Significance
Callus induction is an initial step for typical plant in vitro regeneration, and recent studies show that auxin-induced callus formation in multiple organs occurs from the pericycle or pericycle-like cells via a root developmental pathway. We demonstrate here that very-long-chain fatty acids (VLCFAs) or their derivatives act as the critical signal in restricting the callus-forming capacity of the pericycle and thus the regeneration capability in Arabidopsis. Our work not only discloses an unidentified role of VLCFAs in defining the regeneration capacity, but also sheds light on the signals that govern the cell states in plant organs. Our findings also may have relevance for investigating the possible role of VLCFAs in the regulation of cell states in animals.
Keywords: VLCFA, pericycle, callus formation, regeneration
Abstract
The already differentiated organs in plants have a remarkable capacity to regenerate new individuals under culture conditions. Plant in vitro regeneration practically starts with the induction of a pluripotent cell mass, the callus, from detached organs on auxin-rich callus-inducing medium (CIM), which is generally required for subsequent regeneration of new bodies. Recent studies show that CIM-induced callus formation occurs from the pericycle or pericycle-like cells through a root developmental pathway, whereas the signals involved in governing callus-forming capacity of pericycle cells remain unknown. Here we report that very-long-chain fatty acids (VLCFAs) play a critical role in confining the pericycle competence for callus formation and thus the regeneration capacity of Arabidopsis. By genetic screening, we identified the callus formation-related 1 (cfr1) mutant, which bypasses the inhibition of callus-forming capacity in roots by solitary-root (slr/iaa14). We show that CFR1 encodes 3-ketoacyl-CoA synthase 1 (KCS1), which catalyzes a rate-limiting step of VLCFA biosynthesis. Our biochemical and genetic analyses demonstrate that VLCFAs restrict the pericycle competence for callus formation, at least in part, by regulating the transcription of Aberrant Lateral Root Formation 4 (ALF4). Moreover, we provide evidence that VLCFAs act as cell layer signals to mediate the pericycle competence for callus formation. Taken together, our results identify VLCFAs or their derivatives as the confining signals for mediating the pericycle competence for callus formation and thus the regeneration capacity of plant organs.
In plants, the already differentiated organs or tissues have a remarkable capability to regenerate new organs or entire individuals under appropriate culture conditions (1, 2). The initial step of a typical plant in vitro regeneration often starts with the induction of a pluripotent cell mass known as a callus from detached organs (explants) on auxin-rich callus-inducing medium (CIM), which is generally required for the subsequent regeneration of new organs or whole plant bodies (3, 4). Thus, callus formation has long been considered a process through which already-differentiated somatic cells acquire regenerating capability (2).
The molecular events of callus formation have begun to be described only recently (2, 5). Studies with Arabidopsis explants of multiple organs, including roots, hypocotyls, and petals, have revealed that the CIM-induced callus formation occurs from pericycle or pericycle-like cells, and that the derived calli resemble some characteristics of root meristem by ectopically expressed root meristem genes (6, 7). The recent findings that the four Arabidopsis lateral organ boundary domain (LBD) transcription factors play key roles in directing CIM-induced callus formation, and that the root meristem PLETHORA (PLT) genes are required for subsequent regeneration, supports the idea that CIM-induced callus formation from the pericycle follows a root developmental program (8, 9). In contrast, wound-induced callus formation has been shown to be directed by the AP2 transcription factors WOUND INDUCED DEDIFFERENTIATIONs (WINDs). The wound-induced callus does not exhibit the expression of root meristem genes, and its formation is not blocked in the mutant defective in lateral root formation (10), suggesting that WIND-mediated callus formation likely represents a cell dedifferentiation program in plants (5).
Because CIM-directed callus formation occurs from pericycle or pericycle-like cells, the appropriate competence of pericycle or pericycle-like cells appears to be critical for CIM-induced callus formation and thus the regeneration capacity of various organs. Indeed, specific ablation of the pericycle function in Arabidopsis by the pericycle-specific transactivation of a diphtheria toxin chain A effector indeed abolishes both lateral root formation and CIM-induced callus formation (11, 12). The Arabidopsis Aberrant Lateral Root Formation 4 (ALF4), which encodes a nuclear protein expressed in multiple organs and was initially shown to modulate lateral root formation and other developmental processes (13), was recently demonstrated to be involved in the regulation of pericycle competence for CIM-induced callus formation. Disruption of ALF4 leads to the loss of callus-forming capability in multiple organs, including roots, cotyledons, and petals (7). The protoplasts prepared from alf4-1 plants fail to reinitiate cell division (14), suggesting that ALF4 may be required for pericycle and possibly other cell types to enter the regeneration programs. However, the signals governing ALF4-mediated pericycle competence remain unclear.
The very-long-chain fatty acids (VLCFAs) generally include fatty acids with an acyl chain length of ≥18 carbons, which are biosynthesized by the fatty acid elongase complex that sequentially adds two carbons into the acyl chain (15). The fatty acid elongase complex in plants consists of ketoacyl-CoA synthase (KCS), ketoacyl-CoA reductase (KCR), 3-hydroxy acyl-CoA dehydratase (HCD, also known as PASTICCINO 2, or PAS2), and enoyl-CoA reductase (ECR) (16–19). Recent studies suggest that the VLCFAs or their derivatives, such as cuticular lipids, phospholipids, and sphingolipids, are not only components of protective barriers or cell membranes, but also may act as signaling molecules to mediate various biological processes. In mammals, VLCFAs have been shown to play important roles in cell apoptosis and cell differentiation, as well as in termination of cell proliferation (20–22). In plants, the Arabidopsis loss-of-function mutants pas2 and kcr1 are embryo-lethal, whereas their leaky alleles exhibit enlarged shoot apical meristems, fused rosette leaves, and altered lateral root branching (17, 18, 23). VLCFAs are also known to regulate programmed cell death during plant–pathogen interactions, to promote cell elongation in cotton fibers by activating ethylene biosynthesis, and to act as a cell layer signal to regulate cell proliferation in the Arabidopsis shoot apex by suppressing cytokinin biosynthesis (24–27).
Here we report that VLCFAs play a crucial role in restricting the competence of the pericycle for callus formation and thus the regeneration capacity in Arabidopsis. We provide evidence that VLCFAs act as cell layer signals to confine the pericycle competence for callus formation, at least in part, by inhibiting ALF4 transcription. Our findings indicate that VLCFAs or their derivatives serve as critical signals in mediating CIM-directed callus formation and hence the regeneration capacity in plants.
Results
cfr1 Bypasses the Inhibition of Callus-Forming Capacity by solitary-root.
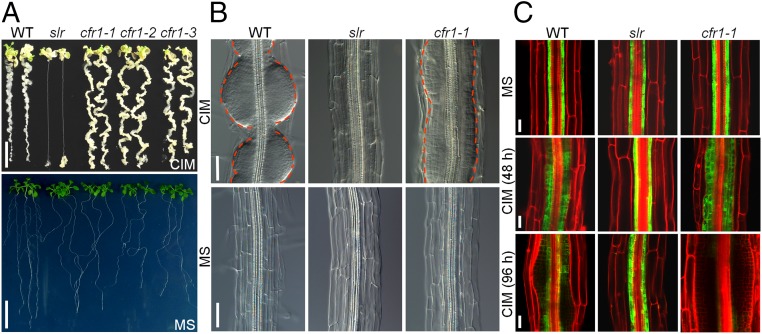
We previously demonstrated that the four Arabidopsis LBD transcription factors act downstream of auxin response factor (ARF) 7 and ARF19 to direct CIM-induced callus formation (8). To further explore the molecular basis of plant regeneration, we performed a genetic screen with ethyl methanesulfonate (EMS)-mutagenized solitary-root (slr, also known as iaa14) plants (28), based on the knowledge that the primary roots of slr, with the exception of root apical meristems, are incapable of forming calli on CIM (Fig. 1A). This screen allowed us to identify several mutants, termed callus formation-related (cfr), by their apparently callus-forming phenotype in their primary roots (Fig. 1A). Three of the cfr mutants displayed a similar phenotype, and genetic analyses showed that they resulted from a recessive mutation in a single gene and were genetically allelic to each other; thus, they were named cfr1-1, cfr1-2, and cfr1-3 (Fig. 1A).
Fig. 1.
cfr1 enhances the callus-forming capacity of pericycle. (A) Callus-forming phenotype (Upper) and morphology (Lower) of WT, slr, and three alleles of cfr1 seedlings. (Scale bars: 1 cm.) (B) Cytology of callus formation (Upper) and cellular organization (Lower) of the mature zone in WT, slr, and cfr1-1 roots. (Scale bars: 50 µm.) (C) Expression of the pericycle marker J0121 in the mature zone of WT, slr, and cfr1-1 roots on MS or CIM. (Scale bars: 25 µm.)
The cfr1 seedlings exhibited a strong callus-forming phenotype throughout the primary roots when incubated on CIM, which restored the defect in callus formation of the slr roots (Fig. 1A and Fig. S1A). However, when grown on the medium lacking plant hormones, cfr1 seedlings were still defective in lateral root initiation and gravitropism, as were the slr roots (Fig. 1A and Fig. S1 B and C). Moreover, like slr, cfr1 mutants still displayed hyposensitivity to exogenous auxin in initiating the lateral roots (Fig. S1B), suggesting that the overall auxin responses are not altered in cfr1 plants. In addition, the cfr1 and slr plants grown in soil had a similar morphology, including small rosette leaves, short inflorescence stems, and enhanced apical dominance (Fig. S1 D–F). These observations demonstrate that the mutation in CFR1 could bypass the callus-forming capacity inhibited by slr.
Fig. S1.
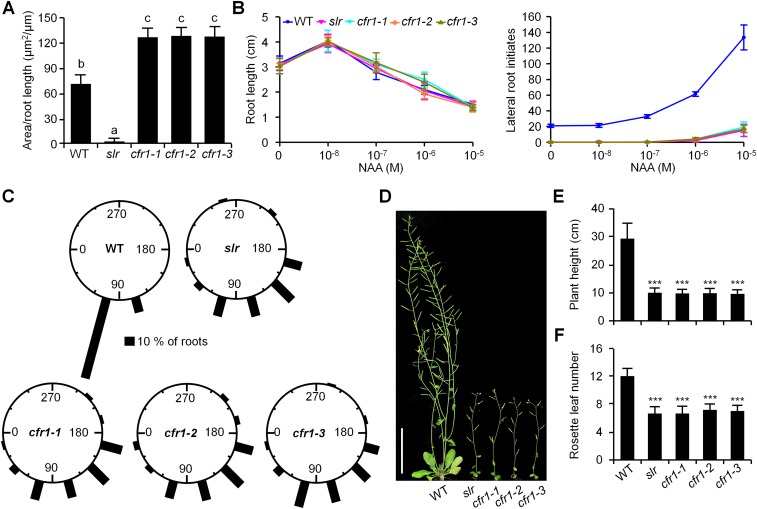
Phenotypic characterization of cfr1 mutants. (A) Area of pericycle or pericycle-derived callus layer in longitudinal direction of the WT, slr, and three cfr1 allele roots incubated on CIM for 4 d. n = 18. Error bars are SD. Significance was determined by one-way ANOVA with Tukey's test. *P < 0.05. (B) Primary root length and lateral root initiates of WT, slr, and three alleles of cfr1 incubated on the MS containing different concentrations of NAA. n = 18. Error bars are SD. (C) Gravitropism of WT, slr, and three alleles of cfr1 roots. Here 4-d-old seedlings subjected to gravitropic assay and reorientation of roots for 50 seedlings in each genotype after 24 h were assigned to 1 of 12 30° sectors. (Scale bar: 10%.) (D and E) Morphology (D) and plant height (E) of 50-d-old seedlings of WT, slr, and cfr1 alleles. n = 19. Error bars are SD. Significance was determined by Student’s t test. ***P < 0.001. (Scale bar: 5 cm.) (F) Rosette leaf numbers of the WT, slr, and cfr1 plants before bolting. n = 19. Error bars are SD. Significance was determined by Student’s t test. ***P < 0.001.
cfr1 Enhances Pericycle Competence for Callus Formation.
We then used differential interference contrast (DIC) microscopy to compare the cytological characteristics of mature region of primary roots in WT, slr, and cfr1-1 plants before and after incubation on CIM. Before being transferred to CIM, the cfr1-1, slr, and WT roots had the same arrangement of cell layers with a normally organized structure (Fig. 1B); however, after seedlings were incubated on CIM, the callus formation in WT occurred at regular intervals from pericycle cells with a structure of lateral root-like initials, whereas the pericycle cells of cfr1-1 proliferated along the entire roots, leading to formation of a continuous callus layer without an apparent interval structure (Fig. 1B). We next visualized the expression of J0121, a widely used pericycle identity marker (11), in WT, slr, and cfr1-1 roots before and after incubation on CIM. Similarly, the expression of J0121 in cfr1-1 roots was similar to that in both WT and slr roots before incubation on CIM (Fig. 1C). After seedlings were incubated on CIM, J0121 expression was exclusively maintained in pericycle cells of slr primary roots but gradually decreased in the WT pericycle cells in which the initial structures developed (Fig. 1C). In contrast, J0121 signals became disappeared in the entire cfr1-1 pericycle where the extensive cell proliferation occurred (Fig. 1C). These observations indicate that the callus in cfr1 originates from pericycle cells, and that the pericycle cells of cfr1 have a high competence to enter the callus-forming program.
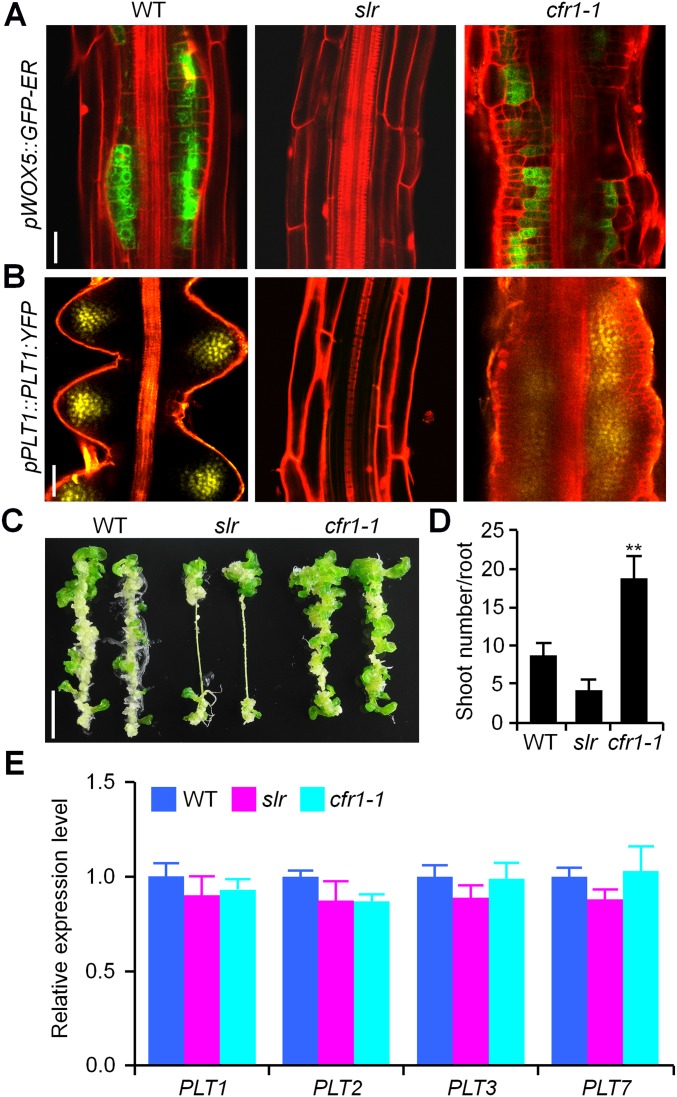
To determine whether the calli derived from cfr1 have root meristem characteristics, we monitored the expression of pWOX5::GFP-ER and pPLT1::PLT1:YFP, two markers expressed in the root meristem and recently shown to be characteristic markers of CIM-induced calli derived from multiple organs (7, 29, 30). The fluorescent signals were detected in both WT and cfr1-1 calli, but not in slr pericycle cells (Fig. S2 A and B), indicating that the callus derived from cfr1 has the property of root meristems. We incubated cfr1-1 seedlings on CIM for 12 d and then transferred them to shoot-inducing medium (SIM), and observed that adventitious shoots regenerated efficiently from cfr1-1 calli (Fig. S2 C and D).
Fig. S2.
Characteristic and regeneration capability of calli derived from cfr1. (A and B) Expression of WOX5 (A) or PLT1 (B) in the mature zone of WT, slr, and cfr1-1 roots incubated on CIM. Here 7-d-old seedlings carrying a pWOX5::GFP-ER (A) or pPLT1::PLT1:YFP (B) construct were incubated on CIM for 4–6 d. (Scale bars: 25 µm.) (C) Shoot regeneration of the calli derived from WT, slr, and cfr1-1. (Scale bar: 1 cm.) (D) Number of adventitious shoots regenerated on the root-derived callus of WT, slr, and cfr1-1. n = 18. Error bars are SD. Significance was determined by Student’s t test. **P < 0.01. (E) Expression of PLT1, PLT2, PLT3, and PLT7 genes in the calli of WT, slr, and cfr1-1. n = 3. Error bars are SD.
To test whether cfr1 has an effect on the shoot-regenerating capability of the callus, we compared the expression of PLT genes, which is considered to reflect the shoot-regenerating competence of calli (9), in WT, slr, and cfr1-1 root-derived calli. We observed that expression levels of these PLT genes were comparable among the three genotypes (Fig. S2E), suggesting that cfr1 might affect mainly the callus-forming capacity of the pericycle rather than the regenerating capability of derived calli.
CFR1 Encodes the 3-Ketoacyl-CoA Synthase 1 (KCS1).
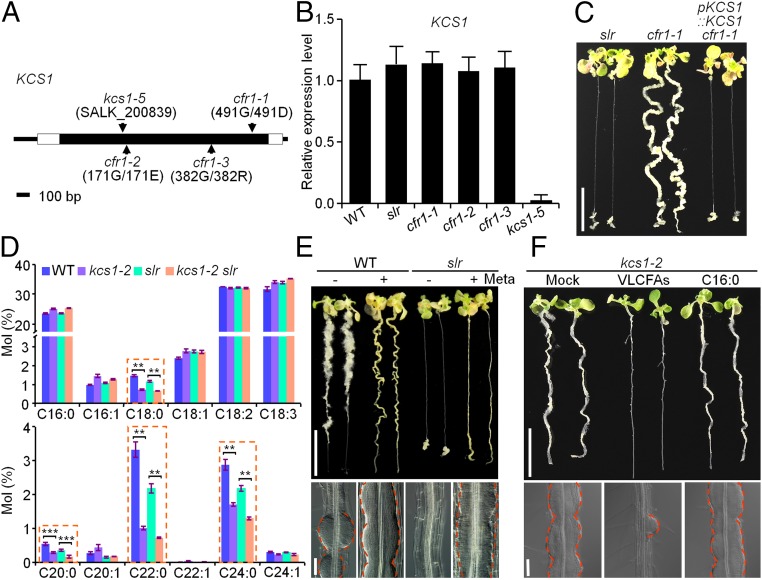
Using an F2 population of cfr1-1 crossed with the Arabidopsis Landsberg erecta (Ler) ecotype, we finely mapped CFR1 to a region of ∼110 kb on chromosome 1. Sequencing of genes in this region in the cfr1-1 genome enabled the identification of a G-to-A transition at a position +1,472 bp from the start codon of KCS1 that resulted in an amino acid substitution of 491Gly to 491Asp in KCS1 (Fig. 2A). Further sequencing of KCS1 in cfr1-2 and cfr1-3 validated that the coding regions of KCS1 contained allelic mutations (Fig. 2A). Expression analysis showed that the KCS1 transcript levels in the three cfr1 alleles were comparable to those in WT and slr plants (Fig. 2B). We then introduced the WT KCS1 genomic DNA (∼4.5 kb, including a 2.7-kb promoter region) into cfr1-1, and found that the callus-forming phenotype in cfr1-1 roots was fully blocked in these transgenic plants (Fig. 2C), indicating that the KCS1 mutation confers the enhanced callus-forming phenotype observed in the cfr1 plants.
Fig. 2.
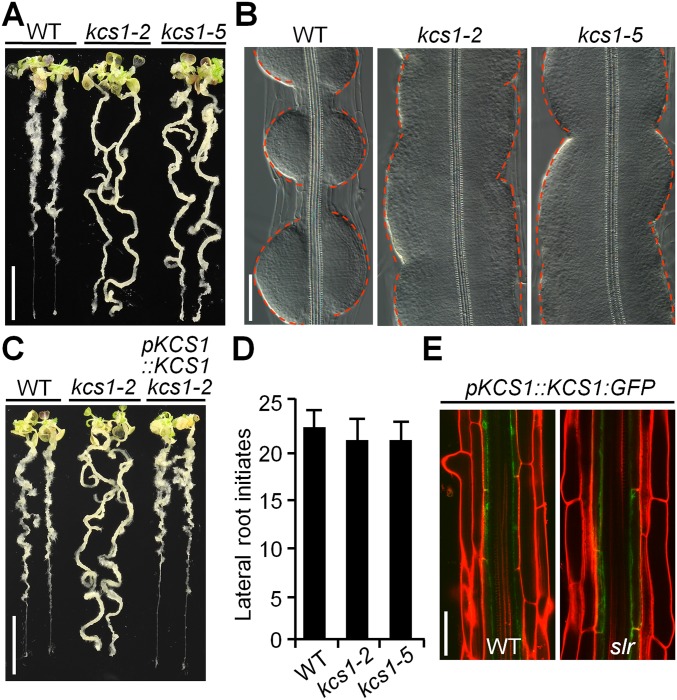
VLCFAs restrict the pericycle competence for callus formation. (A) Map-based cloning and sequencing showing the mutation sites of cfr1 in the coding region of KCS1. The T-DNA insertion site of kcs1-5 is also indicated. (B) The expression level of KCS1 in WT, slr, the cfr1 alleles, and kcs1-5. n = 3 biological replicates. Error bars are SD. (C) Callus-forming phenotype of slr, cfr1-1, and transgenic cfr1-1 seedlings carrying pKCS1::KCS1. (Scale bar: 1 cm.) (D) Total fatty acid composition of WT, kcs1-2, slr, and kcs1-2 slr roots. n = 3 biological replicates. Error bars are SD. Significance was determined by Student’s t test. **P < 0.01; ***P < 0.001. (E) Effect of metazachlor (Meta) on pericycle competence for callus formation. WT and slr seedlings were incubated on CIM supplemented with (+) or without (−) exogenous application of 5 µM metazachlor. (Scale bars: 1 cm in Upper, 50 µm in Lower.) (F) Exogenous VLCFAs inhibit callus-forming capacity in the kcs1-2 pericycle. kcs1-2 seedlings were incubated on CIM supplemented with VLCFAs (a mixture of C18:0, C20:0, C22:0, and C24:0) or C16:0 fatty acids; the tert-butyl methyl ether for dissolving VLCFAs served as a control (Mock). (Scale bars: 1 cm in Upper, 50 µm in Lower.)
We then crossed cfr1 mutants with WT and obtained kcs1 mutants that lacked the slr mutation. Because the kcs1-1 mutant has been previously characterized in Arabidopsis (31), we designated the newly identified alleles as kcs1-2, kcs1-3, and kcs1-4 and the cfr1 allele as kcs1 slr. We also obtained a T-DNA insertion mutant (SALK_200839), kcs1-5, from the Arabidopsis Biological Resource Center (ABRC), in which a T-DNA sequence was inserted in the KCS1 coding region and KCS1 mRNA was undetectable (Fig. 2 A and B). The kcs1-2 and kcs1-5 seedlings incubated on CIM still exhibited the enhanced callus-forming phenotype in their primary roots, with an additive morphology of WT and kcs1 slr roots by flattened initial structures (Fig. S3 A and B). Further introduction of WT KCS1 genomic DNA into kcs1-2 fully restored the callus-forming phenotype of kcs1-2 to the morphology observed in WT (Fig. S3C). Moreover, lateral root formation in kcs1-2 and kcs1-5 was not altered (Fig. S3D), and KCS1 accumulation in slr roots was similar to that in WT (Fig. S3E). These results support the idea that the KCS1-mediated pericycle competence for callus formation is independent of SLR-modulated lateral root formation.
Fig. S3.
Enhanced callus-forming capacity of pericycle in kcs1 mutants. (A and B) Callus-forming phenotype (A) and cytological characteristics (B) in the roots of WT, kcs1-2, and kcs1-5 seedlings on CIM. (Scale bars: 1 cm in A, 50 µm in B.) (C) Callus formation in WT, kcs1-2, and kcs1-2 plants carrying a pKCS1::KCS1 construct. Here 7-d-old seedlings were incubated on CIM for 12 d. (Scale bar: 1 cm.) (D) Lateral root initiates in 10-d-old WT, kcs1-2, and kcs1-5 seedlings. n = 18. Error bars are SD. (E) Accumulation of KCS1 in the roots of transgenic pKCS1::KCS1:GFP and pKCS1::KCS1:GFP slr plants. (Scale bar: 50 µm.)
VLCFAs Play an Inhibitory Role in Confining Pericycle Competence for Callus Formation.
KCS1 is a part of the fatty acid elongase complex and catalyzes a rate-limiting step in VLCFA biosynthesis (Fig. S4A) (15, 31). To test whether the VLCFAs are responsible for altered pericycle competence for callus formation in kcs1 and kcs1 slr, we first compared the total fatty acid levels in roots of WT, kcs1-2, slr, and kcs1-2 slr. In agreement with the known function of KCS1 in VLCFA biosynthesis, the saturated VLCFA levels for C18:0, C20:0, C22:0, and C24:0 in kcs1-2 and kcs1-2 slr were only approximately 30–60% of those in WT and slr plants (Fig. 2D).
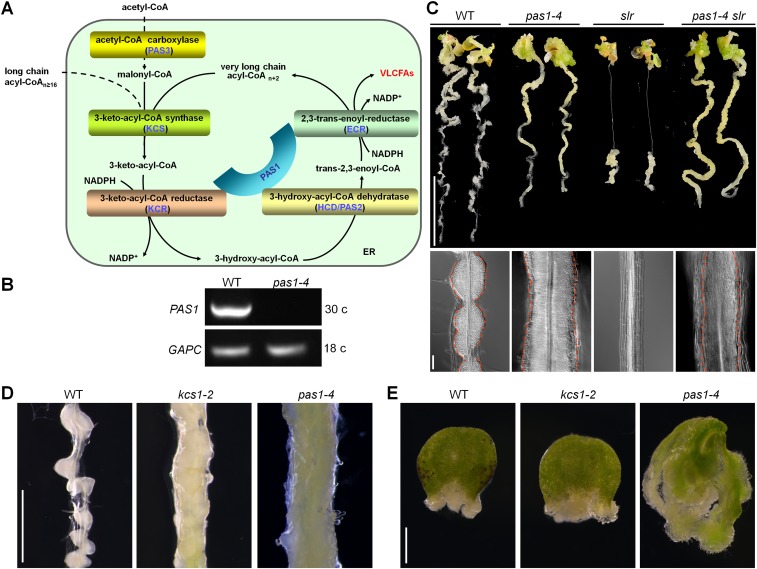
Fig. S4.
Callus formation in VLCFA-deficient mutants. (A) Schematic representation of the fatty acid elongase complex. VLCFAs are biosynthesized by the elongase complex composed of KCS, KCR, HCD (also known as PAS2), and ECR. (B) RT-PCR analysis of PAS1 expression in WT and pas1-4 (SALK_051324) plants. (C) Callus-forming phenotype of WT, pas1-4, slr, and pas1-4 slr plants. Phenotypic and cytological characterizations were performed with 7-d-old seedlings incubated on CIM for 12 d (Upper) or 4 d (Lower), respectively. (Scale bars: 1 cm in Upper, 50 µm in Lower.) (D and E) Callus-forming phenotype in the hypocotyl (D) and cotyledon (E) of WT, kcs1-2, and pas1-4 plants. (Scale bars: 1 mm.)
We next incubated WT and slr seedlings on CIM supplemented with metazachlor, a known inhibitor of VLCFA biosynthesis that acts by inhibiting the activities of KCS1 and other KCS enzymes (32). As expected, the WT and slr roots incubated on CIM with metazachlor recapitulated the callus-forming morphology observed in the pericycle of kcs1 and kcs1 slr roots, respectively (Fig. 2E). We then incubated the kcs1-2 seedlings on CIM supplemented with a mixture of VLCFAs (C18:0, C20:0, C22:0, and C24:0) or their precursor C16:0 fatty acids, and observed that the exogenous application of VLCFAs, but not of C16:0 fatty acids, almost fully blocked the callus-forming capacity of kcs1-2 roots (Fig. 2F). We also obtained a T-DNA insertion mutant (SALK_051324), pas1-4, from the ABRC, in which the PASTICCINO 1 (PAS1) that encodes a scaffold protein of the fatty acid elongase complex was disrupted (Fig. S4 A and B) (33). Consistently, pas1-4 seedlings grown on CIM also displayed an enhanced callus-forming phenotype, as did kcs1, and introduction of pas1-4 into slr resulted in bypassed callus formation in pas1-4 slr roots (Fig. S4C). These findings demonstrate that VLCFAs play an inhibitory role in confining pericycle competence for callus formation.
We next tested whether VLCFA deficiency affects the callus-forming capacity of aerial organs by incubating hypocotyls and cotyledons of kcs1-2 and pas1-4 on CIM. As shown in Fig. S4 D and E, although callus formation in the kcs1-2 cotyledon appeared to be slightly enhanced compared with that in WT, the strong callus-forming phenotype was observed in the kcs1-2 and pas1-4 hypocotyls and the pas1-4 cotyledon, implicating that VLCFAs also have an effect on the callus-forming capacity of aerial organs.
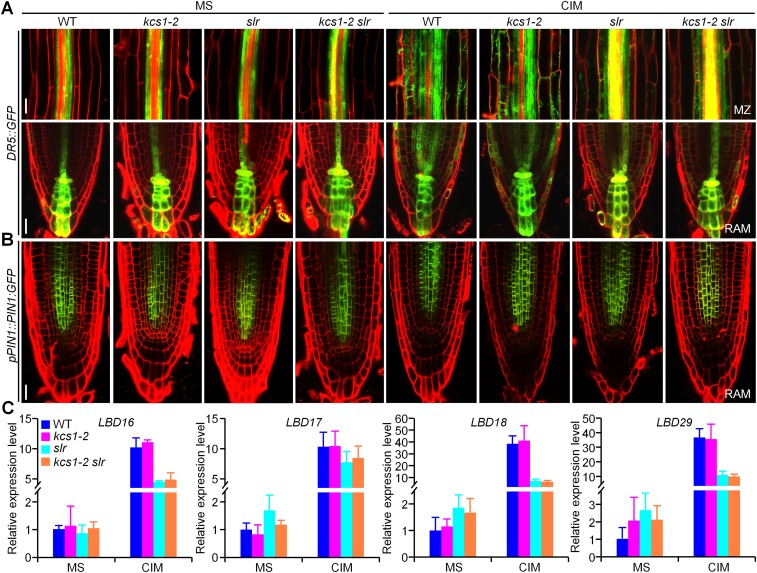
Because auxin plays an essential role in directing callus formation and VLCFAs have been suggested to regulate polar auxin transport on lateral root formation (3, 8, 33), we also explored whether the VLCFA-mediated pericycle competence for callus formation is related to endogenous auxin hemostasis or spatial accumulation. Careful comparison of DR5::GFP and PIN1:GFP in the roots of WT, kcs1-2, slr, and kcs1-2 slr before and after incubation on CIM revealed that the overall auxin accumulation in kcs1-2 or kcs1-2 slr roots was similar to that in WT or slr roots, respectively (Fig. S5 A and B). Likewise, the expression levels of auxin-induced LBD16, LBD17, LBD18, and LBD29, which are targets of SLR-ARF7/ARF19 (8, 34), were comparable in the kcs1-2 and WT roots but were reduced to a similar level in the slr and kcs1-2 slr roots (Fig. S5C). These observations suggest that VLCFA-mediated pericycle competence for callus formation is not attributable to endogenous auxin homeostasis or spatial accumulation.
Fig. S5.
Auxin accumulation and response in kcs1-2 and kcs1-2 slr. (A) Expression of DR5::GFP in the mature zone (MZ) and root apical meristem (RAM) of WT, kcs1-2, slr, and kcs1-2 slr roots before or after incubation on CIM for 6 h. (Scale bars: 25 µm.) (B) PIN1 accumulation in the RAM of WT, kcs1-2, slr, and kcs1-2 slr roots before or after incubation on CIM for 6 h. (Scale bar: 25 µm.) (C) qRT-PCR analysis of LBD16, LBD17, LBD18, and LBD29 expression in WT, kcs1-2, slr, and kcs1-2 slr roots before or after incubation on CIM for 24 h. n = 3. Error bars are SD.
ALF4 Acts Downstream of VLCFAs.
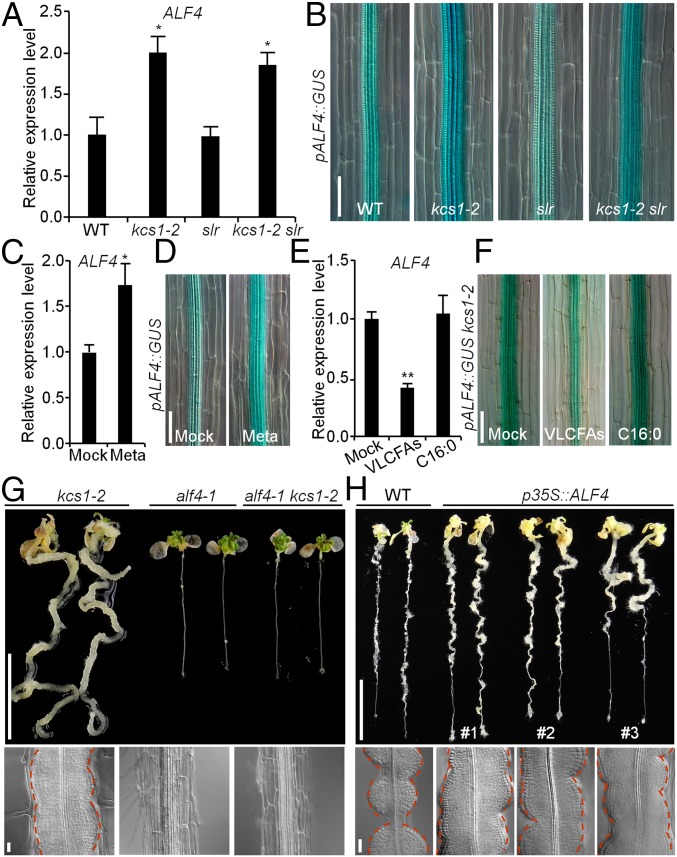
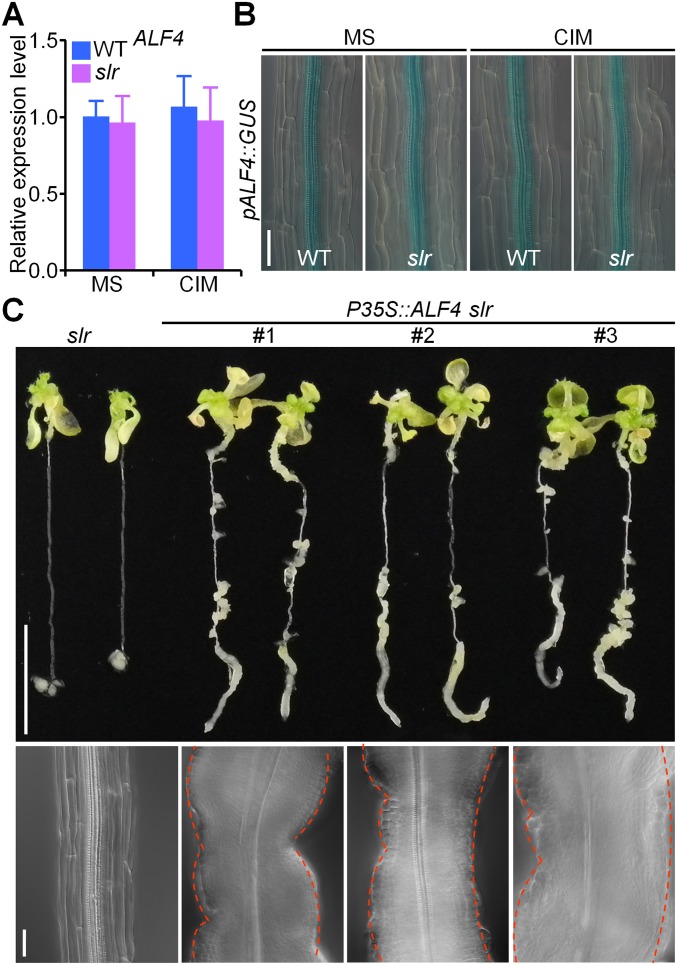
Because Arabidopsis ALF4 has been reported to be necessary for CIM-induced callus formation in multiple organs (7, 14), we speculated that ALF4 may be involved in VLCFA-mediated pericycle competence for callus formation. To test this, we first compared the expression of ALF4 in WT, kcs1-2, slr, and kcs1-2 slr seedlings. Our real-time quantitative RT-PCR (qRT-PCR) analysis showed that the ALF4 expression was indeed elevated by approximately twofold in kcs1-2 and kcs1-2 slr compared with that in WT and slr (Fig. 3A). This elevation was further validated by GUS staining assayed with the primary roots of transgenic plants harboring a pALF4::β-glucuronidase (GUS) construct (Fig. 3B). In contrast, ALF4 expression was found to be comparable in the WT and slr seedlings on either MS or CIM (Fig. 3 A and B and Fig. S6 A and B), implicating that the alteration of ALF4 transcription caused by kcs1 is independent of the slr mutation. Moreover, treatment with metazachlor resulted in an elevation of ALF4 expression in WT roots (Fig. 3 C and D), whereas exogenous application of the VLCFA mixture, but not the C16:0 fatty acids, caused decreased ALF4 transcription in kcs1-2 roots (Fig. 3 E and F). These results demonstrate that VLCFAs could suppress ALF4 transcription.
Fig. 3.
ALF4 acts downstream of VLCFAs. (A and B) qRT-PCR and GUS staining analyses of ALF4 expression in WT, kcs1-2, slr, and kcs1-2 slr. n = 3 biological replicates. Error bars are SD. Significance was determined by Student’s t test. *P < 0.05. (Scale bar: 50 µm.) (C and D) ALF4 expression in WT or pALF4::GUS seedlings treated with or without (Mock) 5 µM metazachlor (Meta). n = 3 biological replicates. Error bars are SD. Significance was determined by Student’s t test. *P < 0.05. (Scale bar: 50 µm.) (E and F) ALF4 expression in kcs1-2 or pALF4::GUS kcs1-2 seedlings treated with VLCFAs or C16:0 fatty acids. n = 3 biological replicates. Significance was determined by Student’s t test. **P < 0.01. (Scale bar: 50 µm.) (G and H) Callus-forming phenotype (Upper) and cytological morphology (Lower) of kcs1-2 alf4-1 plants (G) and transgenic p35S::ALF4 plants (H). (Scale bars: 1 cm in Upper, 20 µm in Lower.)
Fig. S6.
Ectopic expression of ALF4 partially rescues the callus-forming capacity of slr roots. (A and B) qRT-PCR (A) and GUS staining (B) analyses of ALF4 expression in WT and slr roots on MS and after incubation on CIM for 6 h. n = 3. Error bars are SD. (Scale bar: 50 µm.) (C) Callus-forming phenotype (Upper) and cytological characteristic (Lower) of slr and transgenic p35S::ALF4 slr plants. Seedlings incubated on CIM for 10 d and 6 d were used for morphological and cytological characterization, respectively. (Scale bars: 1 cm in Upper, 50 µm in Lower.)
To further examine the possibility that ALF4 acts downstream of VLCFAs, we generated alf4-1 kcs1-2 double-mutant plants by crossing kcs1-2 with alf4-1−/+ plants, and examined the callus-forming capacity of these plants when incubated on CIM. As shown in Fig. 3G, disruption of ALF4 completely blocked pericycle cells from forming callus in kcs1-2. Furthermore, the transgenic plants overexpressing ALF4 on CIM recapitulated the enhanced callus-forming phenotype observed in kcs1-2 (Fig. 3H), and the overexpression of ALF4 partially rescued the callus-forming defect in slr roots (Fig. S6C). These findings suggest that the inhibition of pericycle competence for callus formation by VLCFAs occurs, at least in part, through the regulation of ALF4 transcription.
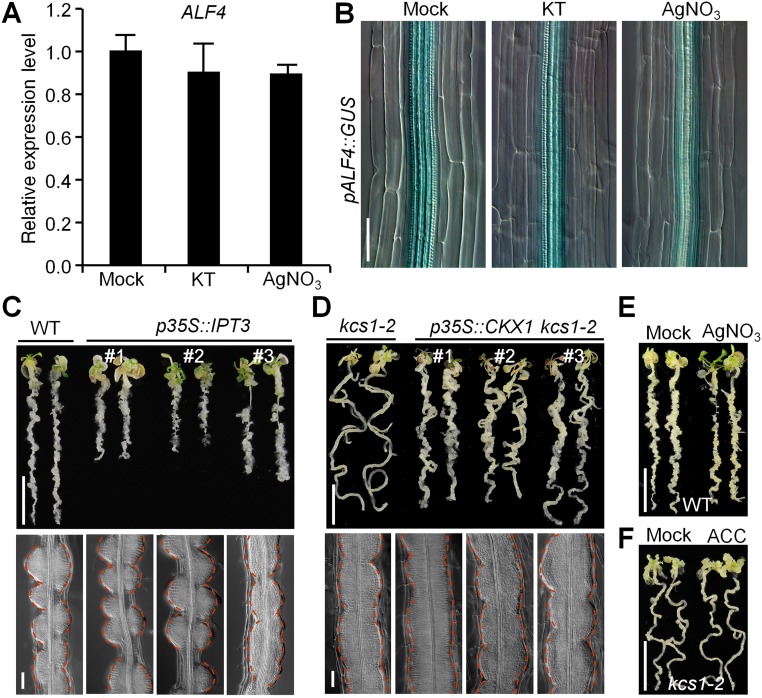
Because previous work has also shown that VLCFAs can repress cytokinin biosynthesis but activate ethylene biosynthesis (26, 27), we investigated whether VLCFA-mediated pericycle competence for callus formation is associated with cytokinin or ethylene homeostasis. Both qRT-PCR and GUS staining assays showed that ALF4 expression was not affected by the exogenous application of either kinetin or the widely used ethylene signaling inhibitor AgNO3 (35), demonstrating that ALF4 does not transcriptionally respond to cytokinin or ethylene (Fig. S7 A and B). The transgenic plants overexpressing ISOPENTENYLTRANSFERASE 3 (IPT3), a gene that encodes an enzyme that catalyzes a rate-limiting step in cytokinin biosynthesis (36), did not recapitulate the callus-forming phenotype observed in kcs1-2, and ectopic expression of Cytokinin Oxidase 1 (CKX1), which results in a cytokinin deficiency in transgenic plants (37), did not block or attenuate the callus-forming phenotype of kcs1-2 (Fig. S7 C and D). Similarly, application of AgNO3 to WT plants or the treatment of kcs1-2 with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) had no obvious effect on callus formation (Fig. S7 E and F). These observations suggest that VLCFA-modulated pericycle competence for callus formation does not rely on the alteration of cytokinin or ethylene homeostasis.
Fig. S7.
VLCFA-mediated pericycle competence does not rely on cytokinin or ethylene homeostasis. (A and B) ALF4 transcription is not responsive to cytokinin or AgNO3 treatment. Here 7-d-old WT and pALF4::GUS seedlings were treated with or without (Mock) 2 µM kinetin or 10 µM AgNO3 for 6 h, respectively. n = 3. Error bars are SD. (Scale bar: 50 µm.) (C and D) Callus formation (Upper) and cytological characteristics (Lower) of transgenic p35S::IPT3 (C) and p35S::CKX1 kcs1-2 (D) seedlings on CIM. (Scale bars: 1 cm in Upper, 50 µm in Lower.) (E and F) Effect of AgNO3 or ACC on pericycle competence for callus formation. Callus-forming phenotype of 7-d-old WT seedlings on CIM with or without (Mock) 10 µM AgNO3 (E) or kcs1-2 seedlings on CIM with or without 10 µM ACC (F) for 12 d. (Scale bars: 1 cm.)
VLCFAs as Cell Layer Signals in Confining Pericycle Competence for Callus Formation.
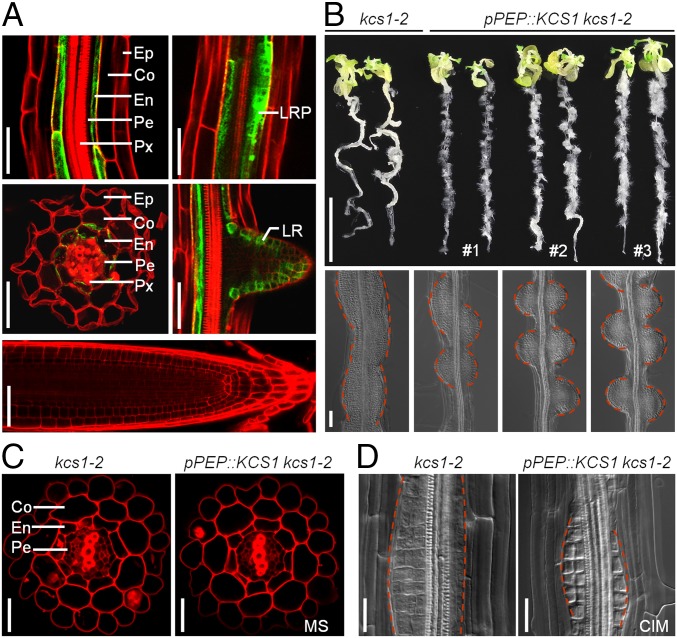
Arabidopsis KCS1 is expressed in almost all organs, including roots, stems, leaves, and flowers (16, 31). To examine whether KCS1 accumulates in the pericycle cells, we visualized KCS1 accumulation in the roots of transgenic kcs1-2 plants harboring a pKCS1::KCS1:GFP construct in which the enhanced callus-forming phenotype was blocked. As shown in Fig. 4A, abundant GFP signals were observed in the endodermis of primary roots, in the proliferating cells of lateral root primordium, and in the emerged lateral root; however, GFP signals were undetectable in the pericycle cells of mature zones and in the meristem region (Fig. 4A). This finding suggests that VLCFAs synthesized in the endodermis may act as cell layer signals to affect ALF4 expression in the pericycle and thus restrict pericycle competence for callus formation.
Fig. 4.
VLCFAs as cell layer signals in confining pericycle competence for callus formation. (A) KCS1 accumulation in roots of kcs1-2 plants harboring a pKCS1::KCS1:GFP construct. (Scale bars: 50 µm.) (B) Callus-forming phenotype (Upper) and cytological characteristics (Lower) in kcs1-2 seedlings carrying a pPEP::KCS1 construct. (Scale bars: 1 cm in Upper, 50 µm in Lower.) (C and D) Cell layer organization and callus origin in the mature zones of kcs1-2 and pPEP::KCS1 kcs1-2 roots. Sectioning and a clearing assay were performed with the roots on MS (C) and on CIM (D), respectively. (Scale bars: 20 µm.) Ep, epidermis; Co, cortex; En, endodermis; Pe, pericycle; Px, protoxylem; LRP, lateral root primordium; LR, lateral root.
To test this, we attempted to express KCS1 in the cortex of kcs1-2 roots by generating transgenic kcs1-2 plants expressing KCS1 driven by the promoter of Plastid Endopeptidase (PEP), which is expressed exclusively in the cortex of elongation and mature zones of roots (38). We observed that the cortex-expressed KCS1 could also suppress the enhanced callus-forming phenotype but did not affect the cell layer organization and callus origin in the kcs1-2 roots (Fig. 4 B–D). These findings support the VLCFAs or their derivatives as cell layer signals in confining the pericycle competence for callus formation and thus the regeneration capacity in plants.
Discussion
The maintenance of varied cell competences in an organ is critical for body construction in both animals and plants, and the properly maintained states of pericycle or pericycle-like cells within plant organs also greatly contribute to their remarkable regeneration capabilities (2, 7). Recent studies have suggested that ALF4 is critical for the pericycle competence for CIM-induced callus formation, whereas the signals and molecular basis that govern the pericycle competence for regeneration are unclear. Here we have demonstrated that a deficiency of VLCFAs in Arabidopsis enhances the callus-forming capacity of pericycle cells, whereas exogenous VLCFAs inhibit pericycle cells from forming the callus. We also provide evidence that VLCFAs act as cell layer signals to restrict the pericycle competence for callus formation partially through regulation of ALF4 transcription. These findings thus identify the VLCFAs or their derivatives as important signal molecules for mediating pericycle competence for the regeneration capacity of plant organs. More importantly, the signals that stringently maintain the differentiated states of the cells under developmental progression remain unclear (39), and thus it is likely that VLCFA-mediated ALF4 signaling is also necessary to maintain the optimal states of pericycle or pericycle-like cells and thereby prevent excess callus formation in response to external cues. Our findings may shed light on how plant cell states are stringently maintained during normal growth and development.
VLCFAs are components of the cellular membrane in animal and plant cells, and are present mainly in the form of sphingolipids and phospholipids (15). Increasing evidence suggests that VLCFAs or their derivatives are likely bioactive signals that mediate a variety of developmental processes and environmental responses. In plants, VLCFAs have been shown to mediate development as well as biotic and abiotic responses, including hypersensitive cell death and defense (15, 24–27, 40). In yeast and animals, VLCFAs also serve as precursors of bioactive lipid signaling molecules that regulate cell proliferation and apoptosis (20, 41, 42). Recent studies also suggest that VLCFAs or their derivatives participate in the regulation of animal cell differentiation or organ regeneration processes, such as activation of quiescent muscle stem cells known as satellite cells to proliferate in the process of skeletal muscle regeneration after injury (43, 44). Given that a large number of diverse metabolites are derived from VLCFAs (15, 21, 45), which VLCFA-derived molecule(s) act as signals and what signaling components are involved in regulation of specific biological events in both plants and animals remain unknown. Therefore, it is of interest to further define the molecules and signaling components involved in the regulation of pericycle competence for callus formation and to explore whether they have a similar role in animal cells. Any new knowledge gained through such work also would benefit the manipulation of cell pluripotency in both kingdoms.
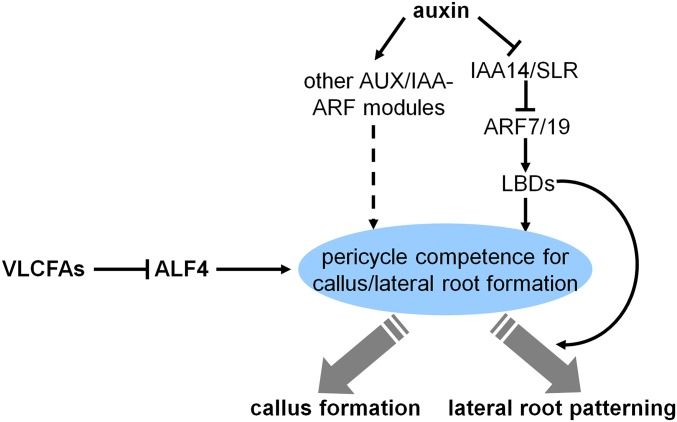
Our finding that the SLR-mediated lateral root initiation is not necessarily required for CIM-induced callus formation in kcs1 slr mutant also raises a question regarding the extent to which the lateral root formation and CIM-induced callus formation programs overlap. In Arabidopsis, the root pericycle is responsible for lateral root initiation and CIM-induced callus formation (6, 7, 11, 12), and the CIM-induced callus formation follows a root developmental pathway (6, 7). Indeed, several mutants defective in lateral root initiation, including slr, arf7 arf19, p35S::LBD16-SRDX, and alf4-1, display a compromised or blocked callus-forming phenotype on CIM (7, 8, 13, 28, 34). Surprisingly, we found that the enhanced callus-forming capacity in kcs1 is independent of slr mutation. Moreover, the severe mutant or transgenic plants deficient in VLCFA biosynthesis, such as pas1 or KCR1-RNAi plants, have been reported to exhibit retarded lateral root formation (17, 33). Thus, the VLCFA-mediated pericycle competence for callus formation is through a pathway independent of SLR-modulated lateral root formation (Fig. S8). Because the enhanced callus-forming phenotype of kcs1 slr and kcs1 is observed only on CIM, and the resulting calli still have root meristem characteristics, it is likely that the molecular events of the induction of pluripotent cells with root meristem characteristics by auxin are shared for both callus formation and lateral root initiation, whereas the other differentiation programs directed by SLR are still necessary for lateral root patterning (Fig. S8).
Fig. S8.
Proposed model for the roles of VLCFAs and auxin in callus and lateral root formation. The VLCFAs or derivatives serve as the signals to restrict the pericycle competence for callus and/or lateral root formation by repressing the ALF4 transcription, whereas the auxin-directed SLR-ARF7/19 and other IAA-ARF modules (e.g., IAA12-ARF5, IAA28-ARFs) play critical roles in directing the pericycle to form callus or lateral roots. The SLR pathway is still required for the lateral root patterning program. Thus, the pericycle cells with enhanced competence caused by deficiency of VLCFAs could bypass the inhibition of slr to form callus on CIM.
Finally, because auxin is a key phytohormone in directing pericycle-derived lateral root initiation and callus formation (3, 8, 46), and because previous studies have suggested that a deficiency of VLCFAs in the pas1 mutant results in an alteration of polar auxin distribution (33), the extent to which the VLCFA-mediated pericycle competence for callus formation is associated with auxin responses remains unclear. Although we observed that the overall auxin distribution and response in kcs1 and kcs1 slr plants are not obviously altered and previous work has also shown that ALF4 expression and subcellular localization of ALF4 are not regulated by auxin (13), the enhanced callus-forming phenotype in kcs1 and kcs1 slr is observed only on CIM, which contains excess amounts of the nontransportable auxin analog 2,4-dichlorophenoxyacetic acid (2,4-D) (47). Moreover, a recent study has suggested that the perturbed graft formation in alf4-1 occurs along with the decreased auxin responsiveness (48). Therefore, we could not exclude the possibility that VLCFA-mediated pericycle competence for callus formation is related to the alteration of auxin response or sensitivity of pericycle cells. Further work is still needed to clarify whether the pluripotent states of cells are closely associated with their auxin responsiveness or sensitivity in plants.
Materials and Methods
Plant Materials and Growth Conditions.
The cfr1 mutant was identified from an EMS-mutagenic population of the slr mutant. The slr and alf4-1 mutants, as well as the J0121, pWOX5::GFP-ER, pPLT1::PLT1:YFP, DR5::GFP, and pPIN1::PIN1:GFP marker lines, have been described previously (11, 13, 28–30, 49, 50). T-DNA insertion mutants kcs1-5 (SALK_200839) and pas1-4 (SALK_051324) were obtained from the ABRC. pKCS1::KCS1, pKCS1::KCS1:GFP, pALF4::GUS, pPEP::KCS1, and OE lines of ALF4, IPT3, and CKX1 were generated in this experiment. Regeneration assays were performed on CIM and SIM as described by Valvekens et al. (4).
Cytological Analyses.
For histological analysis, roots were fixed and cleared according to a previously described method for DIC microscopy (51). Thin sections were created as described by Wang et al. (52), and confocal microscopy was performed using a Leica SP5 confocal microscope. GFP and YFP signals were detected by excitation with an argon laser at 488 nm and a spectral detector set at 505–550 nm for the emission. The propidium iodide (PI) signal was visualized by excitation with an argon laser at 488 nm and a spectral detector set at >585 nm for the emission.
Analysis of Fatty Acids.
Total fatty acids of the roots were methylated and extracted for lipid analysis according to the method described by Browse et al. (53).
More detailed information on the experimental methods is provided in SI Materials and Methods. The primers used in this study are listed in Table S1.
Table S1.
Primers used in this study
| Primer name | Sequence (5′-3′) | Note |
| pKCS1:KCS1-KpnI F | ggtaccATGCGATCATCCTCAAAACTC | Construct |
| pKCS1:KCS1 R | TTGCTTTATCATTTATGGGTGTG | Construct |
| pKCS1::KCS1:GFP-PmeI F | gtttaaacATGCGATCATCCTCAAAACTC | Construct |
| pKCS1::KCS1:GFP-AscI R | ggcgcgccATTGCACAACTTTAACCG | Construct |
| pALF4::GUS-BamHI F | ggatccCAGAAAACCATGCTCAATCAAAC | Construct |
| pALF4::GUS-SmaI R | cccgggAACGCCAAAACTTTTCAATTTC | Construct |
| p35S::ALF4-XbaI F | tctagaATGGAATCGTCCATTGAAG | Construct |
| p35S::ALF4-XbaI R | tctagaCTAATGACTTTTCAACTTTTCTTC | Construct |
| p35S::IPT3-XhoI F | ctcgagATGATCATGAAGATATCTATGGCT | Construct |
| p35S::IPT3-EcoRI R | gaattcTCACGCCACTAGACACCG | Construct |
| p35S::CKX1-BglII F | agatctATGGGATTGACCTCATCCTTAC | Construct |
| p35S::CKX1-StuI R | aggcctTTATACAGTTCTAGGTTTCGGCAG | Construct |
| pPEP-SacI F | gagctcCATTCGATGTTCACCATGCAAAA | Construct |
| pPEP-KpnI R | ggtaccGGTTTTGGCTAATGTGATTGTGTA | Construct |
| KCS1cds-KpnI F | ggtaccATGGAGAGAACAAACAGCA | Construct |
| KCS1cds-SalI R | gtcgacTCATTGCACAACTTTAACC | Construct |
| LBb1.3 | ATTTTGCCGATTTCGGAAC | T-DNA |
| alf4-1 F | TTCTCCTCGTTTCGGGTTTG | SSLP |
| alf4-1 R | AGTCTTGAAATCCTCCGGCTT | SSLP |
| KCS1-qRT F | TCTCCTGCTACAAACCGGAA | qRT-PCR |
| KCS1-qRT R | CGTGTCATCGGTGAATGAT | qRT-PCR |
| PAS1 RT F | GCTTAAAATGGGAGAGTGGAGG | RT-PCR |
| PAS1 RT R | TCTGAGTCTACAGTGCCGTTGG | RT-PCR |
| GAPC F | CACTTGAAGGGTGGTGCCAAG | RT-PCR |
| GAPC R | CCTGTTGTCGCCAACGAAGTC | RT-PCR |
| ACTIN7 F | TGGCCGATGGTGAGGATATT | qRT-PCR |
| ACTIN7 R | AACCAGCCTTCACCATTCCA | qRT-PCR |
| PLT1-qRT F | TGAGGAAGAAGCAGCAGAAGC | qRT-PCR |
| PLT1-qRT R | CGGTTGATCTCGAAGTTGGTC | qRT-PCR |
| PLT2-qRT F | CGTGGTGCATCCATGTATCG | qRT-PCR |
| PLT2-qRT R | TGCTTCTTCCTCCGTGCTG | qRT-PCR |
| PLT3-qRT F | TCAGGAGGAAGAGTAGCGGTT | qRT-PCR |
| PLT3-qRT R | CTTTGTTCCCAGCAACTCGG | qRT-PCR |
| PLT7-qRT F | ATCGTGGAGTCACCCGACAT | qRT-PCR |
| PLT7-qRT R | TCCTTTTCTGGCTTGACCTTC | qRT-PCR |
| LBD16-qRT F | ACCCTGTTTATGGATGTGTCTCTC | qRT-PCR |
| LBD16-qRT R | TGCCTTCATTTGCATGACTTG | qRT-PCR |
| LBD17-qRT F | GCGTTTCTCATATCTTTTCCCTC | qRT-PCR |
| LBD17-qRT R | TCATGCTTTGTGTTGCTTGTTG | qRT-PCR |
| LBD18-qRT F | GTCGCTCACATCTTTGCTCTTC | qRT-PCR |
| LBD18-qRT R | AGGTAGCTCTAGTGATGCCAAATG | qRT-PCR |
| LBD29-qRT F | GCTAGGCTTCAAGATCCCATC | qRT-PCR |
| LBD29-qRT R | TGTGCTGCTTGTTGCTTTAGA | qRT-PCR |
| ALF4-qRT F | CTCCTTCAATGACAGCAATCCT | qRT-PCR |
| ALF4-qRT R | CGTATCAACAGCCGCACAAT | qRT-PCR |
SI Materials and Methods
Plant Materials and Growth Conditions.
Unless specified otherwise, the Arabidopsis thaliana ecotype Columbia-0 (Col-0) served as the WT in this study. The slr and alf4-1 mutants, as well as the J0121, pWOX5::GFP-ER, pPLT1::PLT1:YFP, DR5::GFP, and pPIN1::PIN1:GFP marker lines, have been described previously (11, 13, 28–30, 49, 50). The J0121 line was crossed with Col-0 to eliminate potential biases raised by its C24 background. The T-DNA insertion mutant kcs1-5 (SALK_200839) and pas1-4 (SALK_051324) were obtained from the ABRC and verified by PCR analyses.
Seeds were sterilized and germinated on half-strength Murashige and Skoog medium (1/2 MS) plates (1/2 MS medium, 1% sucrose, 1% plant agar, with pH adjusted to 5.7 using 1.0 M KOH) after stratification for 3–4 d at 4 °C. Seedlings grown vertically on 1/2 MS medium and plants grown in soil in a growth room under a 16-h light/8-h dark photoperiod at 22 °C with an illumination intensity of 80–90 µmol m−2 s−1 were used for the morphological characterization of roots and aerial organs, respectively (8).
Isolation of cfr1 and Positional Cloning of the CFR1 Gene.
Approximately 6,000 individual M1 plants were obtained from 0.2% (vol/vol) EMS-mutagenized slr seeds, and their M2 seedlings were germinated and grown vertically on 1/2 MS medium for 7 d and then incubated for 6–12 d on CIM containing B5 basal medium supplemented with 2.2 µM 2,4-D (Sigma-Aldrich) and 0.2 µM kinetin (KT; Sigma-Aldrich) (4). The candidate cfr mutants were identified by the restored callus-forming phenotype in primary roots. The candidates were recovered on 1/2 MS for 5 d and then transferred into soil for further validation of their progenies. Genetic analyses of cfr1 mutants were performed by assessing the segregation ratio in the F2 population of cfr1 homozygotes back-crossed with slr. Allelic tests of cfr1-1, cfr1-2, and cfr1-3 were carried out by examination of the callus-forming phenotype in the primary roots of F1 plants crossed with each other. Map-based positional cloning of CFR1 was conducted using a mapping population generated by crossing cfr1-1 with Ler. The genomic DNA of 1,005 F2 individuals with the cfr1-1 phenotypes were used for analyzing cosegregation with various SSLP and CAPS molecular markers. DNA sequencing analyses were performed within a 110-kb region on chromosome 1 in cfr1-1 and for the KCS1 locus in slr, cfr1-2, and cfr1-3.
Plasmid Construction and Plant Transformation.
A genomic KCS1 DNA fragment containing a 2,736-bp promoter and a 1,587-bp coding region was amplified by PCR and cloned into the pCAMBIA1300 plasmid (Cambia) to generate a complementation construct for the cfr1 and kcs1-2 mutants. The pKCS1::KCS1:GFP construct was generated by fusion of amplified KCS1 in-frame with GFP in the pMDC83 plasmid. To generate the pALF4::GUS construct, the 2,607-bp promoter region of ALF4 was amplified and cloned upstream of the GUS gene in the pCAMBIA1391 plasmid (Cambia). To generate the p35S::ALF4, p35S::IPT3, and p35S::CKX1 constructs, cDNA fragments of ALF4, IPT3, and CKX1, respectively, were amplified by RT-PCR and then cloned into the pVIP96 binary vector. To generate the pPEP::KCS1 construct, the 1,671-bp promoter region of PEP was amplified and cloned upstream of the KCS1 gene in the pCAMBIA1300 plasmid for the kcs1-2 mutant. The primers used for generating the constructs of this study are listed in Table S1.
Arabidopsis transformations were performed by the floral-dip method, using the EHA105 or GV3101 Agrobacterium strains. At least 10 independent transgenic lines with a single T-DNA insertion were generated, and T3 homozygotes of at least three lines were used for subsequent experiments for each construct.
Gene Expression Analysis.
Total RNA was extracted with TRIzol reagent (Invitrogen). cDNA was prepared from 1 µg of total RNA that was reverse-transcribed with SuperScript II Reverse Transcriptase (Invitrogen). RT-PCR and qRT-PCR were conducted as described previously (8), and the expression levels of glyceraldehyde-3-phosphate dehydrogenase C subunit (GAPC) and ACTIN7 served as internal controls for RT-PCR and qRT-PCR, respectively. All of the qRT-PCR analyses were performed with three independent biological replicates. The primers used for the RT-PCR and qRT-PCR analyses are listed in Table S1.
The GUS staining assay was performed as follows. Seedlings of transgenic plants were incubated in a 50 mM phosphate buffer (pH 7.0) solution containing 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 0.1% Triton X-100, and 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-Gluc) at 37 °C for several hours. Samples were fixed and cleared according to the method described by Malamy et al. (51), and then visualized by DIC microscopy (Olympus BX51).
Culture Conditions and Regeneration Assays.
To examine callus initiation and the cellular characteristics in roots, 7-d-old seedlings were incubated on solid CIM for 8–12 d and 4 d, respectively. To quantify the callus-forming capacity, the area of pericycle or derived callus layer was determined with the cleared roots by ImageJ software after 4 d of incubation on CIM. To analyze the effect of VLCFAs on ALF4 expression and callus-forming capacity, 5 µM metazachlor (Sigma-Aldrich), 600 µM VLCFAs (a mixture of C18:0, C20:0, C22:0, and C24:0), or 600 µM C16:0 fatty acids (Sigma-Aldrich) were supplemented to MS medium or CIM, respectively.
To test the effects of cytokinin or ethylene on ALF4 expression and callus formation, 2 µM KT, 10 µM AgNO3 (Sigma-Aldrich), or 10 µM ACC (Sigma-Aldrich) were added to MS medium or CIM. To investigate shoot formation capacity, the callused seedlings incubated on CIM for 12 d were transferred to SIM in the B5 medium supplemented with 0.9 µM indole-3-acetic acid (IAA; Sigma-Aldrich) or 24.6 µM N6-Δ2-isopentenyladenine (Sigma-Aldrich), as described previously (4).
Root Morphology and Microscopy.
The primary roots of 10-d-old seedlings were grown vertically and photographed, and lengths were determined with ImageJ software. Roots were fixed and cleared as described previously (51). In brief, samples were transferred into microcentrifuge tubes containing 0.24 N HCl in 20% (vol/vol) methanol and then incubated at 57 °C on a heat block for 15 min. The solution was replaced with 7% (wt/vol) NaOH in 60% ethanol for 15 min at room temperature. Tissues were rehydrated in 40%, 20%, and 10% ethanol for 15 min at each concentration, and finally infiltrated for 30 min in 25% glycerol (vol/vol) diluted in 5% ethanol. Roots were mounted in 50% glycerol on microscope slides, and lateral root initiates were examined and quantified by DIC microscopy (Olympus BX51). Statistical significance was evaluated using Student’s t test.
Thin sections were created as described by Wang et al. (52). For this, 5-d-old seedlings were fixed in 2.5% (wt/vol) paraformaldehyde (PFA; Sigma-Aldrich) and vacuum infiltrated for 30 min, and then stored overnight at 4 °C. Fixed samples were then washed with 10%, 20%, and 30% (wt/vol) sucrose in 1% PFA at pH 7.0 for 20, 20, and 30 min, respectively. Samples were then embedded in 5–7% (wt/vol) LM agarose (Promega) liquid gel at 30 °C and placed at 4 °C. Sections of roots were prepared at 50–80 µm using a Leica VT1200S vibrating-blade microtome.
For confocal microscopy, the root samples were mounted in 10 mg L−1 PI (Sigma-Aldrich) and imaged using a Leica SP5 confocal microscope. GFP or YFP signal was detected by excitation with an argon laser at 488 nm and a spectral detector set at 505–550 nm for the emission. The PI signal was visualized by excitation with an argon laser at 488 nm and a spectral detector set at >585 nm for the emission.
Auxin Response and Gravitropic Assay.
To examine the root response to exogenous auxin, 4-d-old seedlings were transferred to solid MS supplemented with various concentrations of 1-naphthylacetic acid (NAA) for 5 d and photographed. The primary root length was determined using ImageJ software. Seedlings were then cleared, and the number of lateral root initiates was quantified. To investigate the gravitropic response of roots, the roots of 4-d-old seedlings grown vertically were straightened, and plates were then turned 90° and kept in darkness. The root reorientations were determined at 24 h after gravity stimuli using ImageJ software.
Analysis of Fatty Acids.
Total fatty acids in the roots of 10-d-old seedlings were methylated and extracted using a modified version of a previously described protocol (53). In brief, 200 mg of fresh root tissue was collected and ground into a powder under liquid nitrogen. The powder was then transferred to a 20-mL Teflon-lined screw-capped glass tube (Agilent), with heptadecanoic acid (HPLC grade; Sigma-Aldrich) added as an internal standard. Then 6 mL of freshly prepared 2.5% H2SO4 (vol/vol) in methanol was added into the glass tube, and samples fully mounted in the foregoing solution were heated at 80 °C for 1.5 h.
Once the samples cooled, 9 mL of 0.9% (wt/vol) NaCl was added, and fatty acid methyl esters were extracted into 3 mL of heptane. The heptane-containing fatty acid methyl esters were concentrated with a stream of nitrogen. Fatty acid methyl esters were then redissolved in 500 µL of heptane and analyzed on a HP-INNOWax GC column (30 m long, 0.25 mm, 0.25 µm; 19091N-133; Agilent) with a gas chromatograph coupled to a triple quadruple mass spectrometer (7890A/7000B; Agilent). The column was operated with helium as the carrier gas, an injection temperature of 250 °C, and a detector temperature of 260 °C. The oven temperature was increased from 60 °C to 260 °C in increments of 8 °C min−1, and then held for an additional 9 min. The experiments were performed with three independent biological replicates.
Acknowledgments
We thank Dr. Masao Tasaka for the slr mutant, Dr. John L. Celenza for the alf4-1 mutant, Dr. Ben Scheres for pWOX5::GFP-ER and pPLT1::PLT1:YFP, Dr. Gerd Jürgens for DR5::GFP, Dr. Jiří Friml for pPIN1::PIN1:GFP, Dr. Jim Haseloff for the J0121 marker lines, and the Arabidopsis Biological Resource Center for the seeds of SALK_200839 and SALK_051324. We also thank Zhen Xue for the total fatty acid measurements. This work was funded by the Ministry of Science and Technology of China (Grant 2013CB967300) and the National Natural Science Foundation of China (Grants 31230009 and 31371447).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522466113/-/DCSupplemental.
References
- 1.Birnbaum KD, Sánchez Alvarado A. Slicing across kingdoms: Regeneration in plants and animals. Cell. 2008;132(4):697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: Dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011;21(4):212–218. doi: 10.1016/j.tcb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–130. [PubMed] [Google Scholar]
- 4.Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeuchi M, Sugimoto K, Iwase A. Plant callus: Mechanisms of induction and repression. Plant Cell. 2013;25(9):3159–3173. doi: 10.1105/tpc.113.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atta R, et al. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57(4):626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 7.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell. 2010;18(3):463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012;22(7):1169–1180. doi: 10.1038/cr.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kareem A, et al. PLETHORA genes control regeneration by a two-step mechanism. Curr Biol. 2015;25(8):1017–1030. doi: 10.1016/j.cub.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwase A, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol. 2011;21(6):508–514. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Laplaze L, et al. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot. 2005;56(419):2433–2442. doi: 10.1093/jxb/eri236. [DOI] [PubMed] [Google Scholar]
- 12.Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta. 2007;226(5):1183–1194. doi: 10.1007/s00425-007-0565-4. [DOI] [PubMed] [Google Scholar]
- 13.DiDonato RJ, et al. Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 2004;37(3):340–353. doi: 10.1046/j.1365-313x.2003.01964.x. [DOI] [PubMed] [Google Scholar]
- 14.Chupeau MC, et al. Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell. 2013;25(7):2444–2463. doi: 10.1105/tpc.113.109538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bach L, Faure JD. Role of very-long-chain fatty acids in plant development, when chain length does matter. C R Biol. 2010;333(4):361–370. doi: 10.1016/j.crvi.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Joubès J, et al. The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol. 2008;67(5):547–566. doi: 10.1007/s11103-008-9339-z. [DOI] [PubMed] [Google Scholar]
- 17.Beaudoin F, et al. Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 2009;150(3):1174–1191. doi: 10.1104/pp.109.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach L, et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA. 2008;105(38):14727–14731. doi: 10.1073/pnas.0805089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Rowland O, Kunst L. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell. 2005;17(5):1467–1481. doi: 10.1105/tpc.104.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 21.Worrall D, Ng CK, Hetherington AM. Sphingolipids, new players in plant signaling. Trends Plant Sci. 2003;8(7):317–320. doi: 10.1016/S1360-1385(03)00128-6. [DOI] [PubMed] [Google Scholar]
- 22.Weber H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002;7(5):217–224. doi: 10.1016/s1360-1385(02)02250-1. [DOI] [PubMed] [Google Scholar]
- 23.Faure JD, et al. The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development. 1998;125(5):909–918. doi: 10.1242/dev.125.5.909. [DOI] [PubMed] [Google Scholar]
- 24.Liang H, et al. Ceramides modulate programmed cell death in plants. Genes Dev. 2003;17(21):2636–2641. doi: 10.1101/gad.1140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raffaele S, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20(3):752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin YM, et al. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell. 2007;19(11):3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobusawa T, et al. Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 2013;11(4):e1001531. doi: 10.1371/journal.pbio.1001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29(2):153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 29.Haecker A, et al. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131(3):657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- 30.Aida M, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119(1):109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17(2):119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 32.Tresch S, Heilmann M, Christiansen N, Looser R, Grossmann K. Inhibition of saturated very-long-chain fatty acid biosynthesis by mefluidide and perfluidone, selective inhibitors of 3-ketoacyl-CoA synthases. Phytochemistry. 2012;76:162–171. doi: 10.1016/j.phytochem.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 33.Roudier F, et al. Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell. 2010;22(2):364–375. doi: 10.1105/tpc.109.071209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19(1):118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyer EM. A potent inhibitor of ethylene action in plants. Plant Physiol. 1976;58(3):268–271. doi: 10.1104/pp.58.3.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA. 2006;103(44):16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mustroph A, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(44):18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steeves TA, Sussex IM. Patterns in Plant Development. 2nd Ed Cambridge Univ Press; Cambridge, UK: 1989. [Google Scholar]
- 40.Savchenko T, et al. Arachidonic acid: An evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell. 2010;22(10):3193–3205. doi: 10.1105/tpc.110.073858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black PN, Færgeman NJ, DiRusso CC. Long-chain acyl-CoA-dependent regulation of gene expression in bacteria, yeast and mammals. J Nutr. 2000;130(2S Suppl):305S–309S. doi: 10.1093/jn/130.2.305S. [DOI] [PubMed] [Google Scholar]
- 42.Young MM, Kester M, Wang HG. Sphingolipids: Regulators of crosstalk between apoptosis and autophagy. J Lipid Res. 2013;54(1):5–19. doi: 10.1194/jlr.R031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saba JD, de la Garza-Rodea AS. S1P lyase in skeletal muscle regeneration and satellite cell activation: Exposing the hidden lyase. Biochim Biophys Acta. 2013;1831(1):167–175. doi: 10.1016/j.bbalip.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagata Y, Partridge TA, Matsuda R, Zammit PS. Entry of muscle satellite cells into the cell cycle requires sphingolipid signaling. J Cell Biol. 2006;174(2):245–253. doi: 10.1083/jcb.200605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trenkamp S, Martin W, Tietjen K. Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci USA. 2004;101(32):11903–11908. doi: 10.1073/pnas.0404600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavenus J, et al. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18(8):450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Delbarre A, Muller P, Imhoff V, Guern J. Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta. 1996;198(4):532–541. doi: 10.1007/BF00262639. [DOI] [PubMed] [Google Scholar]
- 48.Melnyk CW, Schuster C, Leyser O, Meyerowitz EM. A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol. 2015;25(10):1306–1318. doi: 10.1016/j.cub.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friml J, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426(6963):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 50.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 51.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124(1):33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, et al. The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell. 2014;26(5):2055–2067. doi: 10.1105/tpc.114.123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Browse J, McCourt PJ, Somerville CR. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem. 1986;152(1):141–145. doi: 10.1016/0003-2697(86)90132-6. [DOI] [PubMed] [Google Scholar]