Significance
Even though DNA methylation is known to be correlated with gene repression, it has never been demonstrated that this modification must indeed be removed from a gene in order for it to become activated during cell differentiation in vivo. In this paper, we inactivated the enzymes responsible for the demethylation reaction in the B-cell lineage and in this manner have shown that this epigenetic mark plays a critical role in development, independently of the many specific transcription factors that direct the selection of genes involved in cell differentiation. Our study is the first to our knowledge to causally connect all of the molecular components necessary to prove the link between the Tet enzymes, CpG demethylation, expression, and phenotype.
Keywords: Tet2/Tet3, chromatin, differentially methylated regions, DMRs
Abstract
There is ample evidence that somatic cell differentiation during development is accompanied by extensive DNA demethylation of specific sites that vary between cell types. Although the mechanism of this process has not yet been elucidated, it is likely to involve the conversion of 5mC to 5hmC by Tet enzymes. We show that a Tet2/Tet3 conditional knockout at early stages of B-cell development largely prevents lineage-specific programmed demethylation events. This lack of demethylation affects the expression of nearby B-cell lineage genes by impairing enhancer activity, thus causing defects in B-cell differentiation and function. Thus, tissue-specific DNA demethylation appears to be necessary for proper somatic cell development in vivo.
DNA methylation takes place at almost all stages of development including the early embryo as well as during lineage commitment and is mediated through a combination of active and passive processes. Recent studies have raised the possibility that demethylation can occur through the involvement of the ten-eleven-translocation family (Tet1, Tet2, and Tet3) that catalyzes the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) as a first step in the pathway (1, 2). Removal of this unusual base may then be accomplished either by further oxidation followed by base excision repair (3) or through replication dilution (4–6). Genetic experiments have demonstrated that Tet enzymes are key players during early development, with Tet3-mediated DNA hydroxylation being involved in epigenetic programming of the zygotic paternal DNA (7, 8), whereas combinations of Tet1 and Tet2 play a role in the demethylation process that takes place during embryonic stem cell differentiation in vitro (2, 9–12).
Tet enzymes also contribute to lineage development. Thus, changes in the pattern of 5hmC have been shown to accompany neurogenesis in vivo (13), and Tet knockdowns indicate that this process may be essential for the normal progression of neuronal differentiation (ref. 14, reviewed in ref. 15). In the hematopoietic system, as well, targeted Tet deletions [Tet1 alone (16), Tet2 alone (17–20), or Tet1 and Tet2 together (21)] appear to alter global 5hmC and 5mC distribution, perturb stem cell self-renewal, cause altered differentiation, and predispose to malignancies (refs. 19, 22, reviewed in ref. 23). None of these studies, however, has addressed the key question of whether demethylation itself is actually required for gene activation and proper lineage differentiation. To this end, we generated a Tet2/Tet3 knockout specific to B-lymphoid development, isolated cells at different stages of differentiation, and analyzed their methylation patterns. Because this approach targets the demethylation machinery in an exclusive manner, it allowed us to evaluate the role of this modification independently of the many transcription factors that drive the process of B-cell differentiation.
Results
It has already been shown that both Tet2 and Tet3 are highly expressed in B lineage cells (24). With this in mind, we generated Tet2F/Tet3F mice (18, 23) and crossed them with animals expressing Cre under control of the early B-cell–specific Mb1 promoter (25) to obtain mice with a conditional knockout of these enzymes specifically in the B-cell lineage (Materials and Methods). Reduced representation bisulfite sequencing (RRBS) (26) measures DNA methylation levels at a large number of regulatory regions in the genome with high depth and reproducibility. We wanted to test whether B-cell–specific deficiency of Tet2, Tet3, or a combination thereof would specifically impair DNA methylation during B-cell differentiation.
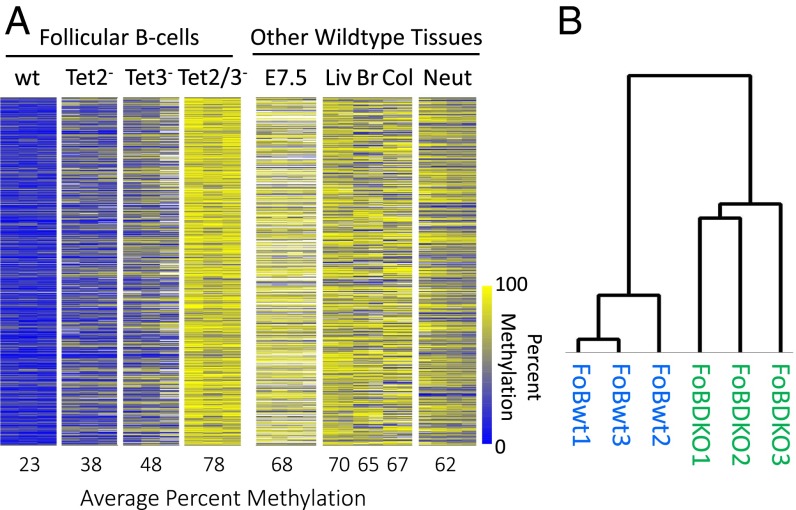
To this end, we isolated mature naïve follicular (Fo) B cells from spleens of 6- to 8-wk-old wild-type and knockout mice by FACS and performed RRBS. Although overall methylation levels were very similar in all samples (Fig. S1A), we identified approximately one thousand four hundred 100-bp tiles that were at least 40% undermethylated in control follicular B cells compared with Tet2/Tet3 double knockouts (DKOs) (P < 10−300, permutation test; Materials and Methods) with lesser effects being observed for each knockout individually. RRBS analysis revealed that these same sites are highly methylated in E7.5 embryos as well as in a variety of adult tissues (Fig. 1). This indicates that they likely become de novo methylated at about the time of implantation and remain modified in most cell types during development, undergoing demethylation exclusively in the B-cell lineage. These results show that Tet2 in combination with Tet3 plays a key role in tissue-specific demethylation.
Fig. S1.
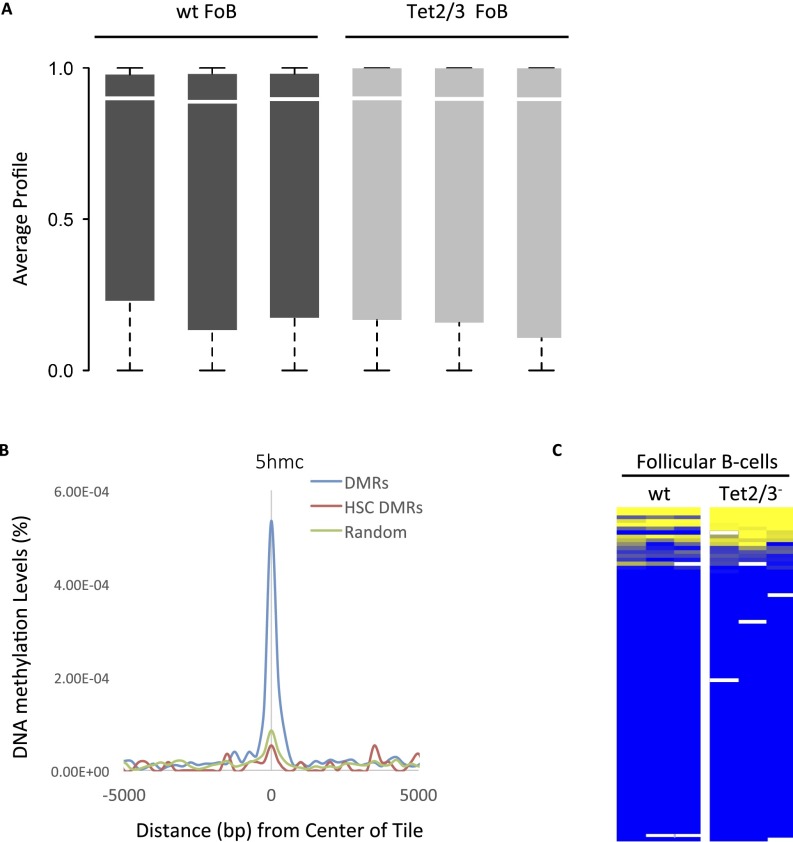
The 5mC and 5hmC methylation distribution. (A) Box plots of global RRBS methylation values for all replicates of wild-type and Tet2/3− follicular B-cell samples. (B) Enrichment of 5hmc in hematopoietic progenitor cells (GSE65895) as a function of distance from the center of each DMR (Fig. 1), compared with a random control. Tiles specifically unmethylated in HSCs are not enriched. The 5hmC levels were determined by HELP-GT (HpaII tiny fragment enrichment by ligation-mediated PCR with β-glucosyl transferase) in Tel1+/+ as opposed to Tet1−/− hematopoietic progenitor cells (70). Average coverage was 10–15× per base. (C) DNA methylation levels of the promoter regions for genes that harbor differentially methylated tiles (n = 356). Only 11 (3%) show a significant difference between wt and DKO. Yellow represents high and blue represents low DNA methylation levels.
Fig. 1.
Effect of Tet knockouts on B cell-specific DNA demethylation. (A) Heatmap of 1,399 tiles comparing DNA methylation levels in follicular B cells (FoB) from wild-type (wt), Tet2–, Tet3–, and Tet2/3– (DKO) samples (n = 3) for each with a difference of at least 40% (P < 10−300, permutation test; Materials and Methods) between the wt and DKO samples (11 tiles showed the opposite methylation ratio). Samples are compared with other wild-type somatic tissues, embryonic day 7.5 (E7.5), liver (Liv), brain (Br), colon (Col),; and neutrophils (Neut) (n = 2–4) (39). Yellow represents high and blue represents low DNA methylation levels. (B) Hierarchical clustering analysis was performed on RRBS tiles from wild-type and Tet2/3– follicular B cells.
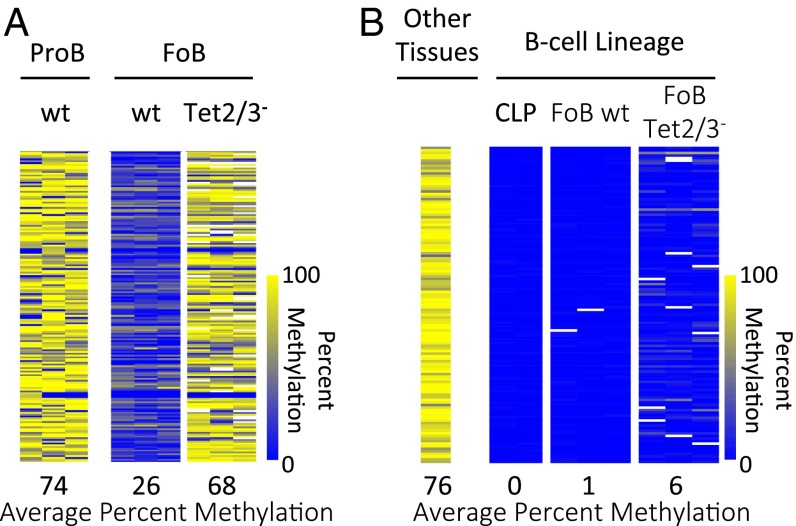
To further pinpoint the role of these enzymes in this demethylation process, we carried out RRBS analysis on pro-B cells, bone marrow-derived B-cell precursors. Examination of tiles that are methylated in wild-type pro-B cells and only undergo specific demethylation during differentiation to mature follicular B cells shows that Tet2/Tet3 double deficiency initiated before the pro–B-cell stage prevents demethylation at over 95% of these sites. Although our assay (RRBS) is not fully genomic, these results strongly suggest that Tet proteins may be responsible for almost all DNA demethylation that occurs at this stage (Fig. 2A). In contrast, examining sequences that have already undergone demethylation in common lymphoid precursors (CLPs), an early stage of development before B-cell commitment, and before the activation of Cre, reveals that the B-cell–specific Tet2/Tet3 knockout has almost no effect on DNA methylation at these sites (Fig. 2B and Fig. S2A). Thus, even though it has already been demonstrated that demethylation occurring at a given stage of B-cell development is retained further on (27), our experiments support the fundamental idea that once established, the undermethylated state is autonomously maintained without the need for continuous demethylase activity (28), thereby constituting a genuine memory mechanism.
Fig. 2.
Tet2/3-mediated stage-specific demethylation. (A) Heatmap of 174 tiles comparing DNA methylation levels between wt Pro-B cells and wt FoB cells which undergo DNA demethylation (>40%) during lineage specification. These tiles fail to undergo demethylation in Tet2/3– FoB cells. (B) Heatmap of 123 tiles specifically unmethylated in common lymphoid progenitor cells (CLPs) compared with average DNA methylation levels from a number of somatic tissues (fat, liver, brain, and heart) (39). These tiles remain unmethylated in the Tet2/3– FoB cells (n = 3) as well. Yellow represents high and blue represents low DNA methylation levels.
Fig. S2.
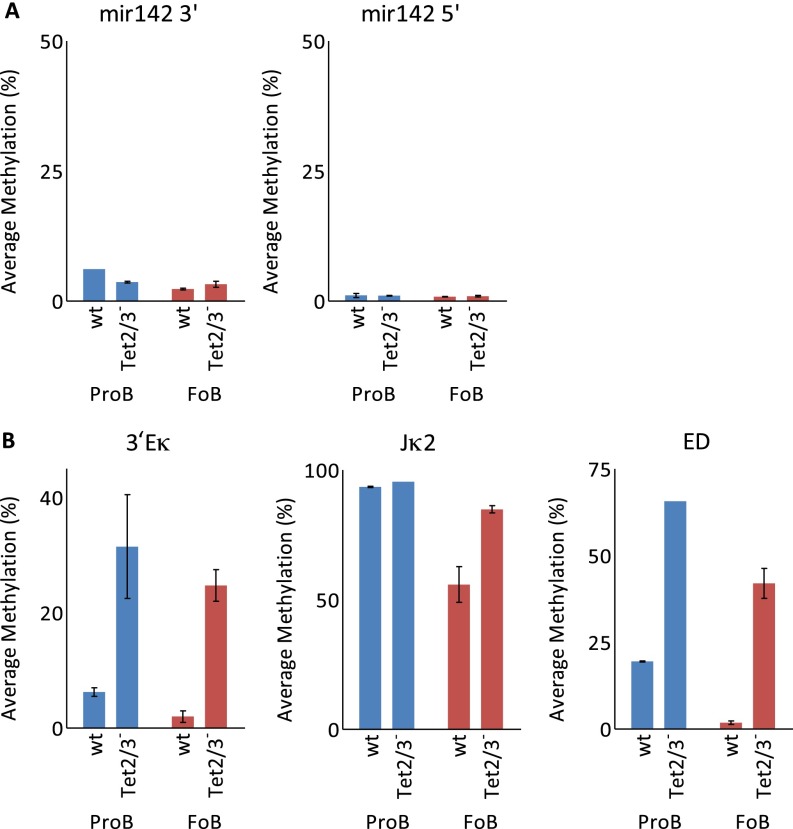
DNA methylation at specific sites. (A) DNA methylation of miR-142 regulatory regions in both Pro-B and FoB wt and DKO cells as determined by single-molecular bisulfite analysis (Miseq). Note that the knockout is initiated after this region has already undergone demethylation demonstrating that the demethylase is not necessary to maintain the undermethylated state. (B) DNA methylation of Igk regions in both Pro-B and FoB wt and DKO cells. Note that in this case, the knockout is initiated before the normal demethylation that occurs at these sites.
To further validate the requirement of the Tet proteins for DNA demethylation, we examined the Igκ locus using specific primers and Bisulfite sequencing to measure DNA modification at sites that are known to undergo specific demethylation during normal B-cell development (29). The results indicated that Tet2 and Tet3 are indeed required for demethylation of key regulatory sites within the Igκ gene locus (Fig. S2B). Taken together, these findings demonstrate that B-cell lineage demethylation takes place through a Tet-dependent biochemical pathway that likely converts 5mC to 5hmC and further oxidation products, which may then be removed by glycosylation and subsequent base excision repair (3). In keeping with this, genome-wide analysis indicates that these same sites are indeed enriched with 5hmC specifically in the B-cell lineage (Fig. S1B) (16).
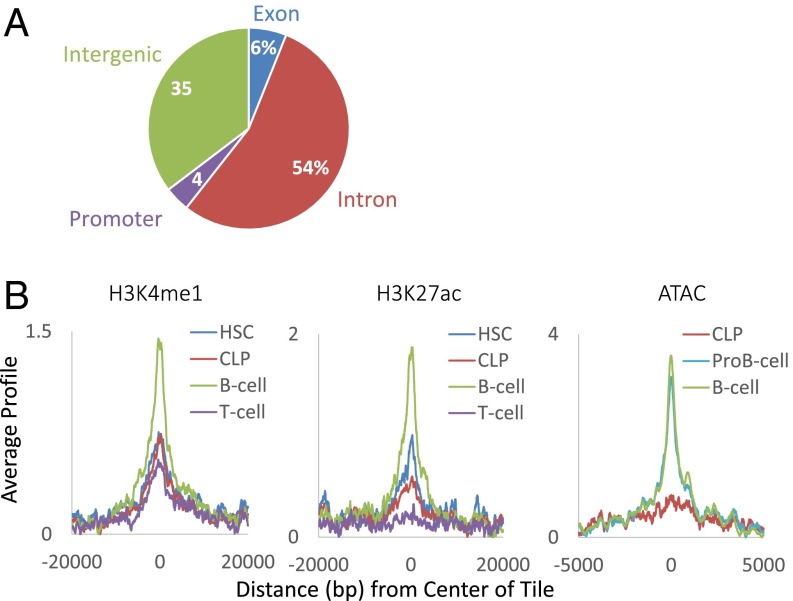
To evaluate the nature of the sites that undergo programmed demethylation during B-cell development, we mapped their locations in the genome, as well as the presence of key chromatin components using published ChIP-Seq data (30–32). Most of the relevant tiles are located within gene coding regions, with only a small percentage being associated with promoters (Fig. 3A). Furthermore, these sites are differentially marked with histone H3K4me1 in B-lineage as opposed to T-lineage cells, with the highest enrichment seen in mature B cells. Because these same tiles are also specifically enriched for histone H3K27Ac and assume a more open configuration as determined by an assay for transposase-accessible chromatin (ATAC) (33) (Fig. 3B), they likely represent enhancer-like elements (34–37) that normally become activated during B-cell development.
Fig. 3.
Characterization of DMRs. (A) Genome distribution of regions demethylated in wt compared with Tet2/3– FoB cells. (B) Left and Center show ChIP-seq of demethylated regions (n = 1,399) as a function of distance from their center for H3K4me1 and H3K27Ac in hematopoietic stem cells (HSC), common lymphoid progenitors (CLP), B cells, and T cells (GSE60103). Right shows ATAC-seq in CLP, pro-B (GSE66978), and B cells (GSE59992). It should be noted that this accessibility marker is not present at early stages of hematopoiesis and only appears in cells that have undergone demethylation at these sites.
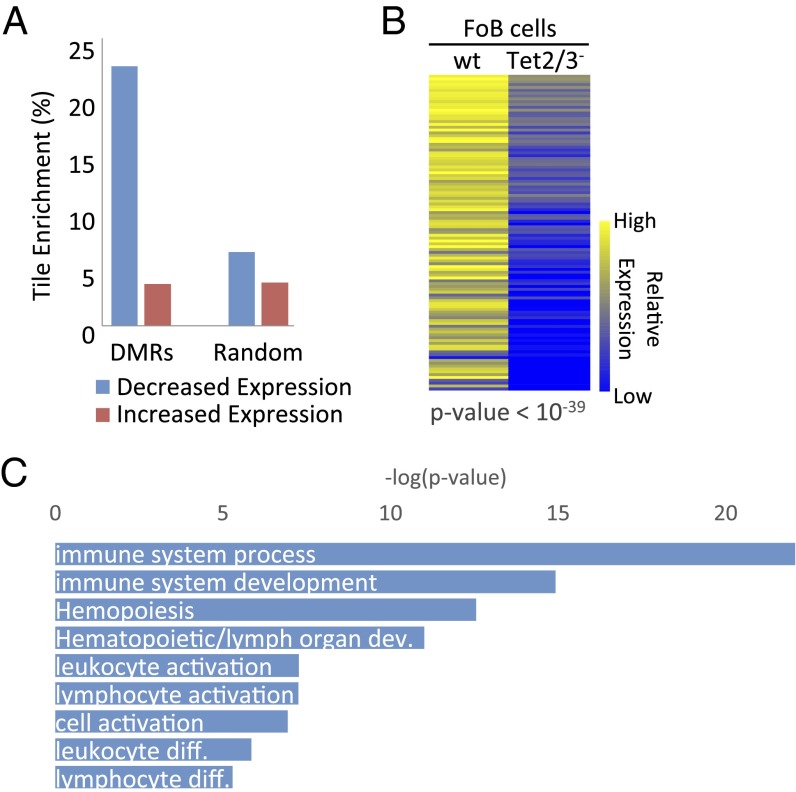
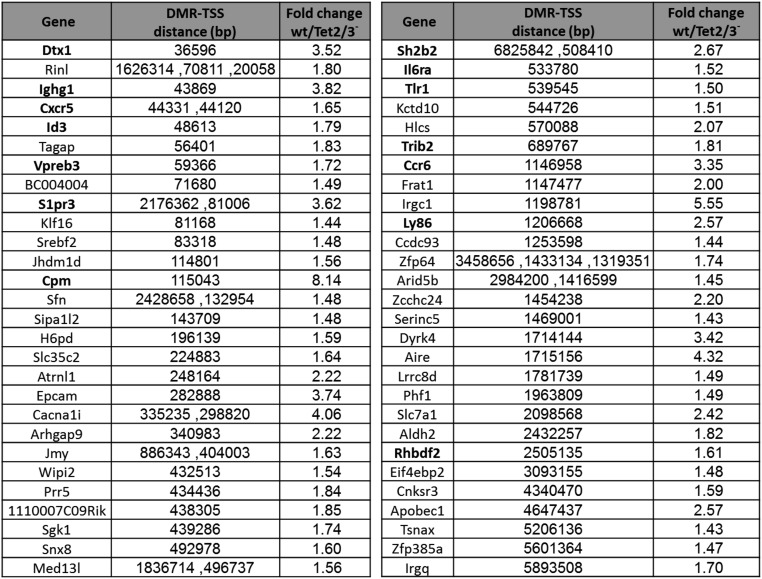
The fact that Tet2/Tet3 deficiency specifically prevents the demethylation that occurs during normal B-cell development presented a unique opportunity to test whether the change in DNA methylation itself plays a role in controlling gene expression in vivo. Because Tet-dependent demethylation seems to take place primarily at putative enhancer elements and not at promoters (Fig. 3), we first restricted our analysis to tiles (n = 814) located within gene domains and compared the expression levels of these genes in the presence or absence of DNA methylation at the enhancer. Strikingly, 23% of these tiles (n = 186), as opposed to a random sample (7%), are located within genes (n = 111) that were found to be inhibited in the Tet2/Tet3 knockout (P < 10−27, z-test of proportions) (Fig. 4A and Fig. S3), with the difference in expression being highly significant (P < 10−39, t test) (Fig. 4B). Furthermore, by then analyzing a published HiC dataset from B-lymphoblastoid cells (38) to detect DMR interactions with distal promoter sequences, we were able to pick up an additional set of 47 genes that are down-regulated in the DKO (Fig. S4).
Fig. 4.
Effect of DNA methylation on expression. (A) Percentage of tiles (n = 814) located in genes with decreased (blue) and increased (red) expression (Tet2/3– < wt FoB cells) associated with DMRs compared with random tiles (P < 10−27, z-test of proportions). (B) Heatmap of relative expression levels (RNA-seq) for genes (n = 111) that harbor putative enhancer sequences. Each column shows average data for three biological replicates. (C) Gene ontology of all DMRs (n = 814) by GREAT analysis (40).
Fig. S3.
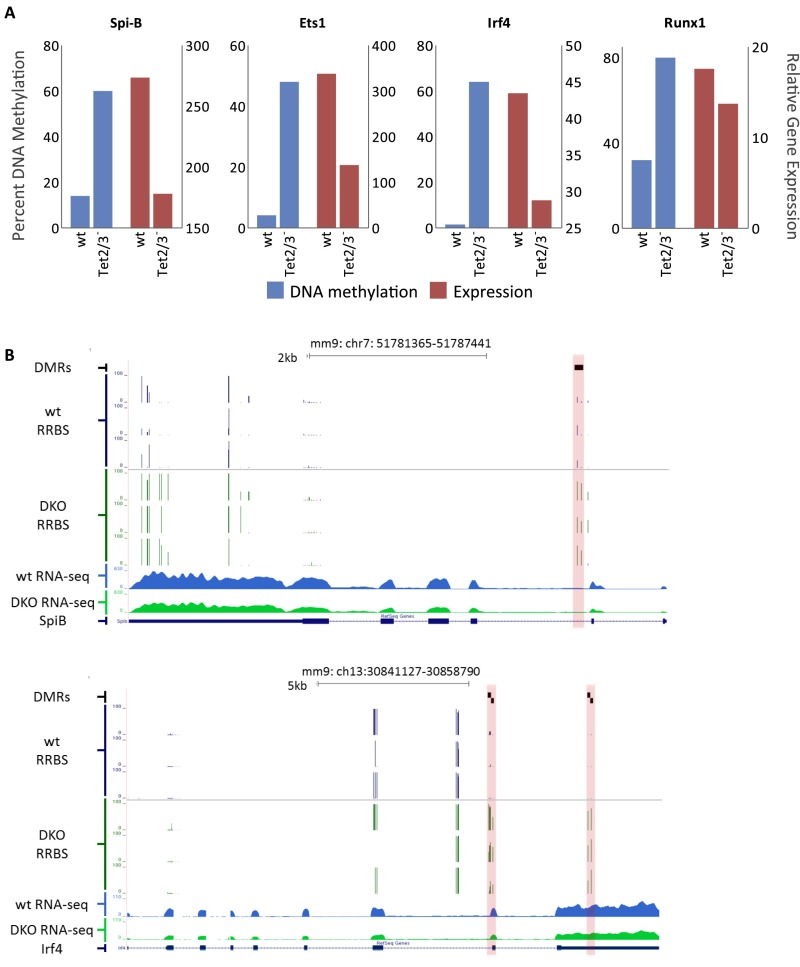
Correlation between DNA methylation and expression. (A) DNA methylation levels (blue) and relative expression levels (red) of selected genes in both wild-type and Tet2/3– FoB cells. (B) RRBS profiles of DNA methylation and RNA-seq analysis at two select loci containing DMRs (highlighted in red) in FoB cells that are associated with genes displaying decreased gene expression in the DKO (average of three replicate experiments).
Fig. S4.
DMR–gene interactions by Hi-C analysis. Genes that interact with distal DMRs whose expression is down-regulated in the DKO. These DMR–promoter contacts were identified from a Hi-C dataset originating from CH12 cells. The table shows the distance of each DMR from its interacting promoter. Some genes interact with more than one DMR. Genes known to be involved in B-cell development or function are noted in bold. Nine of these genes are also associated with intragenic DMRs.
It should be noted that because RRBS only covers a portion of the genome, there are undoubtedly many more, as yet undiscovered, Tet2/3-dependent DMRs that may influence genes in follicular B cells, perhaps explaining the finding that over 1,000 genes are differentially expressed at higher levels in the wild type as shown by RNA-seq (P < 0.05, t test). Furthermore, there are probably other genes that are initially primed by demethylation but still require additional factors to affect expression. Almost all specific genes associated with DMRs have promoters that are completely unaffected by the knockout (Fig. S1C), suggesting that Tet-dependent demethylation represents a regulatory mechanism directed almost exclusively to sequences that most likely have enhancer activity that is sensitive to DNA methylation (39).
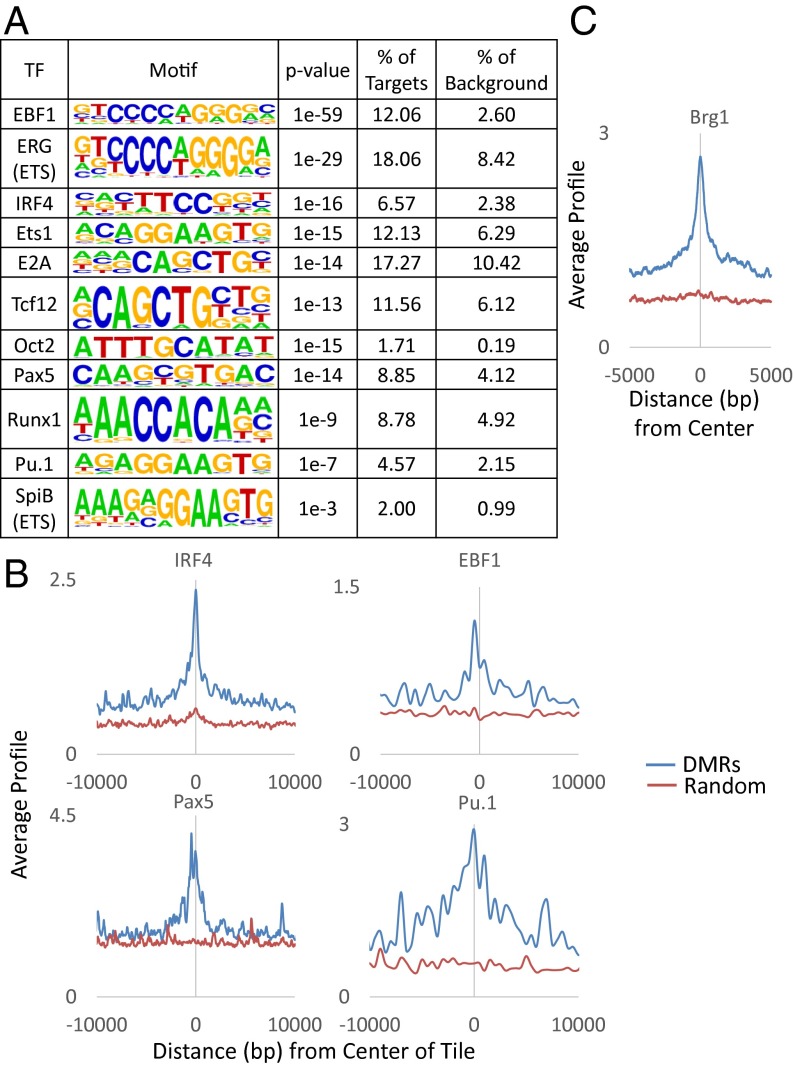
We next asked whether these effects of methylation on gene expression are associated with genes actively involved in B-cell development. Initial gene ontology analysis [Genomic Regions Enrichment of Annotations Tool (GREAT)] (40) indicated that the differentially expressed genes are highly enriched for various aspects of B-cell function, maintenance, and development (Fig. 4C). Many of these genes have also been shown to play a role in hematopoiesis by in vivo genetic analyses (Fig. S5). Furthermore, the presumed regulatory sites themselves harbor a large variety of binding motifs [Hypergeometric Optimization of Motif EnRichment (HOMER)] (41) for key factors known to be important for B-cell development (e.g., Ebf1, Irf4, E2a, Oct2, Pax5, and Pu.1) (Fig. 5A). ChIP analyses adapted from published data (31, 32) indicate that these factors as well as the chromatin remodeler, Brg1 (33), indeed bind to these sites in mature B cells (Fig. 5 B and C).
Fig. S5.
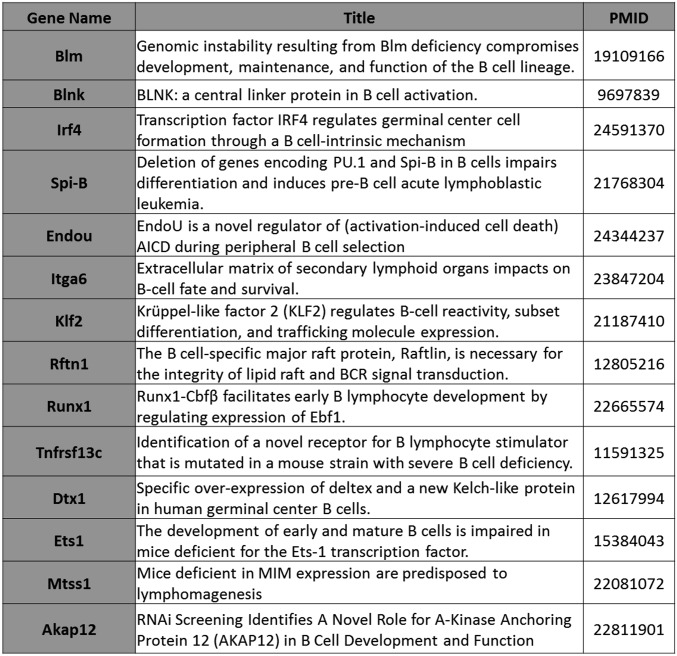
DMR-associated genes that affect B-cell development as determined by genetic analysis.
Fig. 5.
Correlation with expression. (A) Motif analysis of unselected DMRs (n = 1,399) by HOMER (41) showing percent enrichment of target sequences for transcription factors (TFs) compared with background. ChIP-seq of (B) lymphoid transcription factors (IRF4, EBF1, Pax5, and Pu.1) (GSE53595 and GSE38046) and (C) chromatin remodeler Brg1 (GSM1635413) as a function of distance from the center of each DMR compared with a random control.
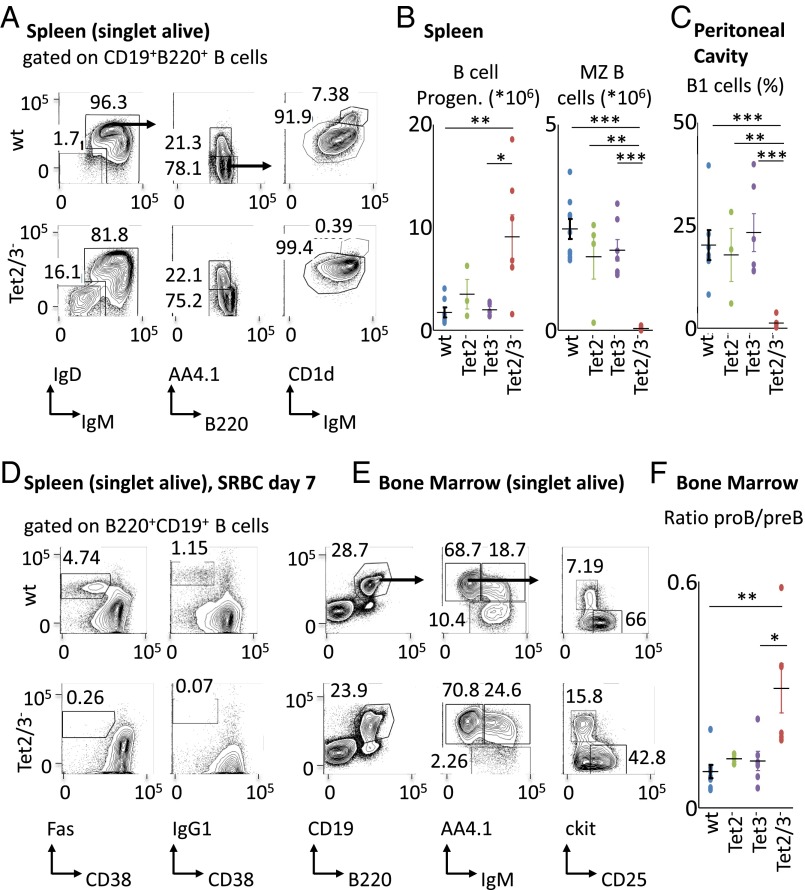
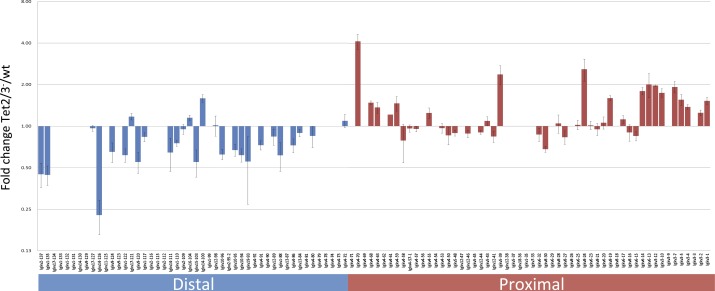
In light of these findings, we suspected that the lack of demethylation might also affect B-cell development and function. To test this we carried out a series of flow cytometry analyses aimed at assessing lymphoid cell composition in wild type, Tet2, and Tet3 single-knockout as well as double-knockout mice. In the bone marrow we detected a significant (threefold) shift in the ratio between pro-B and pre-B cells in the Tet2/Tet3 knockout indicating a partial block in early B-cell development. This was accompanied by an eightfold decrease in the number of mature recirculating B cells (Fig. 6 E and F and Fig. S6 D and E). A similar shift was detected in the spleen where B-cell progenitors were present in abnormally large numbers. Two mature B-cell subsets, namely, Marginal Zone (MZ) B cells in the spleen and B1 cells in the peritoneal cavity, were essentially absent (Fig. 6 A–C and Fig. S6 A and B). Together, these data strongly suggest that demethylation plays a key role in the B-cell maturation process and that its absence leads to an accumulation of more primitive cell types. In addition, RNA-Seq analysis of the Igκ locus in follicular B cells indicated that the DKO causes a change in the light chain repertoire, characterized by a dramatic shift (P < 10−5) to more proximal V region rearrangements (Fig. S7).
Fig. 6.
Population analysis of B cells in Tet2/3 knockouts. Tet2/3 DKO mice display abnormalities during B-cell development. (A) Representative flow cytometry analysis of splenocytes from the indicated genotypes. Numbers adjacent to outlined areas indicate % cells in each plot, and arrows indicate gating strategy. Gates depict IgM+ B cells (B220+CD19+IgM+IgD+/−), IgM− B-cell progenitors (B220+CD19+IgM–IgD–), immature IgM+ B cells (B220+CD19+IgM+AA4.1+), mature B cells (B220+CD19+IgM+AA4.1–), follicular (Fo) B cells (B220+CD19+IgM+AA4.1−CD1dlo), and marginal zone (MZ) B cells (B220+CD19+IgM+AA4.1–CD1dhi). (B) Graphs depict total cell numbers of selected splenic subpopulations as shown in A (n = 4–7). (C) Graph depicts % of B1 cells (B220loCD19+CD5+) among lymphocytes in the peritoneal wash (n = 3–7). (D) Representative flow cytometry analysis of splenocytes from SRBC immunized mice of the indicated genotypes. Numbers adjacent to outlined areas indicate % cells in each plot. Gates depict germinal center B cells [B220+CD19+CD95(Fas)+CD38lo] and class switched IgG1+ B cells (B220+CD19+IgG1+CD38lo). (E) Representative flow cytometry analysis of bone marrow cells from the indicated genotypes. Numbers adjacent to outlined areas indicate % cells in each plot, and arrows indicate gating strategy. Gates depict total B cells (B220+CD19+), mature recirculating B cells (B220+CD19+IgM+AA4.1–), immature IgM+ B cells (B220+CD19+IgM+AA4.1+), pro-B/pre-B cells (B220+CD19+IgM–AA4.1+), pre-B cells (B220+CD19+IgM–AA4.1+CD25+, ckit–), and pro-B cells (B220+CD19+IgM–AA4.1+CD25–, ckit+). (F) Graph depicts the ratio of pro-B/pre-B cells in bone marrow (n = 3–7). All plots are gated on live singlets. Graphs depict mean values and SDs of the respective populations. Significance was calculated by the two-tailed Student t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Fig. S6.
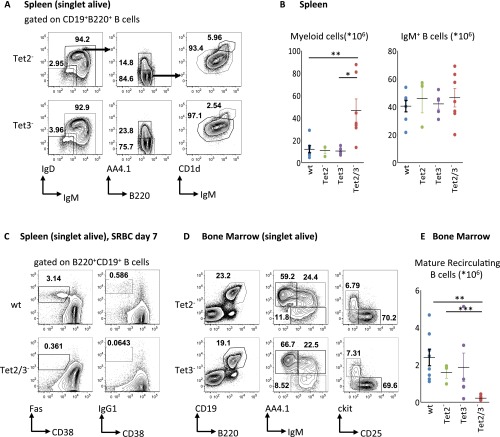
Tet2/3 DKO mice display abnormalities during B-cell development. (A) Representative flow cytometry analysis of splenocytes from the indicated genotypes. Numbers adjacent to outlined areas indicate % cells in each plot, and arrows indicate gating strategy Gates depict IgM+ B cells (B220+CD19+IgM+IgD+/−), IgM− B-cell progenitors (B220+CD19+IgM−IgD–), immature IgM+ B cells (B220+CD19+IgM+AA4.1+), mature B cells (B220+CD19+IgM+AA4.1−), follicular (Fo) B cells (B220+CD19+IgM+AA4.1−CD1dlo), and marginal zone (MZ) B cells (B220+CD19+IgM+AA4.1−CD1dhi). (B) Graphs depict total cell numbers of myeloid cells (CD11b+) and IgM+ B cells (B220+CD19+IgM+IgD+) in the spleen (n = 3–7). (C) Representative flow cytometry analysis of splenocytes from SRBC immunized mice of the indicated genotypes. Numbers adjacent to outlined areas indicate % cells in each plot. Gates depict germinal center B cells [B220+CD19+CD95(Fas)+CD38lo] and class switched IgG1+ B cells (B220+CD19+IgG1+CD38lo). (D) Representative flow cytometry analysis of bone marrow cells from the indicated genotypes. Numbers adjacent to outlined areas indicate % cells in each plot, and arrows indicate gating strategy Gates depict total B cells (B220+CD19+), mature recirculating B cells (B220+CD19+IgM+AA4.1–), immature IgM+ B cells (B220+CD19+IgM+AA4.1+), pro-B/pre-B cells (B220+CD19+IgM–AA4.1+), pre-B cells (B220+CD19+IgM–AA4.1+CD25+, ckit–), and pro-B cells (B220+CD19+IgM–AA4.1+CD25–, ckit+). (E) Graph depicts the total cell number of mature recirculating B cells (B220+CD19+IgM+AA4.1–) in the bone marrow (n = 3–7). All plots are gated on live singlets. Graphs depict mean values and SDs of the respective populations. Significance was calculated by the two-tailed Student t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Fig. S7.
Effect of Tet2/3 DKO on the Igκ rearrangement repertoire. Vκ contribution to the Igκ repertoire was analyzed from RNA-seq data on follicular B cells from three wt and three Tet2/3− mice. RPKM of each Vκ segment was normalized to the sum of all Vκ reads to give a percent of the repertoire. Vκ segments which contribute less than 0.4% of the repertoire in all samples were removed from the analysis to overcome outlier effects of lowly expressed genes. The ratio between the wt and Tet2/3− contribution to the repertoire for each Vκ segment is presented. Error bars represent SEM. The distal V segments (blue) are down-regulated in Tet2/3−, whereas the proximal Vs (red) are up-regulated (P < 10−5).
Despite B-cell developmental defects in Tet2/Tet3 DKOs, the mice displayed normal numbers of mature splenic follicular B cells (Fig. S6). Thus, we asked whether these cells would be functionally impaired and unable to mount an immune response. Indeed, Tet2/Tet3 knockout mice did not respond to T-dependent immunization with sheep red blood cells (SRBCs), as suggested by the absence of germinal center as well as IgG1+ class switched B cells (Fig. 6D and Fig. S6C). Apart from this, the absence of MZ and B1 cells in the DKO animals implies major defects in the production of natural antibodies and immune defense against blood-borne pathogens. It is noteworthy that in our study, no abnormal phenotypes were observed in single Tet2 or Tet3 knockout animals, suggesting that even partial demethylation of B-cell relevant regulatory regions in vivo may allow normal B-cell development and function (Fig. S6). An unexpected phenotype in the DKO animals was an expansion of myeloid cells in the spleen (Fig. S6B), which progressed over time and is the subject of a separate study.
Discussion
As part of the programmed changes in DNA methylation that occur during development, many genes undergo tissue-specific demethylation in association with differentiation, mainly at enhancer-like sequences (42, 43). The physiological role of this demethylation has not yet been elucidated, presumably because the mechanism of this process was not known. In this paper, we demonstrate that almost all demethylation that occurs during B-lineage development requires the specific combination of Tet2 and Tet3 (Fig. 1). Although these enzymes might also exhibit other activities (44–46), the fact that their deletion specifically affects the same tiles that undergo demethylation during B-cell lineage differentiation and are specifically marked with 5hmC (16) (Fig. S1B) provides strong genetic evidence that lineage-specific demethylation is a direct result of Tet activity on its target sequences. These results are in keeping with previous experiments in embryos and tissue culture showing that Tet enzymes indeed bind to enhancer sequences and are required for the generation of 5hmC and subsequent demethylation (47–50) but represent the first demonstration to our knowledge that these enzymes also play a role in tissue-specific demethylation in vivo. This, of course, does not rule out the possibility that other enzymatic components (e.g., AID) may also be necessary for demethylation at specific stages of development in vivo (51).
Despite many decades of research, it is still debated whether DNA methylation plays a causative role in development (52). Thus, even though demethylation of regulatory sequences is often correlated with increases in nearby gene transcription in general (53, 54) and in the B-cell lineage in particular (27), it has not been possible so far to determine whether the removal of methyl groups is indeed required for altering transcription patterns. We have used a Tet2/Tet3 knockout to specifically inhibit demethylation in vivo and in this manner have demonstrated that this process is necessary for the proper control of gene expression and for normal developmental progression. Because demethylation is programmed, it must be mediated by transacting factors that recognize the target sequences. Nonetheless, these factors appear to be insufficient by themselves to fully activate gene transcription. This only occurs once demethylation has taken place, probably because this brings about histone acetylation (55, 56) as well as increased chromatin accessibility (Fig. 3B), which may then allow the binding of additional transcription factors (57, 58).
There is evidence that normal hematopoiesis is very sensitive to changes in DNA methylation metabolism, with knockouts of either Dnmt3a, Tet1, or Tet2 causing defects in differentiation while predisposing mice to hematopoietic malignancies (16, 19, 22, 56, 59, 60). In humans, aberrations in these same genes can be detected in hematopoietic cells of leukemic patients (23) as well as in healthy individuals (61), where they may be selected to become preleukemic (62). Our findings help explain how Tet deficiency may contribute to tumorigenesis by preventing enhancer demethylation at genes critically involved in the control of cellular differentiation, thereby promoting precursor cell proliferation.
Materials and Methods
Mice.
Tet2F (63), Tet3 F (23), and Mb1-cre/+ (wt) (25) mice have been described. All mice are C57BL/6 or have been backcrossed to the C57BL/6 background for more than 10 generations. Mice were bred and housed under specific pathogen-free conditions in microisolator cages in a room illuminated from 0700 to 1900 hours (12:12-h light–dark cycle), with access to water and chow ad libitum. Genotyping of Tet2, Tet3, and Mb1-cre alleles was performed by PCR amplification of genomic DNA purified from ear snips, using the following primers: Tet2 F GCCCAAGAAAGCCAGACCAAGAA, Tet2 R AAGGAGGGGACTTTTACCTCTCAGAGCAA, Tet3 F CAGGTAGGGACGTGAACTGTGG, Tet3 R TGACCAACCCCAACACGGAAC, Mb1-cre F CTGTGGATGCCACCTCTGATGAAGTC, and Mb1-cre R TCTGATTCTCCTCATCACCAGGGACAC. It should be noted that the deletion of Tet2 and Tet3 floxed DNA in the Mb1-cre crosses was extremely efficient in both pro-B and follicular B cells, with no retention of the floxed exons in the Tet3 locus and only 0.5–1% retention in the Tet2 locus as measured by PCR.
Six- to eight-wk-old mice were used for all experiments. For T-dependent immunization experiments, mice were injected once i.v. with 1 × 109 SRBCs (Cedarlane). All animal care followed guidelines of the Max Delbrück Center for Molecular Medicine and the Innsbruck Medical University. Animal care and procedures were approved by the governmental review board (Landesamt für Gesundheit und Soziales Berlin, LaGeSo G0374/13).
Flow Cytometry and Cell Sorting.
Generation of single-cell suspensions was performed by gentle homogenization through nylon mesh filters (70 µM; spleen; BD) or mechanic disruption of cell clusters by pipetting (bone marrow, peritoneal washes). Erythrocytes in bone marrow and spleens were lysed with ACK lysis buffer on ice before staining, and cell numbers were determined using a hemocytometer (Neubauer) and trypan blue exclusion. Single-cell suspensions for flow cytometry were stained with the following antibodies: αB220-BV785 (RA3-6B2); αCD19-BV605 (6D5); αIgD-PerCP/Cy5.5 (11-26c.2a); αCD93-APC and αCD93-PeCy7 (AA4.1); αCD11b-BV711 (M1/70); αCD38-APC (90); αCD117-APC (ACK2); αCD25-PE (PC61) and αCD5-PE (53-7.3) from BioLegend; αCD1d-PE (1B1) from eBioscience; αCD95-PeCy7 (Jo2) and αIgG1-PE (A85-1) from BD; and goat α-mouse IgM, μ chain specific, from Jackson ImmunoResearch. Single-cell suspensions for cell sorting were stained with the following antibodies: αB220-PerCP/Cy5.5 (RA3-6B2); αCD19-BV421 (6D5); αCD93-APC and αCD93-PeCy7 (AA4.1); αCD25-PE (PC61) and αCD117-APC (ACK2) from BioLegend; and goat α-mouse IgM, μ chain specific, from Jackson ImmunoResearch. Samples were acquired on an LSR Fortessa (BD) or sorted on a FACSAria (BD) with FACSDiva software (BD), and data were analyzed with FlowJo software (Tree Star).
Bisulfite Methylation Analysis.
For RRBS, DNA was isolated from snap-frozen tissues or FACS-sorted B cells using the Qiagen DNeasy Blood & Tissue Kit (Cat. 69504) according to the manufacturer’s instructions. RRBS libraries were prepared using 20 ng of DNA as described (26) and run on HiSeq 2500 (Illumina). Results were highly reproducible between replicates as determined by clustering analysis (Fig. 1B).
In general, bisulfite conversion of genomic DNA was carried out using the Qiagen Epitect Bisulfite Kit (Cat. 59104), according to the manufacturer’s instructions. Bisulfite conversion efficiency was always above 99% as judged by conversion of all cytosines not in CG (about 25 million per sample) context to thymines. Specific primers of chosen tiles were designed with the MethPrimer website (64). After PCR amplification, the DNA was extracted from the gel with the Qiagen MinElute Gel Extraction Kit (Cat. 28604) and sequenced by Miseq.
Primers are as follows: regulatory regions of Igκ locus: 3′ Eκ F GTGATTGGTTTGAGAAGATTAA, 3′ Eκ R CATCCCAATACTAAATACAACCT; 3ED F TTAGTTGGTTTGAAGGTGTAGG, 3ED R CCTACACCTTCAAACCAACTAA; Jκ2 F TTTTTGGAGAATGAATGTTAGTGTAATAAT, Jκ2 R TAAAACAATTTTCCCTCCTTAACAC; regulatory regions of miR-142: miR-142 3′ F TTGATATTTGGGGAGATATTATAGT, miR-142 3′ R CCAATAACAAAATCAAACAAAAAC; mir-142 5′ F TAGGGTATGTGAGATGGTTTTT, miR-142 5′ R CCCAAAATTAAAAAACCCTAAT. Targeted tiles were filtered by sequence quality and aligned to the reference sequence using custom scripts in MATLAB. Average coverage of the targeted sequenced regions was ∼25,000 reads per sample. It should be noted that this assay does not distinguish between 5mC and 5hmC. It is therefore possible that the methylation detected in wild-type DNA from pro-B or follicular B cells includes small quantities of 5hmC as an intermediate in the demethylation process.
Gene Expression.
RNA was isolated from snap-frozen FACS-sorted B cells using the Qiagen miRNeasy kit according to the manufacturer's instructions (Cat. 217084). Fifty nanograms of RNA was taken from different samples and reversed to cDNA using the qScript kit (Quanta Biosciences 95047). Expression levels of specific genes were tested and normalized to two different housekeeping genes. For RNA-seq experiments, 20–200 ng of RNA was isolated, and the TruSeq RNA Sample Preparation Kit v2 (Illumina) was used for Library preparation. Primers are as follows: housekeeping genes: UBC F CAGCCGTATATCTTCCCAGACT, UBC R CTCAGAGGGATGCCAGTAATCTA; Gapdh F CCTGGAGAAACCTGCCAAG, Gapdh R CAACCTGGTCCTCAGTGTAGC.
RNA-seq 50-bp single-end reads were obtained from the HiSeq 2500 and analyzed by TopHat2 v2.0.10 with default parameters (65). Differential expression between wild-type and knockout mice was analyzed by CuffDiff v2.1.1 with default parameters (Q value ≤ 0.01) (66) and shown to be reproducible between replicates by clustering analysis (data not shown).
Data Analyses.
All data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE70538, and a portion has been included as Dataset S1. One-hundred-base pair paired-end sequencing reads from RRBS were obtained using the HiSeq 2500. Adapter trimming and quality filtering were performed with the trim galore software (v0.3.3) using default parameters for RRBS analysis with the following command:
trim_galore–rrbs–paired sample_R1.fastq.gz sample_R2.fastq.gz
BSMAP v2.74 (67) was used for read alignment (with genome build mm9) and extraction of single-base resolution methylation levels using the following commands:
bsmap -a sample_R1_val_1.fq.gz -b sample_R2_val_2.fq.gz -d mm9_AllGenome.fa -o bam_file -R -D C-CGG
python methratio.py -i “no-action” -p -g -z -o sample_trim_galore_mm9.fa.meth -d mm9_AllGenome.fa bam_file
One-hundred-base pair tiles and DMRs were calculated with the MethylKit package v0.9.1 (68) using a minimum coverage of 10 per tile, a methylation difference of 40%, and a Q value ≤0.01. The statistical significance of the DMR set was calculated using a permutation test: the above steps were repeated on random permutations of wild-type and knockout samples, and for each permutation the number of DMRs was calculated. The P value was obtained by measuring the quantile of the original DMR size (in our case the original number of DMRs was the highest among all permutations).
Promoter methylation levels were calculated by summing the methylated calls of all single CpGs in the promoter and dividing by the sum of their coverage. Regions with a minimal total coverage of 10 were reported. Specifically, promoter regions were defined as the 2,000 bp upstream to the TSS. For each promoter , let be the list of indices within the promoter region that contain a CpG. Let be the number of methylated calls of index and be the number of unmethylated calls of index . The methylation level of the promoter was defined as
and was reported only if .
Motif analysis was carried out by HOMER v4.7.2 (findMotifs.pl) (41). ChIP-seq data were obtained from the publicly available Gene Expression Omnibus (GEO) database: Pax5 (GSE38046) (32); EBF1, Pu.1, and IRF4 (GSE53595) (31); Brg1 (GSM1635413) (33); 5hmC (GSE65895) (16)—histone modifications H3K4me1 and H3K27Ac; and ENCODE annotation data (GSE31039) (69). Genomic distribution was analyzed using HOMER v4.7.2 (annotatePeaks.pl) (41). Gene ontology analysis was carried out using GREAT (40).
DMRs interacting with genes that show down-regulation in the Tet2/3 DKO were identified from a published Hi-C dataset. Specifically, CH12 high-resolution Hi-C datasets (GSE63525) were analyzed using HOMER, filtering out paired reads that map within 1 kb of each other, map to regions with at least 5× higher than average sequencing coverage or are not within 500 bp of an MboI site. Interactions with a P value <0.01 and a modified Z-score >1.5 were calculated with the analyzeHi-C –interactions tool in the HOMER package, using background models of 10,000, 5,000, 2,000, and 1,000 bp bins. Interactions between promoters and DMRs were identified from this list using the HOMER annotateInteractions.pl tool.
Supplementary Material
Acknowledgments
We thank Amit Lavon for his help with the computational analysis, Claudia Grosse and Janine Cernoch for technical support, and Emmanuel Derudder for valuable discussions. This research was supported by grants from the European Research Council (Grant 268614 to H.C. and Grant 268921 to K.R.), the Israel Cancer Research Fund (H.C. and Y.B.), the US–Israel Binational Foundation (Y.B.), the Rosetrees Foundation (H.C.), the Israeli Centers of Research Excellence (I-CORE) Program of Israel Science Foundation (ISF) (Y.B. and H.C.), Joint Program between the ISF and the National Natural Science Foundation of China (NSFC) (Y.B.), and the Emanuel Rubin Chair in Medical Science (Y.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE70538).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604365113/-/DCSupplemental.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu H, Zhang Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell. 2014;156(1-2):45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Shen L, et al. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15(4):459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peat JR, et al. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Reports. 2014;9(6):1990–2000. doi: 10.1016/j.celrep.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 8.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 9.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccolo FM, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell. 2013;49(6):1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent JJ, et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12(4):470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawlaty MM, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29(1):102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn MA, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Reports. 2013;3(2):291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago M, Antunes C, Guedes M, Sousa N, Marques CJ. TET enzymes and DNA hydroxymethylation in neural development and function - how critical are they? Genomics. 2014;104(5):334–340. doi: 10.1016/j.ygeno.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shide K, et al. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia. 2012;26(10):2216–2223. doi: 10.1038/leu.2012.94. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, et al. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Reports. 2015;13(8):1692–1704. doi: 10.1016/j.celrep.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko M, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263(1):6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103(37):13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle P, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13(10):R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulis M, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47(7):746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 29.Mostoslavsky R, et al. κ chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12(12):1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwickert TA, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15(3):283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Revilla-I-Domingo R, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31(14):3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bossen C, et al. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol. 2015;16(7):775–784. doi: 10.1038/ni.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 35.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantone I, Fisher AG. Unraveling epigenetic landscapes: The enigma of enhancers. Cell Stem Cell. 2011;8(2):128–129. doi: 10.1016/j.stem.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reizel Y, et al. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev. 2015;29(9):923–933. doi: 10.1101/gad.259309.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45(10):1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49(4):645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 45.Mariappa D, Pathak S, van Aalten DM. A sweet TET-à-tête-synergy of TET proteins and O-GlcNAc transferase in transcription. EMBO J. 2013;32(5):612–613. doi: 10.1038/emboj.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi FT, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288(29):20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hon GC, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56(2):286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28(19):2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dominguez PM, et al. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Reports. 2015;12(12):2086–2098. doi: 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci USA. 2015;112(22):6796–6799. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aran D, Hellman A. DNA methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154(1):11–13. doi: 10.1016/j.cell.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 54.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14(3):R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394(6696):842–843. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 56.Rasmussen KD, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29(9):910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4(2):255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 58.Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23(6):1946–1960. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shih AH, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 62.Shlush LI, et al. HALT Pan-Leukemia Gene Panel Consortium Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ko M, Rao A. TET2: Epigenetic safeguard for HSC. Blood. 2011;118(17):4501–4503. doi: 10.1182/blood-2011-08-373357. [DOI] [PubMed] [Google Scholar]
- 64.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 65.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xi Y, Li W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akalin A, et al. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.