Significance
Chronic lymphocytic leukemia (CLL) is the most common human leukemia. We identified two members of the tRNA-derived small RNA (tsRNA) family, ts-3676 and ts-4521, both inactivated in CLL and lung cancer. We further analyzed expression of 120 tsRNAs and found that tsRNAs are dysregulated in CLL and lung cancer. Thus this study uncovers the involvement of this recently identified class of small, non-coding RNAs in hematopoietic malignancies and solid tumors.
Keywords: tsRNAs, ts-4521, ts-3676
Abstract
Chronic lymphocytic leukemia (CLL) is the most common human leukemia, and transgenic mouse studies indicate that activation of the T-cell leukemia/lymphoma 1 (TCL1) oncogene is a contributing event in the pathogenesis of the aggressive form of this disease. While studying the regulation of TCL1 expression, we identified the microRNA cluster miR-4521/3676 and discovered that these two microRNAs are associated with tRNA sequences and that this region can produce two small RNAs, members of a recently identified class of small noncoding RNAs, tRNA-derived small RNAs (tsRNAs). We further proved that miR-3676 and miR-4521 are tsRNAs using Northern blot analysis. We found that, like ts-3676, ts-4521 is down-regulated and mutated in CLL. Analysis of lung cancer samples revealed that both ts-3676 and ts-4521 are down-regulated and mutated in patient tumor samples. Because tsRNAs are similar in nature to piRNAs [P-element–induced wimpy testis (Piwi)-interacting small RNAs], we investigated whether ts-3676 and ts-4521 can interact with Piwi proteins and found these two tsRNAs in complexes containing Piwi-like protein 2 (PIWIL2). To determine whether other tsRNAs are involved in cancer, we generated a custom microarray chip containing 120 tsRNAs 16 bp or more in size. Microarray hybridization experiments revealed tsRNA signatures in CLL and lung cancer, indicating that, like microRNAs, tsRNAs may have an oncogenic and/or tumor-suppressor function in hematopoietic malignancies and solid tumors. Thus, our results show that tsRNAs are dysregulated in human cancer.
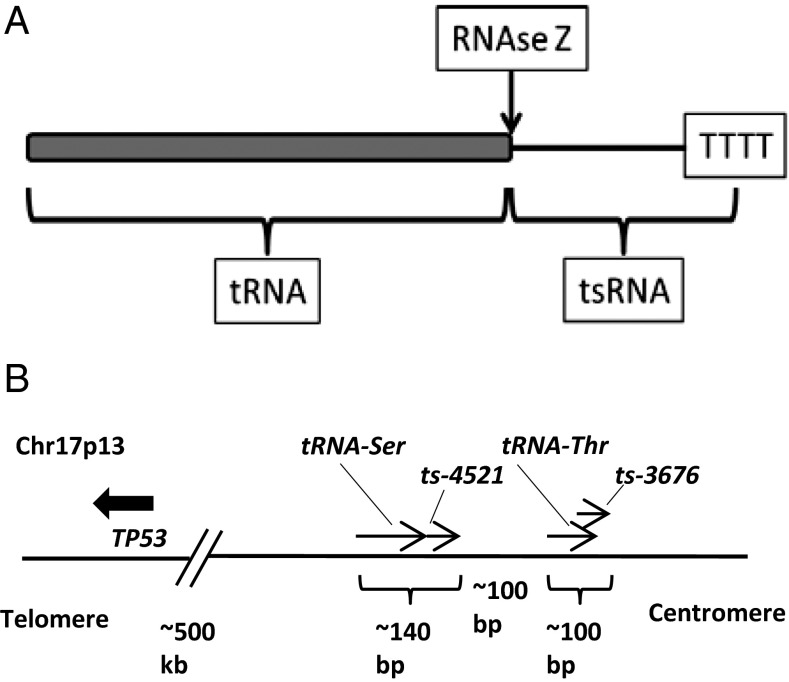
B-cell chronic lymphocytic leukemia (CLL) is the most common human leukemia, occurring as indolent or aggressive disease (1). Aggressive CLLs usually show high levels of Zeta-chain–associated protein kinase 70 (ZAP-70) and unmutated Ig heavy-chain variable region genes (IgVH), whereas indolent CLLs display low ZAP-70 expression and mutated IgVH genes (2). We have shown that deletions and mutations in microRNA 15/16 (miR-15/16) cause the indolent form of CLL (3, 4). To our knowledge, this was the first demonstration that alterations in noncoding genes can cause disease, including cancer (3, 4). MicroRNAs have a very significant role in CLL. Although loss of miR-15/16 leads to the development of the indolent form of CLL (3–5), loss of microRNAs targeting the T-cell leukemia/lymphoma 1 (TCL1) oncogene leads to the aggressive form (6, 7). We also have shown that transgenic mice overexpressing TCL1 in B cells develop the aggressive form of CLL (8, 9). Thus, expression of microRNAs can also serve as markers for CLL onset and progression (5, 10). In addition to microRNAs, another class of small RNAs, P-element–induced wimpy testis (Piwi)-interacting small RNAs (piRNAs), has been studied recently (11–13). Although miRNAs are created through a double-stranded precursor molecule (12), piRNAs biogenesis involves long single-stranded precursor molecules (11). PiRNAs interact with Piwi proteins and appear to cooperate with DNA methyltransferases to affect methylation of genes, suggesting that Piwi/piRNAs complexes are global regulators of chromatin state (14, 15). A few years ago, a new class of small RNAs was reported to derive from tRNAs as a consequence of tRNA processing (16). tRNAs are transcribed from individual tRNA genes by RNA polymerase III. pre-tRNA transcripts are processed by several endonucleases producing mature tRNAs and additional small RNA molecules. The molecules produced from the 3′ end of pre-tRNAs by endonuclease RNase Z represent unique sequences starting exactly at the 3′ ends of tRNAs and ending at the sequence of four consecutive T nucleotides, a stop signal for RNA polymerase III (pol III) (12, 17, 18). In this report we define unique molecules derived from the 3′ ends of pre-tRNAs as tRNA-derived small RNAs (tsRNAs, also known as “3′tRFs”) (Fig. 1A).
Fig. 1.
Ts-3676 and ts-4521 at 17p13. (A) TsRNAs are produced from the 3′ ends of pre-tRNAs. (B) Genomic structure of ts-3676 and ts-4521.
While studying the miR-4521/3676 cluster in CLL, we discovered that these two microRNAs are associated with tRNA sequences (Fig. 1B). We identified miR-3676 as a powerful regulator of TCL1 expression, an important oncogene involved in the development of the aggressive CLLs, and demonstrated that miR-3676 targets three consecutive 28-bp repeats within the 3′ UTR of TCL1 (6). Because mature miR-3676 starts exactly at the end of tRNA-Thr (6) and ends at a transcription termination stop for RNA polymerase III, we could not exclude the possibility that miR-3676 represents a member of a recently identified class of small RNAs, tsRNAs, generated during tRNA processing (17, 19). Here we report that miR-3676 and miR-4521 are indeed tsRNAs and that tsRNAs are involved in cancer pathogenesis.
Results
Short RNAs 3676 and 4521 Are tsRNAs Interacting with Piwi Proteins.
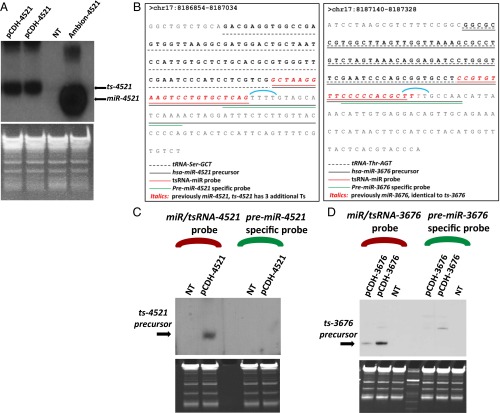
Because short RNAs 3676 and 4521 are associated with tRNA sequences (Fig. 1B), we investigated whether the region containing miR-4521 and miR-3676 expresses tsRNAs or microRNAs. We performed a Northern blot experiment using HEK-293 cells transfected with vectors containing the genomic segment including tRNA-Ser and miR-4521. In the experiment to detect ts-4521, we observed that the band of the exogenous miR-4521 was higher than the band of the miR-4521 commercially purchased from Ambion (Fig. 2A). The difference is compatible with the expected size of a small RNA starting exactly at the 3′ end of the tRNA and ending at the stretch of four Ts located exactly after the predicted sequence of the microRNA (Fig. 2B). Indeed, the stop signal for the RNA pol III is represented by a stretch of four Ts (20). Therefore, the four Ts located immediately after the end of the predicted miR-4521 also represent the end of ts-4521. We then were able to design a set of probes to detect a difference in the hybridization pattern of the ts-4521 precursor molecule and the miR-4521 precursor molecule. Thus, we designed two probes, one complementary to the mature sequence of miR-4521, which is shared by pre–miR-4521 and pre–ts-4521 (shown in red in Fig. 2B), and another starting four bases before within the stretch of four Ts and complementary only to pre–miR-4521 (shown in green in Fig. 2B). As expected, the first probe detected the miR-4521/ts-4521 signal. The second probe failed to produce a signal (Fig. 2C), confirming that this region expresses a tsRNA. This result indicates that the transcription of pre–miR-4521 ends at the four T nucleotides (shown in blue in Fig. 2B), that the last 33 bases of pre–miR-4521 are not transcribed, and therefore miR-4521 cannot be produced, suggesting that miR-4521 is a tsRNA. In the experiment to detect ts-3676, the band of the exogenous miR-3676 is very similar in size to the band of commercially bought miR-3676 (6). In this case, the expected size of a small RNA starting exactly at 3′ end of the tRNA is very similar to the predicted sequence of the microRNA because the stretch of four Ts exactly matches the end of the microRNA sequence (Fig. 2B). Therefore, it was not possible to determine a difference in a hybridization pattern as easily as for ts-4521. Nevertheless, we were able to design two 18-bp probes to detect a difference in the hybridization pattern between the predicted precursor of the ts-3676 and the predicted precursor of the corresponding microRNA (as we did with ts-4521). In the first probe (the tsRNA/miR-probe), all 18 bases covered the sequences shared between pre-microRNA and pre-tsRNA and included the whole mature sequence (which is the same for miR-3676 and ts-3676). In the second probe (the miR-probe) only 12 of the 18 bases covered the region shared by the microRNA and the tsRNA precursors (represented by a part of the mature product); the remaining 6 bases are complementary only to the microRNA precursor (Fig. 2B). We expected no signal (or a much weaker signal) when using the second probe if this region expressed a tsRNA. As shown in Fig. 2D, we obtained no signal from the probe designed to detect the microRNA precursor, but the probe designed on shared regions did show a signal, confirming that this region produces a tsRNA. These data suggest that both miR-3676 and miR-4521 are indeed tsRNAs.
Fig. 2.
The mir-3676/4521 locus produces tsRNAs. (A) Northern blot analysis of endogenous ts-4521 and commercially purchased miR-4521 (Ambion-4521). (B) Sequence of ts-4521 and ts-3676 and the probes used to prove that both small these RNAs are tsRNAs. (C and D) Northern blots showing that small RNAs 4521 (C) and 3676 (D) are tsRNAs. HEK-293 cells were transfected with the indicated constructs. NT, not transfected.
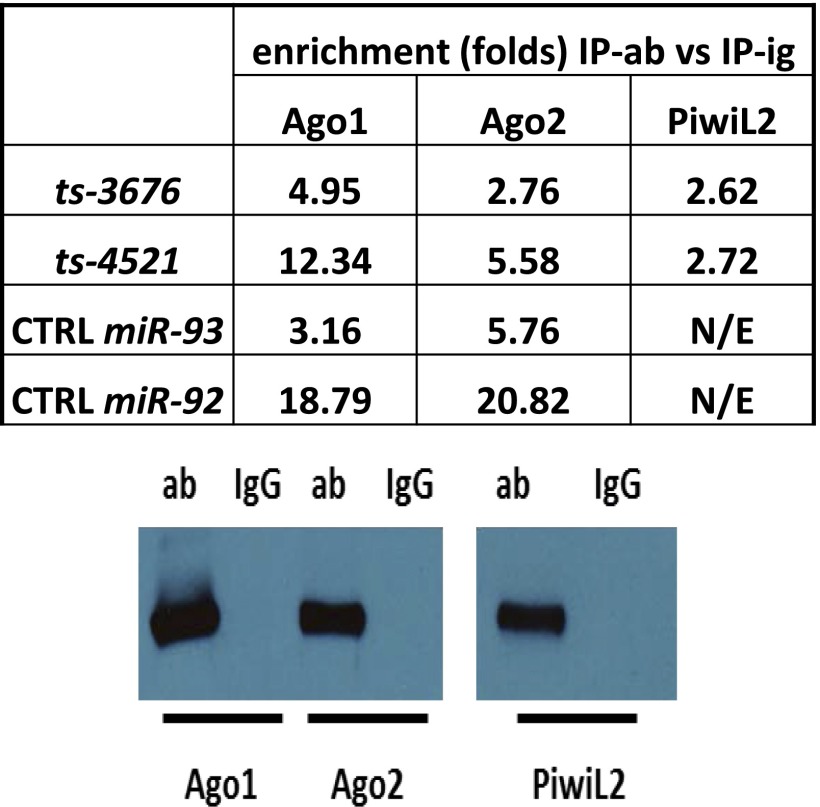
Because we previously demonstrated that ts-3676 can function as a microRNA by targeting the 3′ UTR of TCL1 (6), and tsRNAs are physically similar to piRNAs (both being single-stranded short RNAs containing no stem loop structures), we decided to investigate if the tsRNAs derived from this region can also function as piRNAs. Therefore, we performed an RNA immunoprecipitation experiment (RIP) and found an enrichment of ts-3676 and ts-4521 in the complexes containing tagged Piwi-like protein 2 (PiwiL2) (2.6- to 2.7-fold vs. controls), compared with the load of control microRNAs (mir-92 and miR-93) (Fig. 3). In this experiment we also observed that protein complexes containing Argonaut proteins Ago1 and Ago2 are loaded with control microRNAs (miR-92 and miR-93, enriched 3- to 20-fold vs. controls), but complexes containing PiwiL2 are not, as expected for microRNAs. Instead, ts-3676 and ts-4521 were present in complexes containing Ago1 and Ago2 (enriched 5- to 12-fold vs. controls) and also in complexes containing PiwiL2 (enriched 2.6- to 2.7-fold vs. controls) (Fig. 3). This finding indicates that the miR-4521/3676 locus produces small RNAs, ts-3676 and ts-4521, that can act not only as microRNAs but also as piRNAs by interacting with PiwiL2.
Fig. 3.
ts-3676 and ts-4521 interact with PiwiL2. (Upper) Enrichment of indicated RNA molecules in the complexes of indicated proteins vs. IgG control. N/E, no enrichment. (Lower) Expression of Ago1, Ago2, and PiwiL2 in these experiments, determined by the Western blot using anti-Omni antibody.
Ts-3676 and ts-4521 Are Down-Regulated and Mutated in Cancer.
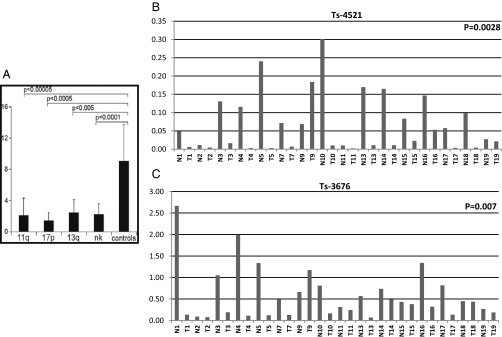
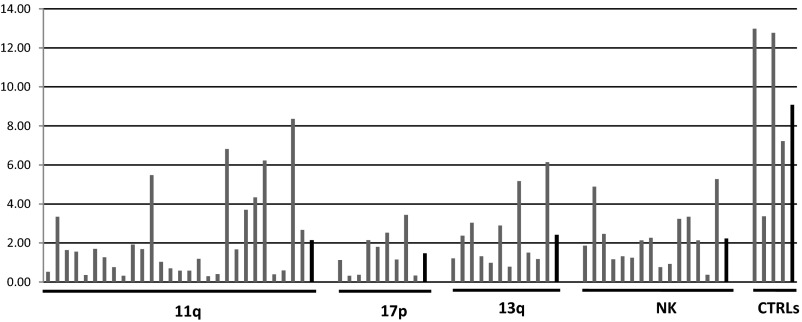
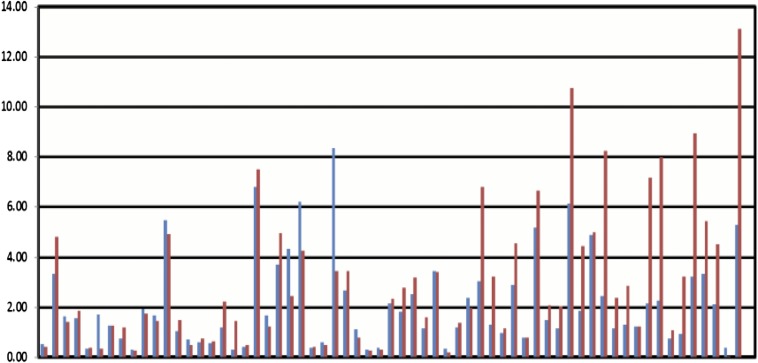
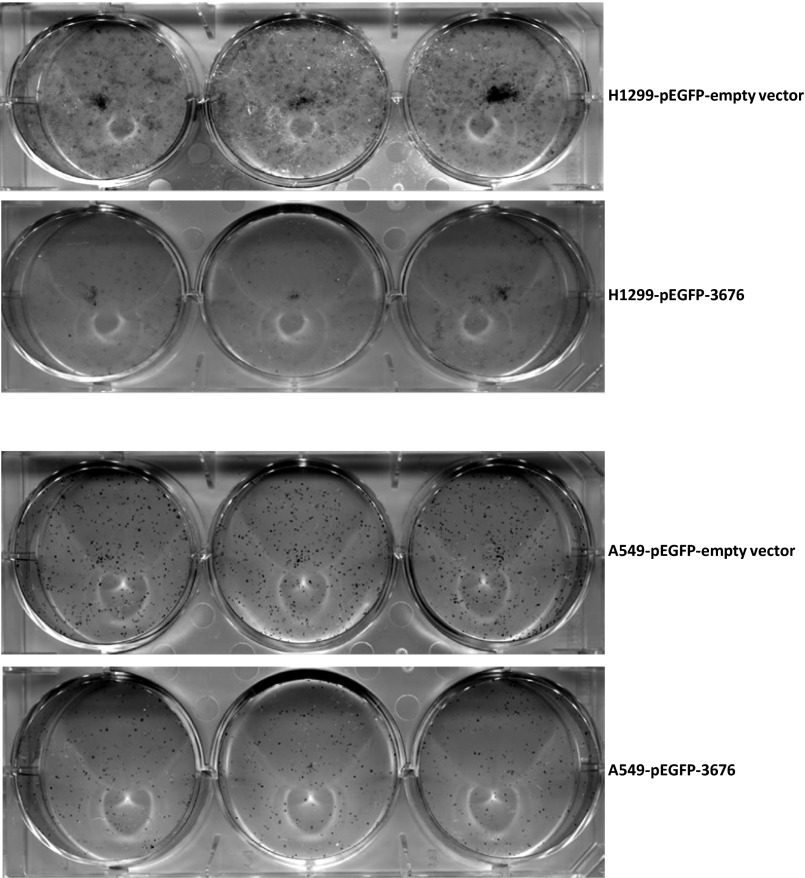
We previously reported that ts-3676 (then described as “miR-3676”) is down-regulated and mutated in CLL (6). We then studied the expression levels of miR-4521 in CLL compared with CD19+ B-cells isolated from peripheral blood mononuclear cells (PBMCs) and cord blood using a set of samples subgrouped by their karyotype [the same set was used to study miR-3676 (6)]. The set included 28 samples with 11q deletions, nine samples with 17p deletions, 11 samples with 13q deletions, and 15 samples with a normal karyotype for a total of 63 CLL samples (6). The real-time RT-PCR data are shown in Fig. 4A and Fig. S1. Ts-4521 expression was statistically significantly down-regulated in CLLs of all karyotypes (three- to eightfold compared with CD19+ B-cells in ∼80% of cases) (Fig. 4A). As we expected, the lowest expression levels of ts-4521 were observed in the 17p-deleted CLLs (these samples contain only one allele of ts-4521) (Fig. 4). Interestingly, ts-4521 showed an expression pattern almost identical to that of ts-3676 in CLL (Fig. S2). To investigate the status of ts-3676 and ts-4521 in lung cancer, we used 17 paired lung cancer samples and matched normal lung tissues. Using real-time RT-PCR, we determined that ts-3676 and ts-4521 are drastically down-regulated in lung cancer samples vs. matched normal lung tissues (Fig. 4 B and C). To determine if ts-3676 and ts-4521 are mutated in lung cancer, we sequenced 300 lung cancer samples. We found six different mutations in six lung cancer samples (∼2%) within in the ts-3676/ts-4521 locus. Three mutations found were germline (50% means that one allele was mutated and one allele was WT), two mutations were somatic [in one case, NYU-1008T, the mutation was identical to a known rare SNP (Rs 368503695) but was not found in PBMCs of the same patient], and in one case PBMCs were not available (Table S1). To determine if tsRNAs can function as tumor suppressors, we carried out a colony assay experiment in H1299 and A549 lung cancer cell lines transfected with ts-3676 or empty vector (Fig. S3). Expression of ts-3676 resulted in a several-fold decrease in colony formation in H1299 and A549 cells, indicating that ts-3676 can function as a tumor suppressor.
Fig. 4.
ts-3676 and ts-4521 are down-regulated in CLL and lung cancer. (A) Down-regulation of ts-4521 expression in four cytogenetic groups of CLL. (B and C) Expression of ts-4521 (B) and ts-3676 (C) in 17 paired lung cancer samples (T) and matched normal lung tissues (N).
Fig. S1.
Expression of ts-4521 in four different cytogenetic groups of CLL and CD19+ B-cell controls. Averages are shown as black bars.
Fig. S2.
ts-3676 (red bars) and ts-4521 (blue bars) show similar expression patterns in CLL.
Table S1.
ts-3676 and ts-4521 are mutated in lung cancer
| ID | Nucleotide change | Mutated allele, %* | Genomic coordinates† |
| NYU-998 | G→A | 50 | chr17:8186895–8186895 |
| 998 PBMCs | 50 | ||
| NYU-1008T | C→T | 50 | chr17:8187250–8187250 |
| 1008T PBMCs | Somatic | 0 | |
| NYU-335 | C→G | 50 | chr17:8090313–8090313 |
| 335-PBMCs | Somatic | 0 | |
| NYU-1179 | T→A | 25 | chr17:8090257–8090257 |
| 1179 PBMCs | 50 | ||
| NYU-303 | T→A | 70 | chr17:8187220–8187220 |
| 303 PBMCs | 50 | ||
| NYU-574 | A→C | 50 | chr17:8187220–8187220 |
| PBMCs were not available |
50% means that one allele is mutated and the other allele is WT.
Coordinates are shown using human GRCh38/hg38 assembly.
Fig. S3.
Colony assay. H1299 and A549 lung cancer cell lines were transfected with the expression vector expressing ts-3676 or empty vector. Five thousand transfected cells were plated in each well; 5 d later, colonies were colored with brilliant blue.
TsRNAs Are Dysregulated in Cancer.
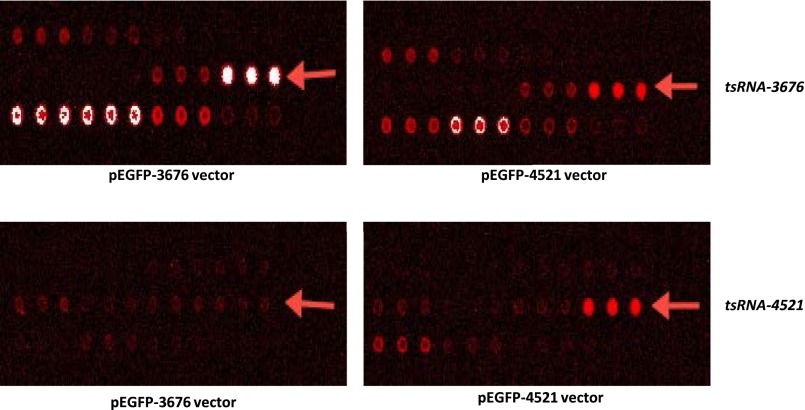
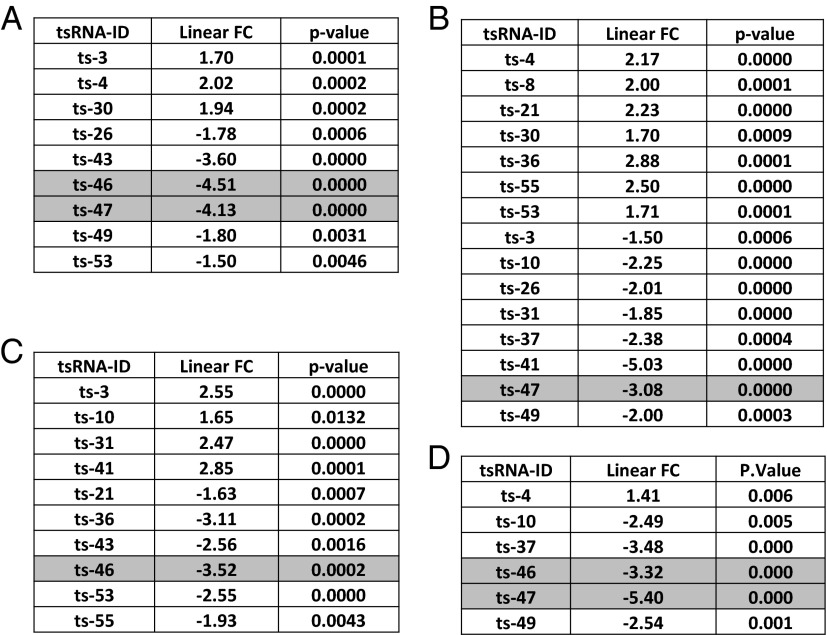
Because we determined that ts-3676 and ts-4521 are inactivated in samples from CLL and lung cancer, we investigated whether other tsRNAs can be up- or down-regulated in cancer. We first created a list of all tsRNAs by retrieving all tRNA sequences (including tRNAs themselves and following unique sequences ending with a four-T termination signal) and isolated unique sequences starting from the 3′ end of tRNAs and ending with the four-T termination signal. We selected only sequences of 16 bp or longer, because shorter sequences cannot easily be detected by microarray or real-time PCR (Table S2). We identified 120 potential tsRNA sequences and designed a microarray chip to study the expression of these tsRNAs in normal and tumor tissues. The chip was designed exactly as previously described for microRNAs (21). To confirm the specificity of our chip, we transfected HEK-293 cells with a vector containing tRNA-ser/ts-4521 or tRNA-Thr/ts-3676 (in pEGFP-N1). Fig. S4 shows the overexpression of ts-3676 and ts-4521 in corresponding hybridization spots of the chip. Thus these results suggest that this microarray chip displays good specificity and sensitivity to detect tsRNAs in human samples. To determine if tsRNAs are dysregulated in CLL, we selected 23 RNA samples from 11 patients with indolent CLL and 12 patients with aggressive CLL and from CD19+ B cells of eight healthy donors (Table S3) and carried out the tsRNA microarray experiment. We found that nine tsRNAs are differentially expressed in aggressive CLL vs. normal CD19+ B cells (Fig. 5A). We found that 15 tsRNAs are significantly differentially expressed in indolent CLL vs. aggressive CLL (Fig. 5B) and that 10 tsRNAs are differentially expressed in indolent CLL vs. normal CD19+ B cells (Fig. 5C). To determine if tsRNAs are dysregulated in lung cancer, we used seven lung cancer samples and five normal adjacent lung tissue samples for microarray experiments and identified six tsRNAs differentially expressed in lung cancer vs. normal lung tissue (Fig. 5D). Interestingly, ts-46 and ts-47 were strongly down-regulated in aggressive CLL samples vs. normal CD19+ B cells, and the same two tsRNAs were strongly down-regulated in lung cancer samples vs. normal lung tissue (Fig. 5), indicating that ts-46 and ts-47 are potential tumor suppressors.
Table S2.
tsRNA sequences used for generation of the tsRNA microarray chip
| ID | Name | Extended sequences | Chromosome | Start | End | Strand | Length of tsRNA (bp) |
| ts-104 | chr6.tRNA99-GlnCTG | acctTTCATTTCTCTCCTTTtgcct | chr6 | 28909358 | 28909382 | − | 16 |
| ts-113 | chr6.tRNA37-ValAAC | aacaGAGCGTAGTTTCGTTTttttg | chr6 | 27203357 | 27203381 | + | 16 |
| ts-13 | chr6.tRNA149-LysTTT | ggcgCTAAAAATGCAACTTTtactt | chr6 | 27302749 | 27302773 | − | 16 |
| ts-15 | chr6.tRNA144-AspGTC | ggagACGAAGCAGTTGCTTTtgatt | chr6 | 27551216 | 27551240 | − | 16 |
| ts-18 | chr6.tRNA138-ArgACG | ctcgCTTGTGCTCTCACTTTttcct | chr6 | 27638324 | 27638348 | − | 16 |
| ts-2 | chr1.tRNA35-GlyGCC | tgcaGCACGCCCTCCCATTTtggtg | chr1 | 161413161 | 161413185 | + | 16 |
| ts-2 | chr1.tRNA37-GlyGCC | tgcaGCACGCCCTCCCATTTtggtg | chr1 | 161420534 | 161420558 | + | 16 |
| ts-2 | chr1.tRNA39-GlyGCC | tgcaGCACGCCCTCCCATTTtggtg | chr1 | 161427965 | 161427989 | + | 16 |
| ts-2 | chr1.tRNA41-GlyGCC | tgcaGCACGCCCTCCCATTTtggtg | chr1 | 161435325 | 161435349 | + | 16 |
| ts-2 | chr1.tRNA43-GlyGCC | tgcaGCACGCCCTCCCATTTtggtg | chr1 | 161450423 | 161450447 | + | 16 |
| ts-23 | chr6.tRNA66-AlaTGC | tccaATAGGTATTAAGGTTTtagcg | chr6 | 28611290 | 28611314 | + | 16 |
| ts-25 | chr6.tRNA71-LysTTT | ggcgAAATATTTGTGTGTTTtactc | chr6 | 28715590 | 28715614 | + | 16 |
| ts-28 | chr6.tRNA82-GlyGCC | tgcaACATGGCATCCCATTTtggct | chr6 | 142578843 | 142578867 | + | 16 |
| ts-30 | chr9.tRNA7-HisGTG | ggcaGGAGGGGAGAAGGTTTttgga | chr9 | 14433918 | 14433942 | − | 16 |
| ts-31 | chr9.tRNA2-ArgTCG | gtcgTGGAGGGGTTATATTTtgttg | chr9 | 112960872 | 112960896 | + | 16 |
| ts-48 | chr16.tRNA2-ArgCCT | ggtaAAGAAAGGCCGAATTTtagtg | chr16 | 3202970 | 3202994 | + | 16 |
| ts-6 | chr1.tRNA56-ThrTGT | gcctCCTGTTGGCTTACTTTtattt | chr1 | 222638416 | 222638440 | + | 16 |
| ts-60 | chr4.tRNA3-CysGCA | ccctCTGTGCTCTGGAGTTTtgccg | chr4 | 124429985 | 124430009 | − | 16 |
| ts-68 | chr6.tRNA121-ThrCGT | gcctTAGGGTGTGCGTGTTTttttt | chr6 | 28615964 | 28615988 | − | 16 |
| ts-70 | chr6.tRNA96-PheGAA | ggcaTGAGAGCGCTCGGTTTtttgt | chr6 | 28949429 | 28949453 | − | 16 |
| ts-78 | chr21.tRNA2-GlyGCC | tgcaGCACGAAAATGTGTTTtggac | chr21 | 18827087 | 18827111 | − | 16 |
| ts-92 | chr16.tRNA6-ProCGG | gcccTAGAAGTGGTTACTTTtccct | chr16 | 3222117 | 3222141 | + | 16 |
| ts-99 | chr3.tRNA12-PseudoGCA | ggatTTTCTCACTTCCATTTtaaag | chr3 | 17741372 | 17741396 | − | 16 |
| ts-109 | chr16.tRNA1-ArgCCG | gtcgTTGCCATGTTAACGTTTtcttc | chr16 | 3200744 | 3200769 | + | 17 |
| ts-33 | chrX.tRNA3-PseudoCTC | ggtaTGAGTCCTTTCTGGTTTtatat | chrX | 51306091 | 51306116 | − | 17 |
| ts-38 | chr12.tRNA5-AspGTC | ggagGTGTGTAGCTGCACTTTtttgg | chr12 | 98897349 | 98897374 | + | 17 |
| ts-42 | chr15.tRNA11-GluTTC | ggaaTGACTGGACCTTTCTTTtggaa | chr15 | 26327360 | 26327385 | − | 17 |
| ts-55 | chr17.tRNA15-CysGCA | ccctCGGGTTTCTTTAAGTTTttttc | chr17 | 37023966 | 37023991 | + | 17 |
| ts-59 | chr4.tRNA4-GlnTTG | gtcaAAATGTTTCCAATGTTTtgtat | chr4 | 40908722 | 40908747 | − | 17 |
| ts-63 | chr6.tRNA171-MetCAT | gctaTATGGCCGCATATATTTtactt | chr6 | 26313331 | 26313356 | − | 17 |
| ts-7 | chr3.tRNA2-ValAAC | aacaCACTAAAAAATCCCTTTttttt | chr3 | 169490087 | 169490112 | + | 17 |
| ts-71 | chr7.tRNA22-CysGCA | ccccCTTTGTCACTATCCTTTtacac | chr7 | 149074580 | 149074605 | − | 17 |
| ts-76 | chr15.tRNA10-SerGCT | gtcgATGTGGTGGCTTACTTTtgctt | chr15 | 40886002 | 40886027 | − | 17 |
| ts-81 | chr1.tRNA116-GluCTC | ggaaAGAGTAACATCTTCTTTtactg | chr1 | 145399212 | 145399237 | − | 17 |
| ts-82 | chr7.tRNA9-TyrGTA | ccctTCTGTTAAGTTTAATTTtggta | chr7 | 149255202 | 149255227 | + | 17 |
| ts-86 | chr6.tRNA84-GlnTTG | acctTTCAAAGGAGTACATTTtggcc | chr6 | 145503927 | 145503952 | + | 17 |
| ts-89 | chr17.tRNA18-ArgCCT | ggtaGAGGTGAAAGTTCCTTTtacgg | chr17 | 73030070 | 73030095 | + | 17 |
| ts-9 | chr6.tRNA158-IleAAT | gccaCTTCCGTGGGTTTGTTTttttt | chr6 | 27144973 | 27144998 | − | 17 |
| ts-90 | chr6.tRNA39-PseudoAAT | gccaCAGTGGGTGTTAGCTTTtcatc | chr6 | 27251934 | 27251959 | + | 17 |
| ts-98 | chr6.tRNA64-GlnTTG | acctTTCAAAGGTGAACGTTTtacag | chr6 | 28557224 | 28557249 | + | 17 |
| ts-12 | chr6.tRNA36-ArgACG | ctcgGGTGTTAATCTTGGCTTTttttt | chr6 | 27183021 | 27183047 | + | 18 |
| ts-16 | chr6.tRNA56-PheGAA | gacaGCTGTTTCCGGACTCTTTtgtac | chr6 | 27632617 | 27632643 | + | 18 |
| ts-21 | chr6.tRNA133-ValCAC | aacaTTGTATTGTCCTCAGTTTtgtct | chr6 | 27696305 | 27696331 | − | 18 |
| ts-34 | chr10.tRNA6-ValTAC | accaTGGTGTGGTCTGTTGTTTtcctg | chr10 | 5895652 | 5895678 | − | 18 |
| ts-39 | chr1.tRNA23-PseudoCTG | acctTGGTGTCTGCTGGCCTTTtactc | chr1 | 147825757 | 147825783 | + | 18 |
| ts-4 | chr1.tRNA51-PseudoCTT | ggcgCTGTGTTTCCCAGCATTTtggga | chr1 | 165566219 | 165566245 | + | 18 |
| ts-65 | chr6.tRNA8-ArgACG | ctcgGTGTAAGCAGGGTCGTTTtacaa | chr6 | 26537795 | 26537821 | + | 18 |
| ts-67 | chr6.tRNA59-IleAAT | gccaAGTATTCTCTGTGGCTTTtatca | chr6 | 27656037 | 27656063 | + | 18 |
| ts-69 | chr6.tRNA119-AlaCGC | tccaAGGCGATCACGTAGATTTtgttt | chr6 | 28641591 | 28641617 | − | 18 |
| ts-75 | chr14.tRNA20-ThrTGT | gcctCGGCTGTAGGAATACTTTtccca | chr14 | 21099297 | 21099323 | − | 18 |
| ts-97 | chr6.tRNA62-SerGCT | gtcgGCTGCATAGCAAGCCTTTtgttc | chr6 | 28180893 | 28180919 | + | 18 |
| ts-105 | chr11.tRNA16-ValTAC | accaCGGCGTGATTCATACCTTTttctt | chr11 | 59318437 | 59318464 | − | 19 |
| ts-36 | chr10.tRNA2-SerTGA | tgcgGAAGCGGGTGCTCTTATTTtttct | chr10 | 69524339 | 69524366 | + | 19 |
| ts-74 | chr12.tRNA10-AspGTC | ggagGCCGGGTACTTTCGTATTTttaaa | chr12 | 125424170 | 125424197 | − | 19 |
| ts-26 | chr6.tRNA74-LeuCAA | gacaCAACTATCTTATTCTCCTTTtactc | chr6 | 28908931 | 28908959 | + | 20 |
| ts-32 | chrX.tRNA4-ValTAC | accaTAGCCGTAAGGCGGCTGTTTttgct | chrX | 18693005 | 18693033 | − | 20 |
| ts-37 | chr11.tRNA17-ValTAC | accaTGAGATGTTACCTAGCGTTTtgtga | chr11 | 59318078 | 59318106 | − | 20 |
| ts-53 | chr17.tRNA8-ThrAGT | gcctCCGTGTTTCCCCCACGCTTTtgcca | chr17 | 8090548 | 8090576 | + | 20 |
| ts-58 | chr2.tRNA13-AlaCGC | tccaAATCAGCCGACGTTTGCTTTttatt | chr2 | 157257349 | 157257377 | + | 20 |
| ts-50 | chr16.tRNA27-LeuTAG | gccaCACCTCAGAAGGTCTCACTTTtctat | chr16 | 22207007 | 22207036 | − | 21 |
| ts-47 | chr15.tRNA4-ArgTCG | gtcgAAGGGAGGTTATGATTAACTTTtagtt | chr15 | 89878373 | 89878403 | + | 22 |
| ts-61 | chr5.tRNA17-PseudoAAG | gccaACTTGTTGTGATTTCTCCATTTtgtcc | chr5 | 180591538 | 180591568 | − | 22 |
| ts-62 | chr5.tRNA16-LeuAAG | gccaGCTTGTTGTGGTTTGCCCATTTtgtcc | chr5 | 180601018 | 180601048 | − | 22 |
| ts-8 | chr5.tRNA19-LeuAAG | gccaGCTTGTTGTGATTCCTCCATTTttgtc | chr5 | 180524448 | 180524478 | − | 22 |
| ts-8 | chr5.tRNA3-LeuAAG | gccaGCTTGTTGTGATTCCTCCATTTttgtc | chr5 | 180528918 | 180528948 | + | 22 |
| ts-96 | chr1.tRNA86-ArgTCT | gatgGTCACCTGGCAGGTGCCTCTTTtattt | chr1 | 159111375 | 159111405 | − | 22 |
| ts-10 | chr6.tRNA157-ValCAC | TTCTTTACTCGTTTGGTAGATTT | chr6 | 27173844 | 27173866 | − | 23 |
| ts-20 | chr6.tRNA135-ThrAGT | TGATTTCTGTACCTTACACATTT | chr6 | 27652451 | 27652473 | − | 23 |
| ts-107 | chr6.tRNA95-AlaAGC | GTTTCTTGTCTGGTTTATGTCTTT | chr6 | 58141853 | 58141876 | − | 24 |
| ts-29 | chr8.tRNA5-TyrGTA | GAGACACCCCCCCCCCCATTATTT | chr8 | 67026312 | 67026335 | + | 24 |
| ts-56 | chr17.tRNA26-CysGCA | CTGTGCTCCGGAGTTACCTCGTTT | chr17 | 37310720 | 37310743 | − | 24 |
| ts-101 | chr17.tRNA7-SerGCT | GCTAAGGAAGTCCTGTGCTCAGTTT | chr17 | 8090266 | 8090290 | + | 25 |
| ts-11 | chr6.tRNA35-SerCGA | CTTCTCGGTACCTCTCTCTCTCTTT | chr6 | 27177710 | 27177734 | + | 25 |
| ts-110 | chr1.tRNA44-PseudoGTC | GTCACGGGAAATTGTGTGAGGATTT | chr1 | 161493007 | 161493031 | + | 25 |
| ts-3 | chr1.tRNA48-AspGTC | GTTACGGAAAATTGTGTGGGGATTT | chr1 | 161574660 | 161574684 | + | 25 |
| ts-91 | chr17.tRNA36-ThrAGT | TTATTCAATTGAAACAGCGTGATTT | chr17 | 8129528 | 8129552 | − | 25 |
| ts-40 | chr12.tRNA14-PseudoCTC | CGACTCCACATCTCTTGCCTCAGTTT | chr12 | 114386519 | 114386544 | − | 26 |
| ts-41 | chr12.tRNA7-AspGTC | CCATATAATATTAAGAAAAAAAATTT | chr12 | 122860995 | 122861020 | + | 26 |
| ts-112 | chr6.tRNA175-SerGCT | CTCGGCTTTCCCTGCTAACTGGGCTTT | chr6 | 26305691 | 26305717 | − | 27 |
| ts-79 | chr1_gl000192_random.tRNA2-PseudoCCC | GCGTGCAGTTTCCGCACAATGCGGTTT | chr1_gl000192_random | 466052 | 466078 | − | 27 |
| ts-85 | chr7.tRNA14-CysGCA | GATGCCACTTAGCTGTGGGTTCTCTTT | chr7 | 149361987 | 149362013 | + | 27 |
| ts-93 | chr7.tRNA19-CysGCA | GCTGCCACTTACCTGTGGGTTCTCTTT | chr7 | 149292278 | 149292304 | − | 27 |
| ts-106 | chr1.tRNA104-PseudoGTT | GCTGCTGCTCCAAGCCTGCTCCAGATTT | chr1 | 149211920 | 149211947 | − | 28 |
| ts-14 | chr6.tRNA49-GlnCTG | GTCGTTTCTTCTTGCTTCCTCACTATTT | chr6 | 27487380 | 27487407 | + | 28 |
| ts-52 | chr19.tRNA4-ThrAGT | CAACCGAGCGTCCAAGCTCTTTCCATTT | chr19 | 33668037 | 33668064 | + | 28 |
| ts-57 | chr17.tRNA22-Undet??? | CATATCTAGCTTCTCAATGCCTCAGTTT | chr17 | 66390929 | 66390956 | − | 28 |
| ts-87 | chr20.tRNA7-PseudoCTC | AAGGAAAAAAAAAGAAATATTTGGATTT | chr20 | 13970442 | 13970469 | − | 28 |
| ts-66 | chr6.tRNA154-IleAAT | CGAGGGTTCTCACCTTCTCTCTCCGATTT | chr6 | 27205321 | 27205349 | − | 29 |
| ts-1 | chr1.tRNA118-HisGTG | GGTGGTTCTAACTTGCTGGGGTGGCGGTTT | chr1 | 145396851 | 145396880 | − | 30 |
| ts-111 | chr6.tRNA4-ArgTCG | TTTGAGATACTGACTAGTCTGGTGTTATTT | chr6 | 26323119 | 26323148 | + | 30 |
| ts-95 | chr14.tRNA4-ThrTGT | TGCGAAACTACTTTCTTGATTCAGGTGTTT | chr14 | 21149922 | 21149951 | + | 30 |
| ts-24 | chr6.tRNA118-PseudoTTT | TAAGGCCTTGGCCTTCTGTTGATGAGTGTTT | chr6 | 28660956 | 28660986 | − | 31 |
| ts-51 | chr17.tRNA21-ArgCCT | TCGAGAGGGGCTGTGCTCGCAAGGTTTCTTT | chr17 | 73030495 | 73030525 | − | 31 |
| ts-83 | chr1.tRNA2-GlyCCC | GCAGGTACTTCTTCATTTCATTATGGCCTTT | chr1 | 17053851 | 17053881 | + | 31 |
| ts-19 | chr6.tRNA58-ValCAC | CCTCCTGAGACTATTTCTCCTTCCCAACATTT | chr6 | 27650562 | 27650593 | + | 32 |
| ts-22 | chr1.tRNA112-GlnCTG | TTGTGTTTCATTGGCATGGTAAGGCCGTGTTT | chr1 | 147737350 | 147737381 | − | 32 |
| ts-45 | chr1.tRNA28-GlnCTG | TTCTGTTTAATTAGGAAGGCAATGCCGTGTTT | chr1 | 149186197 | 149186228 | + | 32 |
| ts-64 | chr1.tRNA15-GlnCTG | TTCTGTTTAATTAGGACGGCAATGTTGTGTTT | chr1 | 145963376 | 145963407 | + | 32 |
| ts-64 | chr1.tRNA19-GlnCTG | TTCTGTTTAATTAGGACGGCAATGTTGTGTTT | chr1 | 147505110 | 147505141 | + | 32 |
| ts-72 | chr1.tRNA22-GlnCTG | TTCTGTGTAACTGGGACGATAATGCCGTCTTT | chr1 | 147801009 | 147801040 | + | 32 |
| ts-73 | chr10.tRNA5-PseudoCTG | AATTATTTCAAATCCTAGAAATAAGAGCATTT | chr10 | 20036576 | 20036607 | − | 32 |
| ts-46 | chr15.tRNA1-HisGTG | TTGTGGAAACAATGGTACGGCAAGGGCCTCTTT | chr15 | 45493421 | 45493453 | + | 33 |
| ts-27 | chr1.tRNA21-HisGTG | ATGTCGTTAGTCTAGGCTGTCAGCTCTTCCCTTT | chr1 | 147753543 | 147753576 | + | 34 |
| ts-27 | chr1.tRNA111-HisGTG | ATGTCGTTAGTCTAGGCTGTCAGCTCTTCCCTTT | chr1 | 147774811 | 147774844 | − | 34 |
| ts-49 | chr16.tRNA15-ThrCGT | GATATCCAACCTTCAGCTATAGGGTGGAGACTTT | chr16 | 14379822 | 14379855 | + | 34 |
| ts-88 | chr1.tRNA16-HisGTG | AAGTTGCTCGTCTGGGTAGTCAGCTGATCCGTTT | chr1 | 146544845 | 146544878 | + | 34 |
| ts-5 | chr1.tRNA61-PseudoTGG | CTTATAACCTATGTGATTTGGGAAATCCTATGTTT | chr1 | 207178119 | 207178153 | − | 35 |
| ts-94 | chr14.tRNA7-ArgACG | GTGGGGTTCCTCGCAGCTTCGCTGCGTGAGCATTT | chr14 | 23398983 | 23399017 | + | 35 |
| ts-44 | chr15.tRNA8-HisGTG | TTGTGAGGACAATGGCACGGCAAGGGGAGTTTGTTT | chr15 | 45492575 | 45492610 | − | 36 |
| ts-54 | chr17.tRNA11-SupCTA | TTTCCTTCCTGTATAACTCAAGAAAGTCTTGGCTTT | chr17 | 15408759 | 15408794 | + | 36 |
| ts-43 | chr15.tRNA9-HisGTG | TTGTGGGAACAATGGCACGGCAAGGGGCTCGGTATTT | chr15 | 45490767 | 45490803 | − | 37 |
| ts-102 | chr6.tRNA146-GlnCTG | GCTCGAAGATATATGTATATATCCTCTTCCTGGACCTTT | chr6 | 27515492 | 27515530 | − | 39 |
| ts-80 | chr17.tRNA14-ThrCGT | GAGACCCGAGGTAGGGCTTTGGCTGTGGGGAAGTCGGGTTT | chr17 | 29877165 | 29877205 | + | 41 |
| ts-108 | chr5.tRNA23-PseudoTTG | GTTACTAGCTGTGTGACCTTGGGCATATTGTTCAACTTCTTT | chr5 | 77318734 | 77318775 | − | 42 |
| ts-17 | chr6.tRNA57-IleAAT | CTGGATGCCATAGTGGGGCCCTTCACTTACCTGAAGGGAGGCCTTT | chr6 | 27636436 | 27636481 | + | 46 |
| ts-35 | chr10.tRNA3-PseudoCAA | TCACCAGCCTTGTGTGTGGGGCCAGTTAATTAACCTCTTTGTGTTT | chr10 | 34591370 | 34591415 | − | 46 |
| ts-77 | chr16.tRNA12-ArgCCT | AGACAACACCGACCGTAGGTGACTTGGGGTAAGGTATCACGCCTTT | chr16 | 3243991 | 3244036 | + | 46 |
| ts-84 | chr11.tRNA2-LysCTT | CCAGATATCAGGGGAAACCAGGCCCTGATATTCACGTGGGTCCTTT | chr11 | 51359973 | 51360018 | + | 46 |
| ts-100 | chr8.tRNA3-GluCTC | CCAAAAAAATGGAAAAAGAAAATAATAAAAGAAAAAAAAAGACGTTT | chr8 | 59504869 | 59504915 | + | 47 |
| ts-103 | chr19.tRNA8-SeCTCA | GGTAGTAACTAAGCGCCTTAAATCTTACTTATCGAACCGGACTTCTTT | chr19 | 45981812 | 45981859 | − | 48 |
Fig. S4.
Specificity of the tsRNA microarray chip. HEK 293 cells were transfected with pEGFP-3676 (Left) or pEGRP-4521 (Right). Overexpression of ts-3676 (Upper Left) and ts-4521 (Lower Right) confirmed the specificity of the chip.
Table S3.
CLL samples used in tsRNA microarray experiments
| ID | VH | ZAP |
| 1465-I | 93.1 | 1.1 |
| 1466-I | 94.9 | 16.6 |
| 1467-I | 90.5 | 0.77 |
| 1468-I | 94.6 | 0.5 |
| 1469-I | 92 | 0.4 |
| 1470-I | ND | 9.3 |
| 1472-I | 97.3 | 13.1 |
| 1473-I | 94.9 | 2.3 |
| 1474-I | 97.5 | 15.6 |
| 1476-I | 88.5 | 3.8 |
| 1477-I | 94.4 | 2 |
| 1479-A | 100 | 10.8 |
| 1481-A | ND | 80.3 |
| 1482-A | 100 | 0.9 |
| 1483-A | 99.3 | 30 |
| 1485-A | 100 | 42.6 |
| 1487-A | 100 | 24 |
| 1488-A | 100 | 32.5 |
| 1489-A | 100 | 42.8 |
| 1490-A | 99.7 | 23.1 |
| 1491-A | 99.7 | 26.1 |
| 1492-A | 100 | 30.3 |
| 1493-A | 100 | 34.2 |
Fig. 5.
TsRNA signatures in CLL and lung cancer. (A) Aggressive CLL vs. normal CD19+ B cells (negative values indicate down-regulation in CLL). (B) Aggressive CLL vs. indolent CLL (negative values indicate down-regulation in aggressive CLL). (C) Indolent CLL vs. normal CD19+ B cells (negative values indicate down-regulation in CLL). (D) Lung cancer cells vs. normal lung (negative values indicate down-regulation in lung cancer).
Discussion
While studying the miR-4521/3676 cluster in CLL, we found that these two microRNAs are associated with tRNA sequences and represent a recently identified class of small, noncoding RNAs, tsRNAs. Thus, to our knowledge we identified the first two tsRNAs (produced from the 3′ end of pre-tRNAs) involved in cancer. Our data established that ts-3676 and ts-4521 are involved in CLL. Similarly to miR-15/16, the first two microRNAs found altered in cancer and deleted in 13q-CLL cases (3), ts-3676 and ts-4521 are deleted in 17p-CLL cases (6). While miR-15/16 target BCL2, a critical oncogene in CLL (5), ts-3676 targets TCL1, another critical oncogene in CLL (6, 22, 23). In addition to their being down-regulated, we found that ts-3676 and ts-4521 are mutated in low percentage of lung cancer cases (∼2%). Most of the mutations were germline and possibly could represent unknown rare polymorphisms. Their functional significance needs to be elucidated further.
TsRNAs are involved in gene-expression regulation via 3′ UTR targeting of mRNAs. This microRNA-like mechanism was reported in 2013 by Maute et al. (18). In that study, the tsRNA CU1276 was reported to associate with all four human Argonaute proteins and to function as a microRNA. Indeed, CU1276 was shown to repress endogenous genes and was found to be down-regulated in lymphoma cell lines and primary biopsies. However, this tsRNA, is derived from a mature tRNA sequence, is not a unique sequence, and belongs to a different class of tRNA-derived fragments. The data presented here combined with our previous report (6) indicate that tsRNAs derived from the 3′ ends of pre-tRNAs can function as microRNAs (i.e., ts-3676 targets TCL1 expression). On the other hand, tsRNAs do not share the biogenesis mechanism that defines microRNAs but instead are single-stranded RNAs, as are piRNAs. piRNAs interact with Piwi proteins and appear to function in concert with DNA methyltransferases to affect methylation of genes, suggesting that Piwi/piRNA complexes are global regulators of the DNA methylation state and possibly provide sequence specificity for DNA methylation (14, 15). Because of the similarity of tsRNAs to piRNAs, we hypothesized and then proved that these molecules can interact with Piwi proteins (24). Interestingly, in a recent report, the Tetrahymena Piwi protein Twi12, essential for growth, was found to be loaded with mature tRNA fragments (25).
Very recently Zhang et al. (26) reported that a small RNA derived from the 5′ end of tRNA can interact PiwiL2 and PiwiL4 (in that case enrichment was three- to fourfold, vs. the 2.7-fold enrichment we found here). Thus, it is possible that tsRNAs can function as a sequence template for site-specific methylation.
As mentioned above, in this report we studied tsRNAs produced from the 3′ ends of pre-tRNAs by endonuclease RNase Z representing unique sequences starting exactly at the 3′ ends of tRNAs and ending at the sequence of four consecutive T nucleotides. Other tRNA-derived small RNAs are produced from mature tRNA sequences and are not unique sequences (16). Although these other small RNAs also may have a role in cancer, it is technically difficult to study their expression and regulation as a class, because the genomic location from which these molecules came is not clear. On the other hand, our microarray experiments showed that tsRNAs derived from 3′ ends of pre-tRNAs can be studied as a class and may have oncogenic or tumor-suppressor function in cancer.
Methods
CLL Samples.
The study was carried out in accordance with the protocol approved by the Institutional Review Board of the Ohio State University. The set of CLL samples used in sequencing and real-time RT-PCR were obtained from 63 CLL patients enrolled in the CLL Research Consortium (CRC), who provided written informed consent. Samples were subgrouped according to their karyotype: 28 11q-deleted samples, nine 17p-deleted samples, 11 13q-deleted samples, and 15 samples from CLL patients with a normal karyotype. CLL samples were predominantly (>95%) composed of CD5+/CD19+ B cells.
An additional 23 samples from patients enrolled in the CRC were used for tsRNA microarray hybridization. Samples were selected according to their clinical and molecular features as following: 11 samples from patients with clinically indolent disease, mutated IgVH, and low ZAP-70 expression and 12 samples from patients with a clinically aggressive disease, unmutated IgVH, and high ZAP-70 expression. DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen). RNA was extracted using standard TRIzol (Invitrogen) methods. The quality of the RNAs of each sample was analyzed on the Agilent 2100 Bioanalyzer.
We also used 17 samples from patients with nonsmall cell carcinoma of the lung and matched normal lung tissues for real-time PCR and 300 lung cancer samples for sequencing of ts-3676 and ts-4521. These samples were provided by Harvey Pass, Langone Medical Center of New York University, New York.
Sequencing, Real-Time RT-PCR.
Mutations in the region containing ts-4521 and ts-3676 were determined by PCR amplification and sequencing. The mutation rate was evaluated by inspection of fluorescent peak trace chromatograms. For amplification, we used CG-rich high-fidelity advantage 2× polymerase master mix (Clontech) with the primer sequences 4521-F: AGTTCTAGACCTCTCAGCAG and 4521-R: GACAGTACTGGCAAGTACAG. PCR products were purified using ExoSAP-IT (USB) and sequenced. Ts-4521 and tsR-3676 expression was assayed by real-time RT-PCR using Taqman MicroRNA assays hsa-miR-4521 (assay ID 465004_mat, catalog no. 4427975) and hsa-miR-3676-3p (assay ID 464747_mat, catalog no. 4427975), according to the manufacturer’s protocol (Thermo Fisher Scientific) and were normalized using RNU6B (assay ID 001093, catalog no. 4427975; Thermo Fisher Scientific). Additional Taqman MicroRNA assays used were has-miR-92a (assay ID: 000431, catalog no. 4427975) and has-miR-93 (assay ID: 000432, catalog no. 4427975).
DNA Constructs.
pCDH-3676/4521 were described previously (6). To improve ts-3676 and ts-4521 expression, the fragments containing either tRNA8-ThrAGT and ts-3676 or tRNA8-SerGCT and ts-4521 were subcloned upstream of the EGFP gene of the pEGFP-N1 vector (Clontech), obtaining the expression vectors pEGFP-3676 and pEGFP-4521,respectively. For pMAX-piwiL2, pMAX-ago1, and pMAX-ago2, the ORFs of PiwiL2, Ago1, and Ago2 were subcloned in pcDNA4/HisMax (Invitrogen) vector from pCMV6-Entry-mycDDK tagged TrueORF gold expression vector purchased from OriGene [stock-keeping unit (SKU) nos. RC205464, RC209163, andRC218078, respectively].
Northern Blot Analysis.
HEK-293 cells were transfected in T25 flasks with 3 μg of pCDH-4521, pCDH-3676, or pCDH empty vector. Northern Blot analysis was carried out as previously described (6). Probe sequences were CTGAGCACAGGACTTCCTTAGC for the tsRNA/miR-4521 precursor; TTTGATGCTACAAAACTGAG for the miR-4521 precursor; AGCGTGGGGGAAACACGG for the tsRNA/miR-3676 precursor; and TTGGCAAAAGCGTGGGGG for the miR-3676 precursor.
RNA Immunoprecipitation and Western Blot Analysis.
HEK 293 cells were transfected in T75 flasks with 16 μg of pMAX-PiwiL2, pMAX-Ago1, and pMAX-Ago2 using the standard FuGENE protocol (Roche). RIP was performed with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manufacturer’s protocol. Briefly, 48 h after transfection cells were suspended in an equal pellet volume of complete RIP lysis buffer. We used 150 μL of RIP buffer, obtaining a total of volume of 300 μL. Ten microliters of lysate were stored for Western blot input, and 10 μL were stored for real-time input. One hundred microliters of each lysate were immunoprecipitated overnight with anti-max magnetic beads or anti–mouse-IGg magnetic beads (as negative control) in 900 μL of RIP immunoprecipitation buffer. Beads were separated by centrifugation, washed, and suspended in 500 μL of RIP wash buffer. Fifty microliters of the bead suspension were stored to test the efficiency of immunoprecipitation by Western blotting (Fig. 3). RNAs were extracted from all immunoprecipitations and inputs according to specifications provided by the kit manufacturer, checked for quality on a BioAnalyzer platform (Agilent), and tested for enrichment of ts-RNA-3676, ts-4521, control miR-92a, and control miR-93 by using real-time PCR techniques. Ten microliters of each input from RIP lysate and 50 μL of each bead suspension after immunoprecipitation were analyzed by Western blot to assess the expression of Ago1-, Ago2-, and PiwiL2-tagged proteins with Omni-probe (anti-max) mouse monoclonal antibody (sc-7270; Santa Cruz). Western Blot analysis was carried out using standard protocols.
tsRNA Microarray Chip Design and Hybridization.
The tsRNA microarray chip was fabricated as previously described for microRNAs (21). A pilot experiment was performed to assess the specificity of the array by hybridization of 5 μg of total RNA extracted from HEK-293 cells previously transfected with either pEGFP-3676 or pEGFP-4521. Hybridizations were performed by following standard protocols on a Tecan HS4800 hybridization station and were scanned. Microarray images were analyzed using GenePix Pro Software. Then 5 μg of RNA extracted from 23 CLL samples (11 indolent and 12 aggressive cancers) and from eight CD19+ B-cell controls were hybridized following the same protocol.
Statistical Analysis.
Sequences were downloaded from the University of California, Santa Cruz, table browser by extracting 50-nt sequences from the 3′ end of tRNA genes as provided by track “tRNA Genes” of group “Genes and Gene Predictions” for human genome assembly GRCh37/hg19. From these 50-nt sequences we selected those that displayed at least one instance of the pattern “TTTT” and considered as tsRNA the subsequence from the first nucleotide at the 5′ end to the third T of the first “TTTT” pattern, for a total of 120 tsRNA sequences (of which seven had dual copies). The extracted tsRNA sequence set thus is comprised of sequences whose length ranges from a minimum of 16 nt to a maximum of 48 nt. Finally, tsRNA sequences that were not at least 23 nt in length were extended by 4 nt at their 5′ end and by 5 nt at their 3′ end to allow the design of an appropriate probe sequence of sufficient length for the custom microarray.
Custom tsRNA microarray data were analyzed by applying normexp negative background correction, quantile data normalization, and moderated t-statistics for differential expression analysis calculated using the Linear Models for Microarray Data (LIMMA) package from the Bioconductor R project (27). P values were adjusted for multiple testing using the Benjamini and Hochberg method to control the false discovery rate (28). All transcripts differentially expressed with a P value <0.05 were considered statistically significant.
Acknowledgments
This work was supported by National Cancer Institute Grants PO1-CA81534 (to T.J.K. and C.M.C.), UO1-CA152758 (to C.M.C. and H.I.P.), and R35-CA197706 (to C.M.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604266113/-/DCSupplemental.
References
- 1.Sgambati M, Linet M, Devesa S. 2001. chronic lymphocytic leukemia, epidemiological, familial, and genetic aspects. Chronic Lymphocytic Leukemias, ed Bruce Cheson B (Marcel Dekker, Inc. New York), 2nd Ed, revised and expanded, pp 33–62.
- 2.Rassenti LZ, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 5.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balatti V, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2015;112(7):2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 8.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan XJ, et al. B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2006;103(31):11713–11718. doi: 10.1073/pnas.0604564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visone R, et al. miR-181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood. 2011;118(11):3072–3079. doi: 10.1182/blood-2011-01-333484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 12.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA: Novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340(2):201–211. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 13.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26(5):603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113(2):373–380. doi: 10.1002/jcb.23363. [DOI] [PubMed] [Google Scholar]
- 15.Siddiqi S, Terry M, Matushansky I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS One. 2012;7(3):e33711. doi: 10.1371/journal.pone.0033711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesh T, Suresh PS, Tsutsumi R. tRFs: miRNAs in disguise. Gene. 2016;579(2):133–138. doi: 10.1016/j.gene.2015.12.058. [DOI] [PubMed] [Google Scholar]
- 17.Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maute RL, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110(4):1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20(6):710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3(4):563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 22.Pekarsky Y, Zanesi N, Croce CM. Molecular basis of CLL. Semin Cancer Biol. 2010;20(6):370–376. doi: 10.1016/j.semcancer.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palamarchuk A, et al. Tcl1 protein functions as an inhibitor of de novo DNA methylation in B-cell chronic lymphocytic leukemia (CLL) Proc Natl Acad Sci USA. 2012;109(7):2555–2560. doi: 10.1073/pnas.1200003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keam SP, et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014;42(14):8984–8995. doi: 10.1093/nar/gku620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24(24):2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. 2016;196(4):1591–1603. doi: 10.4049/jimmunol.1500805. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. (Translated from English) J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]