state principle. J Cereb Blood Flow Metab. 1995;15:152–165. doi: 10.1038/jcbfm.1995.17. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins RA, Phelps ME, Huang S-C. Effects of temporal sampling, glucose metabolic rates, and disruptions of the blood-brain barrier on the FDG model with and without a vascular compartment: studies in human brain tumors with PET. J Cereb Blood Flow Metab. 1986;6:170–183. doi: 10.1038/jcbfm.1986.30. [DOI] [PubMed] [Google Scholar]
- 21.Carson RE. Parameter estimation in positron emission tomography. In: Phelps ME, Mazziotta JC, Schelbert HR, editors. Positron Emission Tomography and Autoradiography: Principles and Applications for the Brain and Heart. New York, NY: Raven Press; 1986. pp. 347–390. [Google Scholar]
- 22.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. New York, NY: McGraw-Hill; 2003. [Google Scholar]
- 23.Spooren W, Ballard T, Gasparini F, Amalric M, Mutel V, Schreiber R. Insight into the function of group I and group II metabotropic glutamate (mGlu) receptors: behavioural characterization and implications for the treatment of CNS disorders. Behav Pharmacol. 2003;14:257–277. doi: 10.1097/01.fbp.0000081783.35927.8f. [DOI] [PubMed] [Google Scholar]
- 24.Stephan D, Bon C, Holzwarth JA, Galvan M, Pruss RM. Human metabotropic glutamate receptor 1: mRNA distribution, chromosome localization and functional expression of two splice variants. Neuropharmacology. 1996;35:1649–1660. doi: 10.1016/s0028-3908(96)00108-6. [DOI] [PubMed] [Google Scholar]
- 25.Rizzo G, Veronese M, Zanotti-Fregonara P, Bertoldo A. Voxelwise quantification of [C](R)-rolipram PET data: a comparison between model-based and data-driven methods. J Cereb Blood Flow Metab. 2013;33:1032–1040. doi: 10.1038/jcbfm.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen DR, Guo Q, Kalk NJ, et al. Determination of [11C]PBR28 binding potential in vivo: a first human TSPO blocking study. J Cereb Blood Flow Metab. 2014;34:989–994. doi: 10.1038/jcbfm.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di L, Umland JP, Chang G, et al. Species independence in brain tissue binding using brain homogenates. Drug Metab Dispos. 2011;39:1270–1277. doi: 10.1124/dmd.111.038778. [DOI] [PubMed] [Google Scholar]
- 28.Zanotti-Fregonara P, Lammertsma AA, Innis RB. Suggested pathway to assess radiation safety of 18F-labeled PET tracers for first-in-human studies. Eur J Nucl Med Mol Imaging. 2013;40:1781–1783. doi: 10.1007/s00259-013-2512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.