Significance
Measuring direct associations between variables is of great importance in various areas of science, especially in the era of big data. Although mutual information and conditional mutual information are widely used in quantifying both linear and nonlinear associations, they suffer from the serious problems of overestimation and underestimation. To overcome these problems, in contrast to conditional independence, we propose a novel concept of “partial independence” with a new measure, “part mutual information,” based on information theory that can accurately quantify the nonlinearly direct associations between the measured variables.
Keywords: conditional mutual information, systems biology, network inference, conditional independence
Abstract
Quantitatively identifying direct dependencies between variables is an important task in data analysis, in particular for reconstructing various types of networks and causal relations in science and engineering. One of the most widely used criteria is partial correlation, but it can only measure linearly direct association and miss nonlinear associations. However, based on conditional independence, conditional mutual information (CMI) is able to quantify nonlinearly direct relationships among variables from the observed data, superior to linear measures, but suffers from a serious problem of underestimation, in particular for those variables with tight associations in a network, which severely limits its applications. In this work, we propose a new concept, “partial independence,” with a new measure, “part mutual information” (PMI), which not only can overcome the problem of CMI but also retains the quantification properties of both mutual information (MI) and CMI. Specifically, we first defined PMI to measure nonlinearly direct dependencies between variables and then derived its relations with MI and CMI. Finally, we used a number of simulated data as benchmark examples to numerically demonstrate PMI features and further real gene expression data from Escherichia coli and yeast to reconstruct gene regulatory networks, which all validated the advantages of PMI for accurately quantifying nonlinearly direct associations in networks.
Big data provide unprecedented information and opportunities to uncover ambiguous correlations among measured variables, but how to further infer direct associations, which means two variables are dependent given all of the remaining variables (1), quantitatively from those correlations or data remains a challenging task, in particular in science and engineering. For instance, distinguishing dependencies or direct associations between molecules is of great importance in reconstructing gene regulatory networks in biology (2–4), which can elucidate the molecular mechanisms of complex biological processes at a network level. Traditionally, correlation [e.g., the Pearson correlation coefficient (PCC)] is widely used to evaluate linear relations between the measured variables (2, 5), but it cannot distinguish indirect and direct associations due to only relying on the information of co-occurring events. Partial correlation (PC) avoids this problem by considering additional information of conditional events and can detect the direct associations. Thus, PC becomes one of the most widely used criteria to infer direct associations in various areas. As applications of PC to network reconstruction (6), recently Barzel and Barabási (7) proposed a dynamical correlation-based method to discriminate direct and indirect associations by silencing indirect effects in networks, and Feizi et al. (8) developed a network deconvolution method to distinguish direct dependencies by removing the combined effect of all indirect associations. However, PC-based methods including these two works (8) can only measure linearly direct associations and will miss nonlinear relations, which play essential roles in many nonlinear systems, such as biological systems. Analogous to PCC and PC, distance correlation (9, 10) and partial distance correlation (Pdcor) (11) were proposed to measure dependence of random vectors, and these statistics are sensitive to many types of departures from dependence (9). However, Pdcor suffers from the false-positive problem, that is, Pdcor(X;Y|Z) may be nonzero even if X and Y are conditional independent given Z (11).
Based on mutual independence, mutual information (MI) can be considered to be a nonlinear version of correlation that can detect nonlinear correlations but not direct associations or dependencies owing to the information of only joint probability, having the same overestimation problem as PCC (12, 13). As one variant of MI, the maximum information coefficient (MIC) method (3) was proposed to detect both linear and nonlinear correlations between variables. MIC is based on MI but has a few different features on measuring nonlinear associations. However, recently, it has been shown that MI is actually more equitable than MIC (14). By further considering conditional independence from MI, conditional mutual information (CMI) as a powerful tool can quantify nonlinearly direct dependencies among variables from the observed data, superior to PC and also MI, and thus has been widely used to infer networks and direct dependencies in many fields (12, 15–17), such as gene regulatory networks in biology (16). However, CMI-based methods theoretically suffer from a serious problem of underestimation, in particular when quantifying the dependencies among those variables with tight associations among them, which severely limits its applications. For example, when we measure the dependency between variables X and Y given variable Z, CMI cannot correctly measure the direct association or dependency if X (or Y) is strongly associated with Z. For an extreme case, if X and Z are almost always equal [i.e., the conditional probability p(x|z) = 1 provided that the joint probability p(x,y,z) 0], then CMI between X and Y given Z is always zero no matter whether X and Y are really dependent or not (18). Clearly, CMI underestimates the dependency because of this false-negative shortcoming, in stark contrast to MI, which generally overestimates the dependency due to its false-positive drawback (16, 19).
To overcome these problems of MI and CMI, we propose a new measurement, “part mutual information” (PMI), by defining a new type of “partial independence” and also deriving an extended Kullback–Leibler (KL) divergence (SI Appendix, section S1) to accurately quantify the nonlinearly direct associations from the measured variables. We show that PMI not only can theoretically avoid the problems of CMI and MI but also keeps the quantification properties. Similar to CMI, it can also be used to reconstruct various types of networks even with loops or circles, for which Bayesian-based methods generally failed. Specifically, in this paper we first define PMI to measure nonlinearly direct dependencies between variables, and then derive its theoretical relations with MI and CMI. Finally, we use a number of benchmark examples to numerically demonstrate PMI features and, further, real gene expression data to reconstruct gene regulatory networks in Escherichia coli and yeast, which all validated advantages of PMI for quantifying nonlinearly direct associations in networks.
Results
MI and CMI.
Assuming that X and Y are two random variables, MI in information theory represents the information that we need when using Y to code X, and vice versa. MI is defined on the basis of KL divergence D (13). For discrete variables X and Y, MI is calculated as
| [1] |
where is the joint probability distribution of X and Y and and are the marginal distributions of X and Y, respectively. MI is nonnegative, and clearly it equals zero if and only if the two variables are independent. Hence, when MI is larger than zero, the two variables are associated. For this reason, we can use MI to measure nonlinear associations between variables, in contrast to correlation (e.g., PCC, which measures linear associations). If the value of MI between two variables is larger, their association is stronger. Clearly, MI in Eq. 1 is evaluated against the “mutual independence” of X and Y, which is defined as
| [2] |
However, as shown in Fig. 1A, if two independent variables X and Y are both associated with a common random variable Z, MI between X and Y is larger than zero, thus wrongly indicating the direct association between X and Y. The reason is that MI relies on the information of mutual independence in Eq. 1 and cannot correctly measure the direct association, which leads to overestimation (i.e., the false-positive problem) (16). However, CMI can correctly measure the direct association for the case of Fig. 1A by further considering the information of conditional probability or conditional independence. In contrast to partial correlation for linearly direct association, CMI for variables X and Y given Z can further detect nonlinearly direct association and is defined as
| [3] |
which can be also represented as
| [4] |
where is the joint probability distribution of X and Y with the condition Z, p(z) is the marginal probability, and p(x|z) and p(y|z) are conditional marginal probability distributions. Clearly, in contrast to MI in Eq. 1 or the mutual independence of X and Y in Eq. 2, CMI in Eq. 3 is evaluated against the “conditional independence” of X and Y given Z, which is defined as
| [5] |
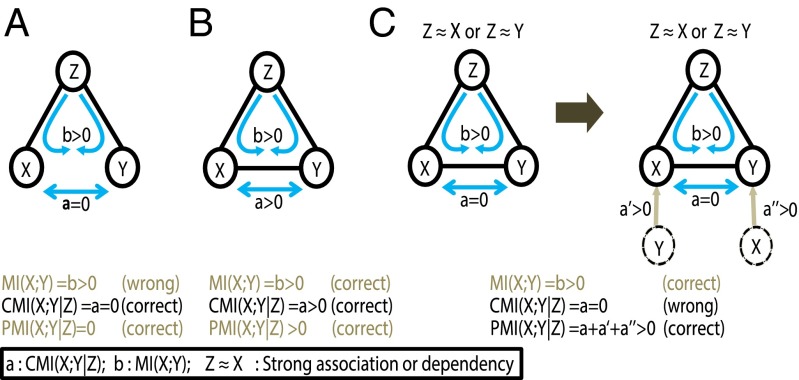
Note that X and Y are two scalar variables, and Z is a vector or (n-2)-dimensional variables (n > 2), and thus Fig. 1 represents a general case for evaluating the direct association of any pair of variables X and Y in a network with n nodes. CMI is also nonnegative, and thus a positive or high CMI implies the dependency of X and Y. In addition to Fig. 1A, as shown in Fig. 1B where X and Y have direct dependency CMI can also correctly measure the nonlinear dependency between X and Y, in particular for those cases where both X and Y have weak associations with Z (e.g., both X and Y are almost independent of Z). However, CMI theoretically suffers from the false-negative or underestimation problem, that is, even when CMI equals zero, X and Y are not necessarily independent (see SI Appendix, Property S6). As shown in Fig. 1C, Left, when X or Y is strongly associated with Z, CMI between X and Y always approaches zero even though X and Y have direct dependency, which means that CMI fails to measure the correct association. In fact, theoretically CMI(X;Y|Z) always suffers from this underestimation problem provided that there are certain associations between X (or Y) and Z (see SI Appendix, Property S6) in a general network. Such cases are general in various natural or engineered systems where variables are associated or interconnected to form a network.
Fig. 1.
A brief Interpretation of differences among MI, CMI, and PMI. (A) X and Y have no direct dependency, but both are associated with Z. For such a case, MI between X and Y is high due to the influence of their common neighbor Z (i.e., MI gives an incorrect result). However, CMI is zero and gives the correct signal. (B) X and Y have direct dependency; CMI between X and Y is high and gives the correct signal, in particular for the cases when X and Y are weakly associated with Z. (C) X and Y have direct dependency, but X or Y is strongly associated with Z. CMI between X and Y approaches zero, thus giving a wrong signal. However, PMI is high and can correctly quantify the dependency for this case. Actually, PMI can give the correct results for all cases (i.e., A–C). a, CMI(X;Y|Z) is the CMI; b, MI(X;Y) is the MI. a′ and a′′ represent the information flow from two virtual nodes of Y and X to the original X and Y, respectively, and they are the second and third terms of PMI in Eq. 10.
PMI.
To overcome those problems of MI and CMI, we propose a novel measure, PMI, to quantify nonlinearly direct dependencies between variables, by introducing a new concept, partial independence. Specifically, in contrast to the conditional independence Eq. 5, partial independence of X and Y given Z is defined as
| [6] |
where p*(x|z) and p*(y|z) are defined (18) as
| [7] |
Then, PMI is defined based on this partial independence Eq. 6.
Definition 1:
X and Y are two scalar variables and Z is an (n-2)-dimensional vector (n > 2). Then, the PMI between variables X and Y given Z is defined as
| [8] |
where p(x,y,z) is the joint probability distribution of X, Y, and Z, and D(p(x,y,z)||p*(x|z)p*(y|z)p(z)) represents the extended KL divergence (see SI Appendix, Definition S1) from p(x,y,z) to p*(x|z)p*(y|z)p(z).
From Eq. 8, it is obvious that PMI is always larger than or equal to zero owing to the extended KL divergence (see SI Appendix, Theorem S1). Eq. 8 can also be rewritten as
| [9] |
Comparing Eq. 4 and Eq. 9, clearly PMI is similarly defined as CMI by replacing p(x|z) p(y|z) in CMI with in PMI, and it is clearly evaluated against the partial independence Eq. 6. In addition, Eq. 9 can be further decomposed as follows:
| [10] |
The second and the third terms in the right-hand side of Eq. 10 correspond to a′ and a′′ in Fig. 1C, respectively. The proof of Eq. 10 is given in SI Appendix. Clearly, CMI(X;Y|Z) is the first term of PMI in Eq. 10, and the second and third terms implicit include the associations between X and Y, respectively, due to Eq. 7.
Next, we show that PMI can overcome the problems of MI and CMI owing to the term of partial independence in Eq. 8. From Eqs. 8–10, we can derive the specific properties of PMI and its relations with CMI and MI, shown in SI Appendix, Table S15 and Properties S1–S9. The properties of the conditional independence Eq. 5 and partial independence Eq. 6 are given in SI Appendix, Properties S7 and S8 and Fig. S1.
First, PMI is symmetric, that is, PMI(X;Y|Z) = PMI(Y;X|Z). Second, PMI is always larger than or equal to CMI, that is, PMI(X;Y|Z) ≥ CMI(X;Y|Z), which implies the potential of PMI to overcome the underestimation problem of CMI. Third, PMI(X;Y|Z) = 0 only if X and Y given Z are independent, and otherwise PMI(X;Y|Z) > 0, which implies that PMI can measure nonlinearly direct dependency between X and Y given Z. All major features of PMI and their related proofs can be found in SI Appendix, Properties S1–S9. When X and Y are independent of Z, that is, and , PMI equals CMI and also MI, that is, PMI(X;Y|Z) = CMI(X;Y|Z) = MI(X;Y). In other words, PMI has features similar to CMI and MI (see SI Appendix, Property S5) when the variables of interest are isolated from others of the system.
Next, we theoretically show that PMI overcomes those problems of CMI and MI. If X and Y are strongly associated, we denote the relation as X ≈ Y, which implies the strongly mutual dependency between X and Y. If X or Y is strongly associated with Z, for example, X ≈ Z or Y ≈ Z, then CMI(X;Y|Z) = 0 regardless of the dependency between X and Y, but PMI(X;Y|Z) relying on the direct dependency of X and Y (see SI Appendix) is generally not necessarily zero. Hence, in Fig. 1C, PMI can detect the direct association, for which CMI failed. Actually, PMI can measure the direct associations correctly for all cases in Fig. 1 (see SI Appendix, Properties S1–S9 for the proofs).
The analyses on the differences between the conditional independence and partial independence are given in SI Appendix, Properties S7 and S8 and Fig. S1. Actually, we can show that both the conditional independence Eq. 5 and the partial independence Eq. 6 hold, that is, p(x|z)p(y|z) = p(x,y|z) and p*(x|z)p*(y|z) = p(x,y|z), if X and Y are conditionally independent given Z (see SI Appendix, Property S7). Thus, both CMI and PMI based on Eq. 5 and Eq. 6 can detect the conditional independence of X and Y given Z (i.e., there is no direct association). However, if X and Y are conditionally dependent given Z (i.e., there is a direct association), the conditional independence Eq. 5 always holds approximately provided that X (or Y) strongly depends on Z, but the partial independence Eq. 6 does not necessarily hold (see SI Appendix, Property S8) and it relies on the direct association of X and Y. Therefore, although both CMI and PMI can give the correct results on the independence of X and Y given Z, PMI can also give the correct results on the dependence of X and Y given Z due to the partial independence Eq. 6, and CMI generally cannot give the correct results on the dependence due to the conditional independence Eq. 5.
Similar to MI and CMI, there are many ways to numerically estimate joint and marginal probabilities (20–22) in PMI directly from the observed data. The most straightforward and widespread method to estimate p(x,y,z) is partitioning the supports of X, Y, and Z into bins of finite size, approximating the continuous distribution by the finite discrete distribution, and converting the integration operators into the finite sum operators. The number of the bins may have some impact on the computing result, but it would not affect the statistical P value so much. Empirically, if Z has n-2 dimensions and there are N data points, the whole n-dimension space will be partitioned into bins with a magnitude of (20–22), where n is the number of total measured variables. However, this approximate method works in the case that N>>n-2. Otherwise, the error of the estimation will be quite large because of the “curse of dimensionality.” If we assume the Gaussian distribution for variables, MI between X and Y can be simplified as below (16, 22):
| [11] |
where is the Pearson correlation coefficient of X and Y. Eq. 11 provides a simple way to calculate MI. By analogy to MI and CMI (16, 22), PMI can be accurately expressed from the covariance matrix of X, Y, and Z.
Theorem 1.
Assume that X and Y are two one-dimensional variables, and Z is an n-2 dimensional (n-2 > 0) vector, and they are defined in an appropriate outcome space. Letting the vector (X,Y,Z) follow multivariate Gaussian distribution, then PMI between X and Y given Z is simplified as follows:
| [12] |
where C, C0, and Σ are all measured matrices given in SI Appendix, section S2 and n is the dimension of vector (X,Y,Z).
The proof of Theorem 1 is given in SI Appendix, section S2. Theorem 1 yields the formula to calculate PMI in a simple but approximate way due to the Gaussian distribution assumption. Besides, we can derive the following relation between PMI and PC:
| [13] |
where is the PC of X and Y when given Z. The proofs can be found in SI Appendix, section S2, Lemmas S1 and S2 and Theorem S2.
Numerical Studies by Simulated Data.
Theoretically, we have shown that PMI is superior to CMI and other linear measurements for quantifying the nonlinearly direct dependencies among variables (SI Appendix, section S1), in particular for those cases that CMI or MI fails. To numerically investigate these, we first used simulated data to study both linear and nonlinear relationships as in the previous work of Barzel and Barabási (7). We compared the performances of PMI with CMI and PC about these direct associations, including linear, parabolic, cubic, sinusoidal, exponential, checkerboard, circular, cross-shaped, sigmoid, and random relationships. Three variables, X, Y, and Z, were simulated, satisfying , where is noise and is noise amplitude. Here we first set that equals zero, that is, we consider noiseless relations. f (X,Z) given Z can be linear, parabolic, cubic, sinusoidal, exponential, checkerboard, circular, cross-shaped, sigmoid, and random functional relations and nonfunctional relations (see SI Appendix, Table S1 and Fig. S2). Various cases, for example X (or Y) is independent of Z or strongly associated with Z, were considered during simulations, and the results for all are displayed in Table 1 and SI Appendix, Tables S4 and S5, where PC(X;Y|Z) is the partial correlation between X and Y given Z and PS(X;Y|Z) is the partial spearman correlation between X and Y given Z. All of the P values are provided in SI Appendix, Tables S2, S3, S6, and S7. We also compared PMI with CMI and PC when X is moderately correlated with Z and results indicated that PMI performs the best, as shown in SI Appendix, Tables S13 and S14.
Table 1.
Comparing PMI, CMI, and PC
| Relation types | PMI(X;Y|Z) | CMI(X;Y|Z) | PC(X;Y|Z) | PS(X;Y|Z) |
| X⊥Z | ||||
| Linear | 1.03* | 0.95* | 1* | 0.98* |
| Quadratic | 0.57* | 0.52* | 0.03 | 0.04 |
| Cubic | 1.27* | 1.23* | 1* | 1* |
| Sinusoidal | 0.88* | 0.83* | 0.78* | 0.78* |
| Exponential | 0.89* | 0.80* | 0.98* | 0.96* |
| Checkerboard | 0.43* | 0.42* | 0.37* | 0.37* |
| Circular | 0.35* | 0.30* | 0.02 | 0.02 |
| Cross-shaped | 0.62* | 0.61* | 0.03 | 0.03 |
| Sigmoid | 0.73* | 0.69* | 0.99* | 0.96* |
| Random | 0.08 | 0.08 | 0.03 | 0.03 |
| X ≈ Z | ||||
| Linear | 2.20* | 0.03* | 1.0* | 0.88* |
| Quadratic | 1.28* | 0.01 | 0.03 | 0.04 |
| Cubic | 1.60* | 0.02 | 0.11* | 0.87* |
| Sinusoidal | 1.33* | 0.01 | 0.03 | 0.02 |
| Exponential | 1.30* | 0.01 | 0.06* | 0.12* |
| Checkerboard | 0.37* | 0.02 | 0.03 | 0.02 |
| Circular | 0.89* | 0.01 | 0.03 | 0.03 |
| Cross-shaped | 1.16* | 0.02* | 0.02 | 0.02 |
| Sigmoid | 1.38* | 0.01 | 0.06 | 0.57* |
| Random | 0.26 | 0.01 | 0.02 | 0.03 |
PS(X;Y|Z) is the partial spearman correlation of X and Y given Z. Asterisks indicate statistically significant P values (see SI Appendix, Tables S2 and S3).
All PMI, CMI, and PC between X and Y were calculated as the means over 1,000 repeats, and the values less than 0.01 were set to zero. The probability distributions were estimated by “bins” -based method (see SI Appendix, section S3) (21). We considered both uniform and normal distributions, shown in Table 1 for uniform distribution and SI Appendix, Tables S4 and S5 for normal distribution, respectively. When Z is independent of X, Table 1 shows clearly that PMI and CMI can detect the true dependency effectively in all relations with high significance (see SI Appendix, Table S2), whereas PC and PS are correct almost only for the linear relation due to its linear nature, and they are also sensitive to cubic, exponential, and sigmoid because these relations are mostly resemble to linear relations in the given interval. However, in Table 1 where X is strongly associated with Z, that is, p(x|z) = 1 for p(x,y,z) 0, CMI completely failed to measure all direct associations and PC and PS only detected the linear relation as expected, whereas PMI correctly identified all direct dependencies. For the cases where Y is strongly associated with Z, that is, p(y|z) = 1 for p(x,y,z) 0, we got the same conclusion. From Table 1, we can find that for the random cases (i.e., when X and Y are actually independent or have no direct dependency) all of PMI, CMI, and PC gave the correct results, which agrees well with our theoretical results. For the normal distribution, as shown in SI Appendix, Tables S4 and S5 and the matched P values in SI Appendix, Tables S6 and S7, we have similar conclusions. In addition to the nonlinear relations in SI Appendix, Table S1, we also conducted the simulations on other types of relations in SI Appendix, Table S8, and, as shown in SI Appendix, Tables S9–S12, all of the results demonstrate the robustness of the PMI.
Comparison for Statistical Power.
As an association measure, statistical power is an important and widely used tool to interpret the results calculated by the measure as well as statistical significance (23). Here, statistical power, or simply “power,” defined similarly as in ref. 14, is the probability that we reject the hypothesis of variables X and Y given Z are independent when X and Y given Z are truly associated. The PMI value shows a significant difference from the value that X and Y given Z are independent. If the false-negative rate, namely a type II error in statistics, is equal to , then the statistical power is . Hence, when the power increases, type II error will decrease.
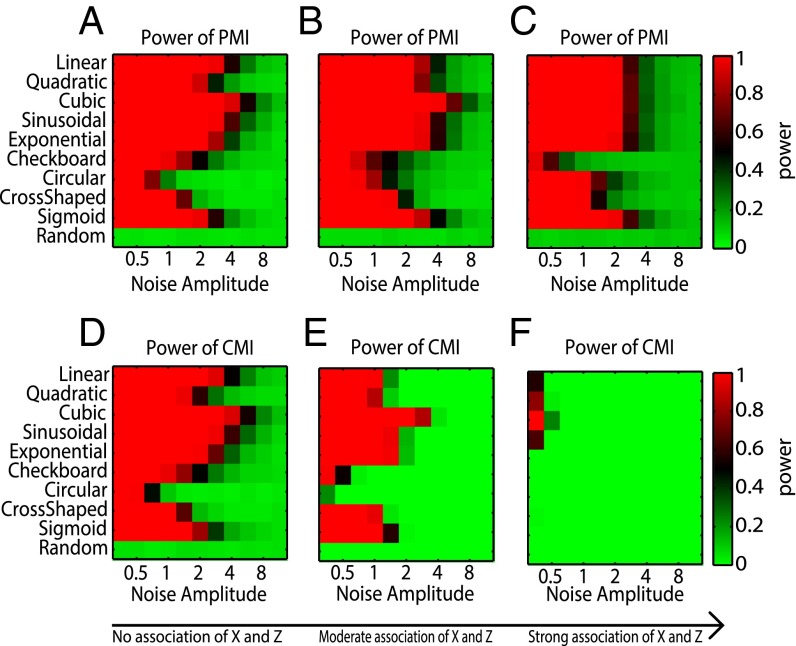
The relationships considered here are the same as in SI Appendix, Table S1. The power of PMI and CMI was presented as a heat map. Fig. 2 A and D show the power of PMI and CMI, where the two measures present almost the same results in the situation when X and Z are independent (i.e., no association of X and Z). This is consistent with the theoretical conclusion that PMI performs as well as CMI when X or Y is not associated with Z. However, in Fig. 2 B and E, when the association between X and Z becomes moderate or stronger, the power of PMI is higher than that of CMI. Also, in Fig. 2 C and F when X is almost always equal to Z (or strongly associated with Z), that is, p(x|z) = 1 for p(x,y,z) 0, the power of CMI is obviously smaller than that of PMI for all of the relationships, especially for the relationships of sinusoid and exponential, which numerically demonstrates that PMI is a more powerful measure than CMI. Actually, we can show that the power of CMI decreases with the increase of the dependency between X (or Y) and Z. In addition, we also tested the equitability (3) of PMI, which demonstrated its superior ability as shown in SI Appendix, section S4 and Fig. S3.
Fig. 2.
Comparison of statistical power between PMI(X;Y|Z) and CMI(X;Y|Z). Ten relationships between X and Y were used Y = f(X,Z) + aη, and the details of the relation are in SI Appendix, Table S1, where “Random” means X and Y have no direct association, η is normally distributed noise with mean 0 and SD 1, and a is noise amplitudes ranging from 0 to 8. Eleven amplitudes were selected, 0.25, 0.35, 0.50, 0.72, 1.00, 1.41, 2.00, 2.83, 4.00, 5.66, and 8.00, which are distributed logarithmically between 0.1 and 8. A and D show the power of PMI and CMI when X and Z are independent, but X and Y are both related with Y; B and E are the results when X is associated with Z moderately (i.e., X = 0. 5X + 0.5Z). C and F are the cases when X are strongly associated with Z, which means X is almost a copy of Z; we set X = 0.1X + 0.9Z.
DREAM3 Challenge Datasets for E. coli and Yeast.
We applied PMI to the widely used DREAM3 challenge datasets for reconstructing gene regulatory networks (24), where the gold standard networks were from yeast and E. coli. The gene expression data were generated with the nonlinear ordinary differential equation systems (19, 25). In this work, we used DREAM3 challenging data with sizes 10, 50, and 100 to construct the gene regulatory network by PMI-based PC (path consistency) algorithm, which is a greedy iteration algorithm for inferring networks and terminates until the network converges (16). Simply by replacing CMI with PMI in the original CMI-based algorithm (i.e., PCA-CMI) (16), we can obtain PMI-based PC-algorithm, called PCA-PMI, which is based on Eq. 12 in Theorem 1. The detailed algorithm of PCA-PMI is given in SI Appendix. The performance of PMI was evaluated by the true-positive rate (TPR) and false-positive rate (FPR). They were defined as TPR = TP/(TP + FN) and FPR = FP/(FP + TN), where TP, FP, TN, and FN represent the numbers of true positives, false positives, true negatives, and false negatives, respectively. TPR and FPR are used for plotting the receiver operating characteristic (ROC) curves and the area under ROC curve (AUC) is calculated. In addition, we compared PMI and CMI by inferring the gene regulatory networks.
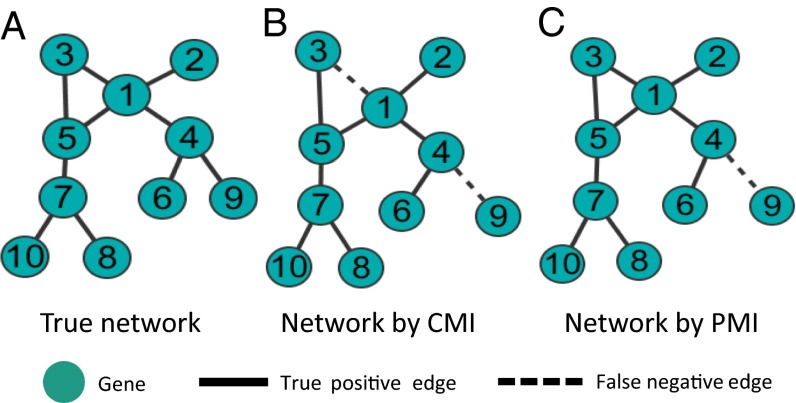
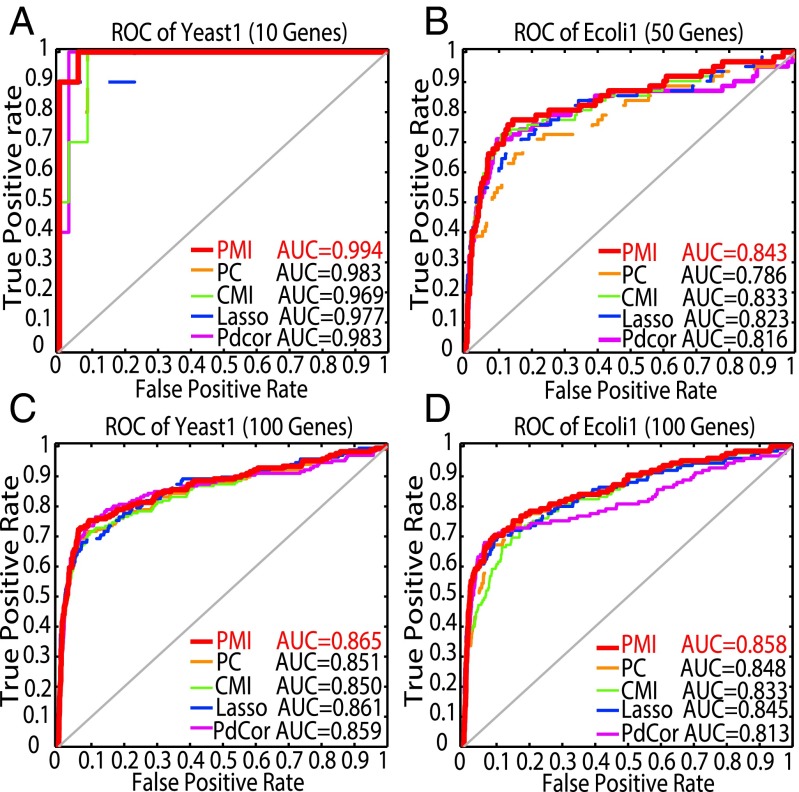
We first analyzed the yeast knock-out gene expression data with 10 genes and 10 samples by PCA-PMI algorithm. If PMI of two genes is less than the threshold, we consider that there is no direct association between these two genes; otherwise, the two genes may have the direct regulation. The joint and marginal probabilities as well as covariance matrices were estimated by Eq. 12 in Theorem 1 with the assumption of the Gaussian distribution due to a large number of variables. Fig. 3A shows the true gene regulatory network, with 10 nodes and 10 edges, and Fig. 3C is the network inferred by PCA-PMI with 10 nodes and 9 edges, where only one edge between G4 and G9 was false-negative. For comparison, we used CMI-based PC-algorithm (PCA-CMI) to construct the same network as shown in Fig. 3B, and the threshold was selected when the accuracy was maximized. Fig. 3B shows that the network predicted by CMI has two false-negative edges, the edge between G1 and G3 and the edge between G4 and G9. Clearly, PMI is more accurate than CMI for this case. To further evaluate the performance of PMI, the AUC was reported. Fig. 4A shows the ROC curve of yeast network with 10 genes, where PMI outperforms CMI apparently in terms of AUC. We also compared PMI with partial correlation, Lasso, and Pdcor, where PMI performed the best.
Fig. 3.
Comparison of yeast gene regulatory network reconstructed by PMI- and CMI-based algorithms. (A) The true network of yeast with 10 genes and 10 edges from DREAM3 challenge datasets. (B) The network predicted by CMI with 10 genes and 10 edges, where two false-negative edges are marked by dashed lines. (C) The network predicted by PMI with 10 genes and 9 edges, where one false-negative edge is marked by a dashed line.
Fig. 4.
ROC curves of the Yeast1 gene regulatory network with gene sizes 10 and 100, and E. coli gene regulatory network with gene sizes 50 and 100 by PMI, CMI, PC, Lasso, and Pdcor. (A–D) The ROC curves of the Yeast1 network with 10 genes and 100 genes and the Ecoli1 network with 50 genes and 100 genes. The red line is for PMI, the green line is for CMI, the orange line is for PC, the blue line is for Lasso, and the pink line is for Pdcor.
Next, we tested the data of yeast gene expression with 100 genes for datasets of 100 samples. We chose different thresholds ranging from 0 to 0.5 for PCA-PMI (see SI Appendix, Fig. S4A). The true network of 100 genes with 166 edges is the real and verified network. We selected the threshold 0.25 with the maximum accuracy 0.97. The ROC curve is shown in Fig. 4C. As a whole, PMI is superior to CMI in terms of ROC curves.
Finally, we performed PMI and CMI computation on two datasets of E. coli network with 50 and 100 genes. Fig. 4 B and D are the ROC curves for these data, where AUC values of PMI are 0.843 and 0.858, respectively, higher than those of CMI. SI Appendix, Fig. S4 A and B show that the accuracy is consistent with changing threshold between 0 and 0.5. As shown in SI Appendix, Fig. S4, with the increase of the network size, the accuracy is improved. We also compared PMI with the partial correlation, Lasso, and Pdcor, and PMI performed the best in all the cases in Fig. 4. In all datasets from the DREAM3 challenge, PMI was verified to be efficient and effective to reconstruct real gene regulatory networks from the measured expression data. Interestingly, when replacing the covariance matrix of the measured variables by the distance covariance matrix (8), we have kPCA-PMI algorithm (see SI Appendix), which has better performance on the above cases than PCA-PMI, shown in SI Appendix, Fig. S5.
Discussion
MI based on mutual independence and CMI based on conditional independence suffer from the problems of overestimation and underestimation, respectively. However, recently, the causal strength CX→Y(X;Y|Z) (see SI Appendix, Property S9) for quantifying causal strength was proposed by Janzing et al. (18). The causal strength (CS) can be used to quantify causal influences among variables in a network. However, CS is asymmetric compared with CMI, and it partially solves the problem of CMI (see SI Appendix, Property S9). In this work, to solve these problems we propose the novel concept of PMI by defining a new “partial independence” in Eq. 6 that can measure nonlinearly direct dependencies between variables with higher statistical power. Owing to this partial independence, PMI can quantify the direct associations effectively by theoretically avoiding the false-negative/-positive problems, in contrast to CMI and also other measures. Moreover, PMI can be also used for reconstructing a network with or without loops. If there is no influence on the two variables of interest from other variables in a network, PMI for measuring the association of these two variables is reduced to MI. Combining with PC-algorithm (16), PMI can efficiently reconstruct various types of large-scale networks from the measured data, such as gene regulatory networks in living organisms. In addition to the theoretical results, both simulated and real data demonstrate the effectiveness of PMI as a quantitative measure for nonlinearly direct associations.
To theoretically define PMI, we introduced partial independence in Eq. 6, in contrast to the conditional independence in Eq. 5 for CMI and mutual independence in Eq. 2 for MI. As shown in SI Appendix, Property S8 and Fig. S1, as well as the computational results, the conditional independence Eq. 5 always holds approximately for the case that X (or/and Y) is strongly associated with Z, even if there is strong dependence of X and Y given Z, but the partial independence Eq. 6 does not necessarily hold. Therefore, when the associations among variables in a network or system are weak, both PMI and CMI can correctly quantify the direct dependencies, but when some associations among variables are moderate or strong (it is actually a general case in a network or system), CMI generally failed due to its false-negative features and actually the error increases with the association strength, whereas PMI can detect their true dependencies due to the effect of the partial independence.
Intuitively, if X (or Y) is strongly associated with Z, CMI(X;Y|Z) = H(X|Z) − H(X|Y,Z) vanishes due to H(X|Z) H(X|Y,Z) 0 because knowing Z leaves almost no uncertainty about X (or Y) from the viewpoint of conditional independence. In other words, strong dependency between X and Z (or between Y and Z) makes the conditional dependence of X and Y almost invisible when measuring CMI(X;Y|Z) only (18). However, PMI(X;Y|Z) can measure correctly for this case because the partial independence makes the conditional dependence of X and Y visible again by replacing p(x|z) p(y|z) with p*(x|z) p*(y|z), where p*(x|z) [or p*(y|z)] implicitly includes the association information between X and Y, different from p(x|z) [or p(y|z)].
We also derived an extended KL divergence (see SI Appendix, Definition S1 and Theorem S1). The difference between PMI and CMI is the two conditional probability distributions [p(x|z)p(y|z) and p*(x|z)p*(y|z)] (see SI Appendix, Fig. S1), and their difference is clearly related to the relation between X and Y (see SI Appendix, Fig. S1). For continuous random variables, PMI can be defined in the same way as Eqs. 7–9, that is,
where and . p(x,y,z) is now the joint probability density function of X, Y, and Z, and all others are the marginal (or conditional) probability density functions of random variables X, Y, and Z.
This work mainly introduced a new information criterion, PMI, to measure direct associations between nodes in a network based on the observed data, but it cannot give the directions of the measured associations, which should be detected by other methods, similar to CMI and MI. Also, PMI is not designed to test the hypothesis on the causal relations for observational studies (26, 27), for which CMI may be appropriate. In addition, how to numerically estimate PMI accurately from a small number of samples without the “bins”-based approximation or Gaussian distribution approximation is a topic for future research. To serve the communities of biology and medicine for inferring gene regulatory networks from high throughput data, all of the source codes and the web tool of our algorithm PCA-PMI can be accessed at www.sysbio.ac.cn/cb/chenlab/software/PCA-PMI/.
Supplementary Material
Acknowledgments
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB13040700 and National Natural Science Foundation of China Grants 61134013, 91439103, and 91529303.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All analysis code reported in this paper is available at www.sysbio.ac.cn/cb/chenlab/software/PCA-PMI/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522586113/-/DCSupplemental.
References
- 1.Whittaker J. Graphical Models in Applied Multivariate Statistics. Wiley; Chichester, UK: 1990. [Google Scholar]
- 2.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 3.Reshef DN, et al. Detecting novel associations in large data sets. Science. 2011;334(6062):1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang YXR, Waterman MS, Huang H. Gene coexpression measures in large heterogeneous samples using count statistics. Proc Natl Acad Sci USA. 2014;111(46):16371–16376. doi: 10.1073/pnas.1417128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alipanahi B, Frey BJ. Network cleanup. Nat Biotechnol. 2013;31(8):714–715. doi: 10.1038/nbt.2657. [DOI] [PubMed] [Google Scholar]
- 7.Barzel B, Barabási A-L. Network link prediction by global silencing of indirect correlations. Nat Biotechnol. 2013;31(8):720–725. doi: 10.1038/nbt.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feizi S, Marbach D, Médard M, Kellis M. Network deconvolution as a general method to distinguish direct dependencies in networks. Nat Biotechnol. 2013;31(8):726–733. doi: 10.1038/nbt.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szekely GJ, Rizzo ML, Bakirov NK. Measuring and testing dependence by correlation of distances. Ann Stat. 2007;35(6):2769–2794. [Google Scholar]
- 10.Kosorok MR. On Brownian distance covariance and high dimensional data. Ann Appl Stat. 2009;3(4):1266–1269. doi: 10.1214/09-AOAS312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szekely GJ, Rizzo ML. Partial distance correlation with methods for dissimilarities. Ann Stat. 2014;42(6):2382–2412. [Google Scholar]
- 12.Frenzel S, Pompe B. Partial mutual information for coupling analysis of multivariate time series. Phys Rev Lett. 2007;99(20):204101. doi: 10.1103/PhysRevLett.99.204101. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber T. Measuring information transfer. Phys Rev Lett. 2000;85(2):461–464. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- 14.Kinney JB, Atwal GS. Equitability, mutual information, and the maximal information coefficient. Proc Natl Acad Sci USA. 2014;111(9):3354–3359. doi: 10.1073/pnas.1309933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunel H, et al. MISS: A non-linear methodology based on mutual information for genetic association studies in both population and sib-pairs analysis. Bioinformatics. 2010;26(15):1811–1818. doi: 10.1093/bioinformatics/btq273. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, et al. Inferring gene regulatory networks from gene expression data by path consistency algorithm based on conditional mutual information. Bioinformatics. 2012;28(1):98–104. doi: 10.1093/bioinformatics/btr626. [DOI] [PubMed] [Google Scholar]
- 17.Hlavackova-Schindler K, Palus M, Vejmelka M, Bhattacharya J. Causality detection based on information-theoretic approaches in time series analysis. Phys Rep. 2007;441(1):1–46. [Google Scholar]
- 18.Janzing D, Balduzzi D, Grosse-Wentrup M, Schölkopf B. Quantifying causal influences. Ann Stat. 2013;41(5):2324–2358. [Google Scholar]
- 19.Zhang X, Zhao J, Hao JK, Zhao XM, Chen L. Conditional mutual inclusive information enables accurate quantification of associations in gene regulatory networks. Nucleic Acids Res. 2015;43(5):e31. doi: 10.1093/nar/gku1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roulston MS. Significance testing of information theoretic functionals. Physica D. 1997;110(1–2):62–66. [Google Scholar]
- 21.Kraskov A, Stögbauer H, Grassberger P. Estimating mutual information. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;69(6 Pt 2):066138. doi: 10.1103/PhysRevE.69.066138. [DOI] [PubMed] [Google Scholar]
- 22.Darbellay GA, Vajda I. Estimation of the information by an adaptive partitioning of the observation space. Ieee T Inform Theory. 1999;45(4):1315–1321. [Google Scholar]
- 23.Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: Detecting and evaluating dependencies between variables. Bioinformatics. 2002;18(Suppl 2):S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- 24.Marbach D, et al. Revealing strengths and weaknesses of methods for gene network inference. Proc Natl Acad Sci USA. 2010;107(14):6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffter T, Marbach D, Floreano D. GeneNetWeaver: In silico benchmark generation and performance profiling of network inference methods. Bioinformatics. 2011;27(16):2263–2270. doi: 10.1093/bioinformatics/btr373. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbaum PR. Observational Studies. Springer; Berlin: 2002. [Google Scholar]
- 27.Hernan MA, Robins JM. Causal Inference. CRC; Boca Raton, FL: 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.