Significance
Many protein factors interact with the ribosome during protein synthesis. Elongation factor 4 (EF-4/LepA) is a widely distributed and highly conserved translational GTPase for which several physiological roles have been proposed. Despite this, the function of EF-4 remains unknown. We have determined a high-resolution crystal structure of the ribosome bound to EF-4 in its GTP-bound state and A-, P-, and E-site tRNAs. Notably, EF-4 induces a distinct conformation of the tRNA bound in the A site, which deviates substantially from that of a canonical A-tRNA. EF-4 interacts with both helical domains of the A-site tRNA, indicating that EF-4 recognizes the L-shaped conformation of tRNA. Our results provide insights into the tRNA remodeling capacity of EF-4 on the ribosome.
Keywords: elongation factor 4, ribosome, tRNA, remodeling, protein–RNA interactions
Abstract
During translation, a plethora of protein factors bind to the ribosome and regulate protein synthesis. Many of those factors are guanosine triphosphatases (GTPases), proteins that catalyze the hydrolysis of guanosine 5′-triphosphate (GTP) to promote conformational changes. Despite numerous studies, the function of elongation factor 4 (EF-4/LepA), a highly conserved translational GTPase, has remained elusive. Here, we present the crystal structure at 2.6-Å resolution of the Thermus thermophilus 70S ribosome bound to EF-4 with a nonhydrolyzable GTP analog and A-, P-, and E-site tRNAs. The structure reveals the interactions of EF-4 with the A-site tRNA, including contacts between the C-terminal domain (CTD) of EF-4 and the acceptor helical stem of the tRNA. Remarkably, EF-4 induces a distortion of the A-site tRNA, allowing it to interact simultaneously with EF-4 and the decoding center of the ribosome. The structure provides insights into the tRNA-remodeling function of EF-4 on the ribosome and suggests that the displacement of the CCA-end of the A-site tRNA away from the peptidyl transferase center (PTC) is functionally significant.
Translation of the genetic information requires protein factors that interact with the ribosome sequentially, regulate its activity, and guide it through the protein synthesis cycle in a concerted manner. Many of those factors are guanosine triphosphatases (GTPases), proteins that use energy from guanosine 5′-triphosphate (GTP) to promote conformational changes that lead to transitions between ribosome functional states (1, 2). In bacteria, for instance, initiation of protein synthesis is largely regulated by initiation factor 2 (IF-2), a GTPase that stabilizes the initiator tRNA in the P site of the ribosome (3). Subsequently, the elongation step is catalyzed by two universally conserved GTPases, elongation factor Tu (EF-Tu) and elongation factor G (EF-G). The ternary complex, consisting of EF-Tu, GTP, and the aminoacyl-tRNA, interacts with the ribosome to decode the codon in the A site of the ribosome. Following accommodation of the aminoacyl-tRNA in the A site of the ribosome and subsequent peptide bond formation, the tRNA–mRNA duplex is translocated by one codon—a process catalyzed by EF-G and GTP (4–6). Termination of protein synthesis is triggered when a stop codon is reached, upon which the newly synthesized protein is released with the help of release factor 3 (RF-3), yet another GTPase (7).
Elongation factor 4 (EF-4/LepA) is a highly conserved protein structurally similar to EF-G (8) and has a ribosome-dependent GTPase activity (9–13). However, despite numerous studies, its function has remained elusive (9–20). Fast kinetic studies showed that EF-4 competes with EF-G during elongation for binding to the pretranslocation (PRE) ribosome, with tRNAs in the A and P sites (17). Despite this, EF-4 was also shown to increase the rate of protein synthesis at high intracellular ionic strength (16), without any effect on translational accuracy (16, 18). Conversely, EF-4 was also reported to bind to the posttranslocation (POST) ribosome and catalyze back-translocation of tRNAs from the E and P sites to the P and A sites, respectively (9–11). Recently, ribosome profiling data suggested that EF-4 reduces ribosomal pausing at certain glycine codons and contributes to translation initiation (13). Because of these sparse and controversial experimental data, combined with limited high-resolution snapshots of EF-4 in complex with the ribosome, the mechanism of action and function of EF-4 have remained unclear.
Recently, the crystal structure of EF-4 with GDP bound to the ribosome was reported (14). In this structure, the ribosome is clockwise ratcheted and the C-terminal domain (CTD) of EF-4 occupies the A site in the 50S subunit, where it reaches into the peptidyl transferase center (PTC) and interacts with the acceptor-stem of the peptidyl-tRNA in the P site. A previous cryo-electron microscopy (cryo-EM) reconstruction of EF-4 bound to the ribosome in the presence of the nonhydrolysable GTP analog GDPNP reported a new conformation of the tRNA bound in the A site, allegedly being an intermediate step trapped in the process of back-translocation (11). However, the low resolution of this cryo-EM reconstruction limits the conclusions that can be drawn from it about the structure and function of EF-4 on the ribosome.
To gain further insights into the function of EF-4, we determined its crystal structure in complex with the Thermus thermophilus 70S ribosome in the presence of the nonhydrolyzable GTP analog, GDPCP, and the A-, P-, and E-site tRNAs. The structure provides a detailed account of the contacts between EF-4, the ribosome, and the A-site tRNA, in particular revealing the network of interactions of the CTD region of EF-4 that stabilize the distorted conformation of the tRNA bound in the A site.
Results
Crystallization of the L9–EF-4 Fusion Protein with the Ribosome.
The crystallization of the wild-type 70S ribosome largely depends on the inter-ribosome packing mediated by ribosomal protein uL9 in the asymmetric unit of the crystal (21). We therefore took advantage of the fact that ribosomes isolated from a Thermus thermophilus strain that carries a truncated endogenous ribosomal protein uL9 (70S:L91–58) altogether lack protein uL9 and do not crystallize under previously published conditions (6, 14). To rescue crystal growth of ribosomes lacking uL9, we incubated the 70S:L91–58 ribosomes with the N-terminal domain of protein uL9, which has been covalently linked to EF-4 and crystallized those ribosomes as described previously (14) (Materials and Methods). Varying the length of the linker between uL9 and EF-4 allows for selection of only those protein fusions that yield crystals, indicating proper docking of EF-4 and uL9 to their respective binding sites on the ribosome. This engineered crystallization approach has recently been used successfully to determine structures of the ribosome bound to EF-4–GDP and EF-G–GDP (6, 14).
Overview of the Structure.
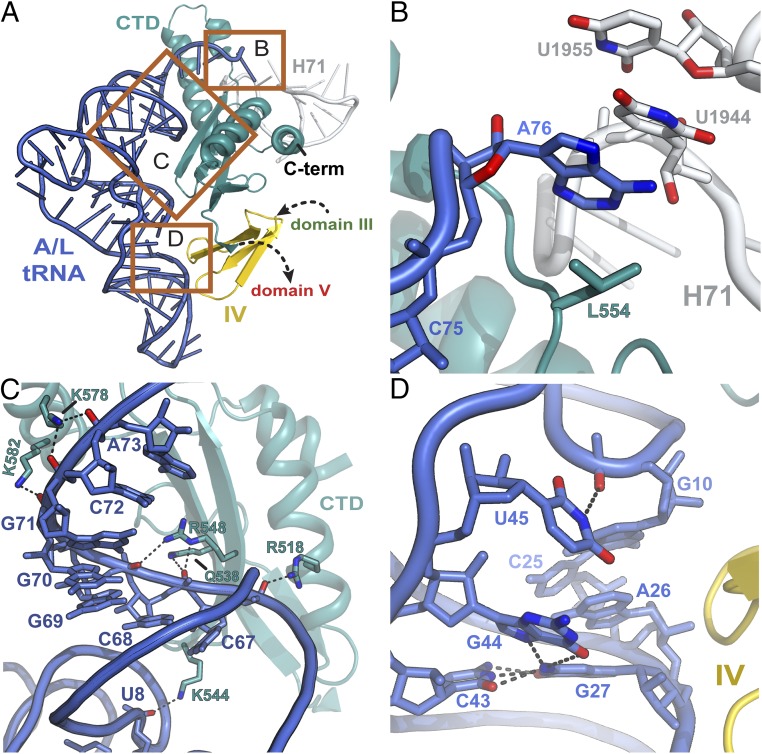
The structure, refined to a resolution of 2.6 Å, contrasts with the previous EF-4–GDP–ribosome structure complex (14) in that it has three tRNAs bound in the A, P, and E sites of the ribosome, thereby mimicking a PRE ribosome substrate (Fig. 1 and Fig. S1A). Although the tip of the CTD (or domain VI) of EF-4 reaches into the PTC and contacts the acceptor-end of the peptidyl-tRNA in the P site as previously observed in the presence of GDP (14), a significant portion of the CTD interacts with the acceptor- and D-stems of the A-site tRNA (Fig. 1). Additional interactions with the A-tRNA are mediated by domain IV of EF-4, which contacts the anticodon-stem region.
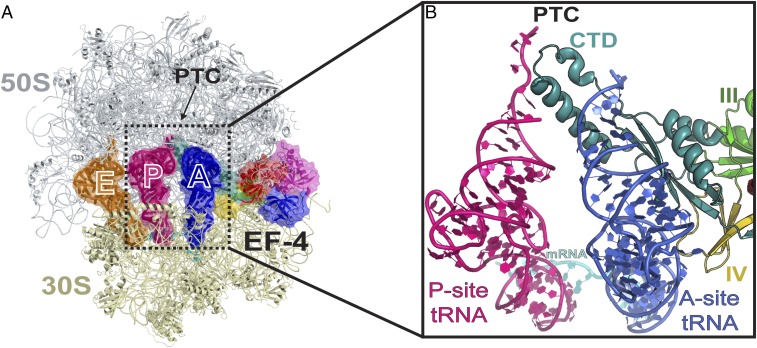
Fig. 1.
The structure of EF-4–GDPCP bound to the ribosome. (A) Overview of EF-4–GDPCP bound to the 70S ribosome. tRNAs in the E, P, and A sites are displayed in orange, pink, and blue, respectively. The 50S and 30S subunits are shown in light blue and yellow, respectively. Portions of the ribosome are omitted for clarity. (B) Close-up view of the C-terminal domain of EF-4 (CTD) (teal) that wraps around the acceptor-stem of the A-site tRNA (blue).
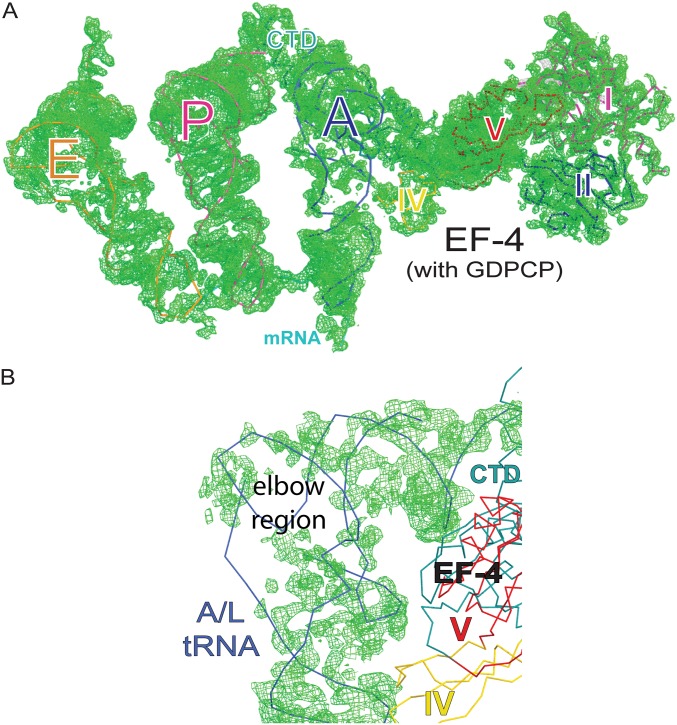
Fig. S1.
Experimental difference Fourier electron density maps. (A) Minimally biased Fobs – Fcalc difference Fourier map of EF-4–GDPCP bound to the 70S ribosome (contoured at ∼2.5σ). tRNAs in the E, P, and A sites and domains of EF-4 are colored as in Fig. 1 of the main text. For clarity, the 50S and 30S subunits of the ribosome are not shown. (B) Close-up view of the elbow region of the A/L-tRNA with the Fobs – Fcalc difference Fourier map shown. The maps were obtained after initial refinement using a vacant ribosome as a starting model.
In this structure, the ribosome is in a classical state of ratcheting. This also contrasts with the previous complex structure of the ribosome bound with EF-4–GDP in which the ribosome is clockwise ratcheted (14). As a result, the conformation of the decoding center is such that it encloses the anticodon-stem loop of the A-site tRNA in the same manner as seen during standard decoding (22, 23), allowing the universally conserved nucleotides A1492 (Escherichia coli nucleotide numbering is used throughout the text) and A1493 in helix 44 (h44), and G530 in h18 of 16S ribosomal RNA (rRNA), to form canonical interactions with the minor groove of the codon–anticodon minihelix of the A-site tRNA (Fig. S2A).
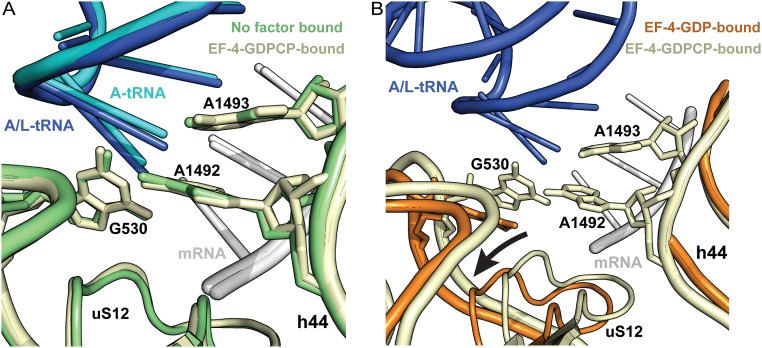
Fig. S2.
Conformation of the decoding center in different ribosome complexes. (A) Superposition of the 70S ribosome structure in the absence of factor (green) [PDB ID code 1VY4 (46)] with the current EF-4–GDPCP complex structure (yellow). The mRNA in both complexes is white, the A/L-tRNA in the EF-4–GDPCP complex is blue, and the A-site tRNA in the ribosome with no factor is cyan. (B) Superposition of the EF-4–GDP–bound 70S ribosome (orange) [PDB ID code 4W2E (14)] with the current EF-4–GDPCP–70S structure (yellow). The A/L-tRNA (blue) and the mRNA (white) are from the current structure complex. In the EF-4–GDP–70S complex, the G530-loop is displaced further away from the codon–anticodon minihelix, effectively opening the decoding center. In both panels, universally conserved nucleotides G530, A1492, and A1493, and ribosomal protein uS12 are shown. Because of the absence of A-site tRNA in the previous EF-4–GDP–70S ribosome complex (orange) [PDB ID code 4W2E (14)], nucleotides A1492 and A1493 in helix 44 (h44) are disordered. In this figure, the structures were superposed by overlapping their 16S rRNAs.
Structural Comparison with the GDP-Bound Form.
The relative interdomain arrangement in EF-4 in this ribosome complex is similar to its GDP-bound form reported previously (14) (Fig. 2A) and to that seen in the previous low-resolution cryo-EM reconstruction of EF-4–GDPNP in complex with the ribosome (11). The latter observation indicates that the chimeric fusion between proteins uL9 and EF-4 does not interfere with the conformation of EF-4 on the ribosome.
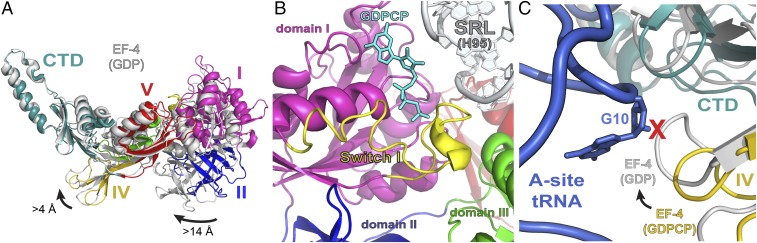
Fig. 2.
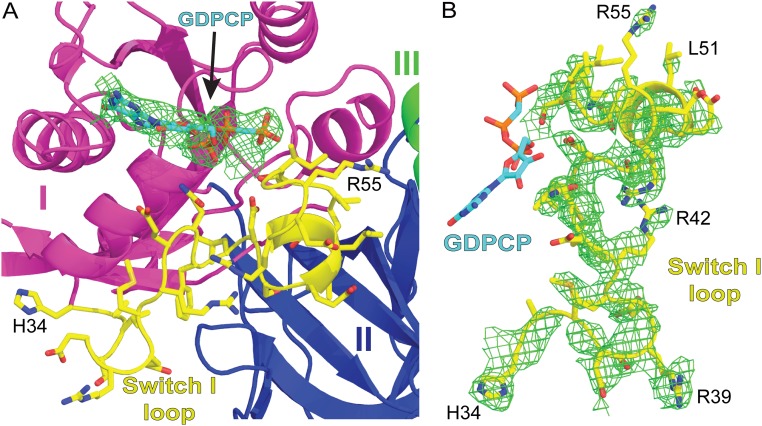
Conformation of EF-4 on the ribosome. (A) Superposition of 23S rRNA in the EF-4–GDP ribosome complex reported previously [PDB ID code 4W2E (14)] with the current EF-4–GDPCP complex reveals the conformational rearrangements in EF-4 that follow GTP hydrolysis. Compared with EF-4–GDPCP (each domain is colored as in Fig. 1), the rearrangements in EF-4–GDP (gray) can be approximated by a rotation around the SRL. (B) Binding of GDPCP (cyan) to EF-4 stabilizes the conformation of residues 35–61 forming the catalytic switch I loop (yellow). (C) The rotation of EF-4 on the ribosome upon GTP hydrolysis results in a collision (red “X” mark) with the A-site tRNA (blue).
Similar to EF-G and RF-3 bound to a GTP analog on the ribosome (24–28), the presence of GDPCP bound to EF-4 stabilizes the conformation of residues 35–61 forming the catalytic switch I loop (Fig. 2B and Fig. S3), consistent with its role in GTP hydrolysis. Switch I loop interacts with the sarcin–ricin loop (SRL) of 23S rRNA and with domain III of EF-4 (Fig. 2B), resulting in a more compact EF-4 structure. Hydrolysis of GTP triggers interdomain rearrangements that result in a more open conformation of EF-4. In agreement with this and compared with the position of EF-4–GDPCP on the ribosome, the overall positioning of EF-4–GDP on the ribosome changes slightly, which can be approximated by a rotation of the G domain around the SRL (Fig. 2A). As a result, domain IV in the EF-4–GDP–ribosome structure shifts by more than 4 Å toward the A site and would collide with the tRNA bound in the A site (Fig. 2 A and C), explaining why the presence of the A-site tRNA is not compatible with the previous EF-4–GDP–ribosome structure (14).
Fig. S3.
Difference Fourier electron density maps of GDPCP and the catalytic switch I loop in EF-4. (A) Minimally biased Fobs – Fcalc difference Fourier map of GDPCP (cyan) in EF-4 bound to the ribosome. Residues forming the catalytic switch I loop are shown in yellow. (B) Same as in A, but showing the Fobs – Fcalc map of the catalytic switch I loop in EF-4. The maps are contoured at ∼2.5σ.
EF-4 Remodels the A-Site tRNA.
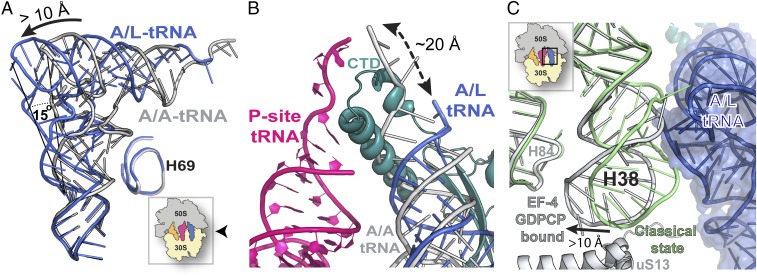
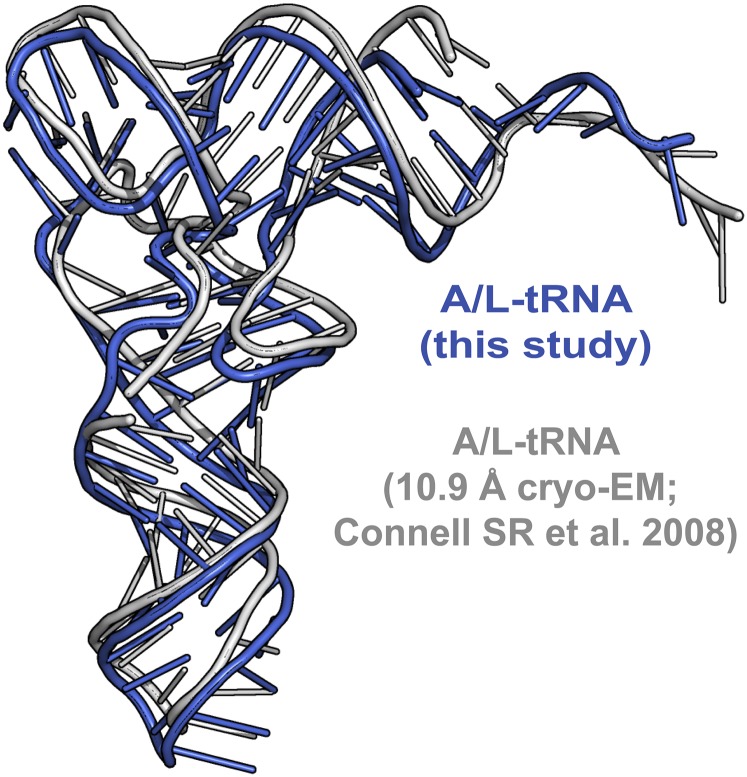
Whereas the tRNA in the P site has the classical P/P conformation, the tRNA in the A site is distorted relative to the position of a canonical A-tRNA (Fig. 3) (Materials and Methods, Note). The overall conformation of the A-site tRNA in the current complex is very similar to the one previously observed in a low-resolution cryo-EM reconstruction with EF-4–GDPNP (Fig. S4) (11), suggesting that it likely represents a similar state. This state of the A-site tRNA binding was heretofore designated A/L, referring to its simultaneous binding to the A site of the ribosome and LepA (EF-4) (11).
Fig. 3.
Distortion of the tRNA bound in the A site. (A) Compared with a tRNA bound to the ribosome in the classical A/A conformation (gray) [PDB ID code 4V51 (45)], the interaction of the A-site tRNA with EF-4–GDPCP alters its conformation (blue). Because of its interaction with EF-4 (LepA), this conformation has previously been named “A/L” (11). The view of the ribosome is indicated by the Inset. (B) The location of the EF-4–CTD (teal) observed in this structure is not compatible with the acceptor-stem of a tRNA bound in the classical A/A conformation (gray). To avoid a collision with the CTD of EF-4, the CCA-end of the A-site tRNA (gray) is displaced away from the PTC, resulting in the A/L conformation (blue). (C) Displacement of the A-site finger (H38) in the 50S subunit of the ribosome along ribosomal protein uS13 yields additional space required to accommodate the elbow region of the A-tRNA in the A/L conformation. The Inset indicates the region of the ribosome shown.
Fig. S4.
Comparison of the A/L conformation of the A-site tRNA. The conformation of the A/L-tRNA in this study (blue) is very similar to the one previously obtained from a low-resolution cryo-EM reconstruction (gray) [PDB ID code 3DEG (11)].
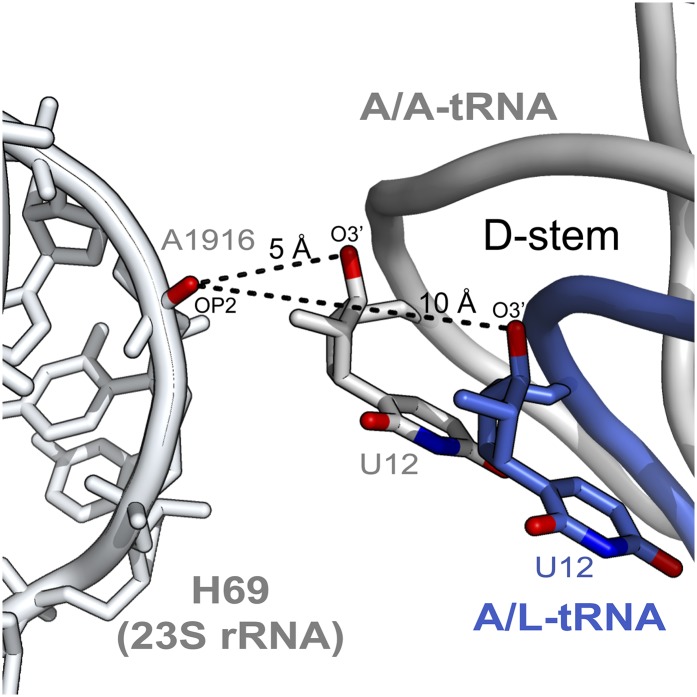
The high resolution of the current complex allows visualizing details of the A-site tRNA distortion mediated by EF-4–GDPCP. Although the anticodon-loop region superposes well with the corresponding part of a tRNA bound in the A/A conformation, the body of the tRNA bends by ∼15° from base pair 28–42 located in the anticodon-helix (Fig. 3A), displacing the D-stem by ∼5 Å further away from helix 69 (H69) of 23S rRNA (Fig. S5). The largest displacement is observed at the top of the tRNA along the T- and acceptor-stem stack (Fig. 3A), where a positional shift of more than 10 Å occurs, moving the CCA-end out of the PTC by about 20 Å (Fig. 3B). The distortion of the tRNA is required to accommodate the CTD of EF-4 in the A site of the ribosome, which would otherwise collide with the acceptor-stem of a tRNA bound in the classical A/A conformation (Fig. 3B). The tip of the A-site finger helix (H38) in the 50S subunit, which contacts ribosomal protein uS13 in the 30S subunit and forms the RNA–protein intersubunit bridge B1a, is displaced by more than 10 Å to accommodate the distorted conformation of the tRNA in the A site (Fig. 3C).
Fig. S5.
Relative distances between the A-site tRNA in two different conformations and helix 69 (H69) of 23S rRNA. The remodeling of the A-site tRNA by EF-4–GDPCP distorts the tRNA conformation, displacing the D-stem of the A/L-tRNA further away from H69 of 23S rRNA. Distances between the phosphate oxygen atom (OP2) of nucleotide A1916 in H69 and the O3′ atom of nucleotide U12 in the D-stem of tRNA are indicated by black dashes.
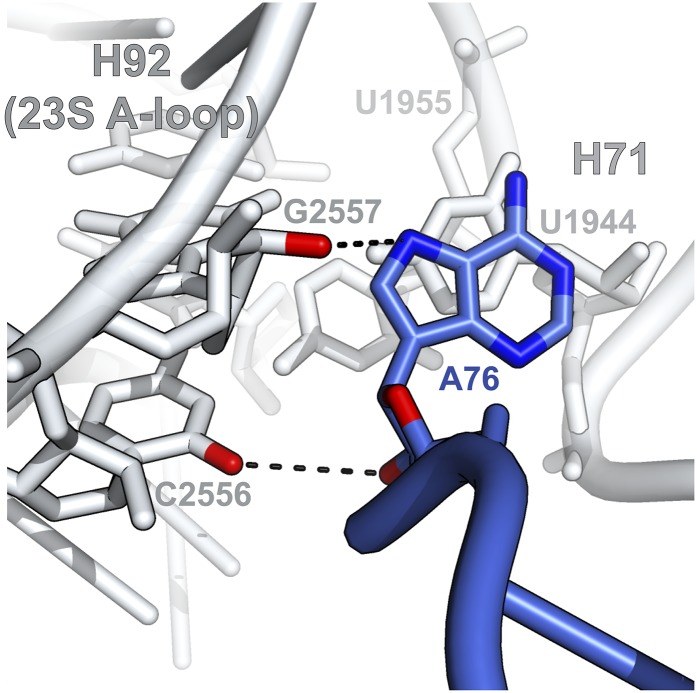
In this structure, the CTD of EF-4 wraps around the acceptor-stem of the distorted A-site tRNA, repositioning the CCA-end such that A76 forms a π-stacking interaction with U1944 in H71 of 23S rRNA and leucine 554 of EF-4, which, with U1955, result in a four-layer stack unit (Fig. 4B). In this position, nucleotide A76 also interacts with the minor groove of H92 of 23S rRNA, which contains the 23S A-loop (Fig. S6). This is consistent with the reported change in chemical protection of this region of 23S rRNA upon EF-4–GDPNP interaction with the ribosome (29).
Fig. 4.
Interactions of EF-4–GDPCP with the A/L-tRNA. (A) Overview of the contacts between EF-4–GDPCP and the A/L-tRNA. Each region is boxed and labeled according to its corresponding panel in this figure. (B) Nucleotide A76 of the A/L-tRNA, together with residues Leu554 of EF-4, U1944, and U1955 of 23S rRNA, are sandwiched to form a four-layer stacking unit. (C) Phosphate backbone-mediated interactions between the CTD of EF-4–GDPCP and the acceptor-stem of the A/L-tRNA. (D) The intrinsic flexibility of nucleotides in the core region of the A/L-tRNA facilitates remodeling of the tRNA in the A site by EF-4. The oxygen and nitrogen atoms are red and dark blue, respectively. Putative hydrogen bonds are shown as black dashes.
Fig. S6.
Interactions of A76 of the A/L-tRNA with 23S rRNA. In addition to participating in a four-layer stacking interaction with H71, nucleotide A76 of the A/L-tRNA also interacts with the minor groove of helix 92 (H92) that contains the 23S A-loop. Putative hydrogen bonds are shown as black dashes.
EF-4 Probes the L Shape of the tRNA.
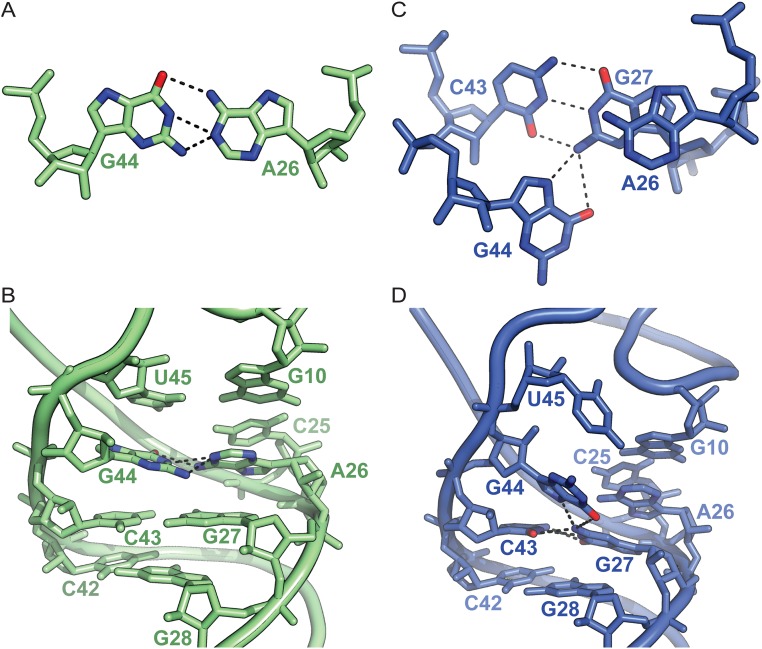
Many residues of the CTD in EF-4 contact the sugar–phosphate backbone of nucleotides 67–73 in the acceptor-stem of the A/L-tRNA (Fig. 4C). Another contact between EF-4 and the A/L-tRNA occurs at the junction of the acceptor- and D-stems, where the sugar–phosphate backbone of nucleotide U8 interacts with lysine 544 of the CTD in EF-4 (Fig. 4C). Domain IV of EF-4 contacts the anticodon/D domains of the A/L-tRNA, in the region of nucleotides G44 and A26 located at the junction between the anticodon- and the D-stems of the A/L-tRNA (Fig. 4 A and D). Remarkably, instead of forming the noncanonical G44–A26 imino base pair usually observed in tRNAPhe (Fig. S7 A and B), G44 bulges out of the helical stem to form a new base triple interaction with the G27–C43 base pair (Fig. 4D and Fig. S7 C and D), disrupting the continuous stacking of the anticodon/D domains. In addition, instead of forming the usual base triple with G10–C25, U45 bulges out and forms a π-stacking interaction with nucleotide G44 (Fig. 4D and Fig. S7 B and D). Several base pair rearrangements in the core of tRNAPhe in addition to bending of ribosome-bound tRNA at the anticodon/D-stem junction have been reported (30–32). The flexibility of nucleotides G44 and U45 observed here suggests that remodeling of the tRNA on the ribosome is facilitated by plasticity in the core of the tRNA body. The elbow region of the tRNA, which interacts neither with EF-4 nor the ribosome, appears to be flexible based on the quality of the electron density of this region (Fig. S1B). Most of the interactions between EF-4 and the A/L-tRNA are backbone mediated, encompass both helical domains of the tRNA, and, therefore, appear to probe the overall L-shaped conformation of the A-site tRNA (Fig. 4).
Fig. S7.
Structural plasticity in the core of the tRNA body. (A) Top view of the G44-A26 imino base pair observed in the A-site tRNAPhe from a previous 70S ribosome structure [PDB ID code 4V51 (45)]. (B) Side view of the junction between the anticodon/D helical domains in the same ribosome-bound tRNAPhe. (C) Top view of the same region within the A/L-tRNA. The EF-4–induced remodeling of the A/L-tRNA is facilitated by nucleotide rearrangements in the core of the tRNA body. Nucleotide G44 bulges out of the anticodon/D helical domains and forms a new base triple with base pair G27–C43. (D) Side view of the A/L-tRNA showing the distortion of the continuous stacking between the anticodon/D helical stems. Putative hydrogen bonds are shown as black dashes.
Discussion
In agreement with previous biochemical data (17), our structure shows that the substrate of EF-4 is likely a PRE ribosome carrying at least the A- and P-site tRNAs, instead of a POST ribosome, which would be required for the previously proposed back-translocation activity of EF-4 (9–11). At this point, however, we still cannot infer with certainty the cellular function of EF-4, although two hypotheses can be reasonably formulated. The hypotheses originate from the observation that EF-4 displaces the CCA-end of the A-site tRNA away from the PTC of the ribosome (Fig. 3B).
The displacement of the CCA-end of the A-site tRNA may lead to its dissociation from the ribosome, which would result in the previous structure of the ribosome in complex with EF-4–GDP (14). In the EF-4–GDP–70S structure, the ribosome is clockwise ratcheted and the decoding center has an open conformation (Fig. S2B) (14), features that would presumably help to disengage the tRNA anticodon stem-loop from the ribosome and may lead to the release of the A-site tRNA. Under stress conditions, such as high magnesium concentration and amino acid starvation, deacylated tRNAs bind to the A site of the ribosome, causing protein synthesis to stall (33–35). In E. coli, the stringent factor RelA senses the presence of a deacyl-tRNA in the A site of the ribosome and triggers a stress response (35–37). EF-4 may provide an alternative solution by clearing the A site of the ribosome to resume protein synthesis.
It is possible that the A/L-tRNA could instead be aminoacylated or carry a nascent polypeptide chain. Based on our structure, there is sufficient space around the displaced CCA-end of the A/L-tRNA for it to be charged with an amino acid (Fig. 4B). It is less likely, however, that EF-4 promotes dissociation of an aminoacyl- or peptidyl-tRNA from the A site. This is because EF-4–mediated dissociation of aminoacyl-tRNA would imply that EF-4 has an effect on translation accuracy, which was shown not to be the case (16, 18). Also, the presence of a nascent polypeptide chain attached to the A-site tRNA would restrict the tRNA conformation. We therefore hypothesize that, if EF-4 pulls the acceptor-stem of a peptidyl-tRNA in the A site backward upon binding to PRE ribosome, the polypeptide chain would also have to travel backward along the peptide exit tunnel. It has been shown that external force exerted on the nascent peptide in the forward direction can alleviate ribosome stalling (38). The physiological significance of pulling backward the peptide chain by EF-4, if any, is not clear. The destabilization of the acceptor-end of the A-site tRNA may help unlock a stalled ribosome, such that tRNAs could be translocated. To this end, the hydrolysis of GTP, which causes domain IV of EF-4 to collide with the A-site tRNA (Fig. 2C), could promote tRNA movement in the forward direction. Another possibility is that a displacement of the CCA-end of the peptidyl-tRNA in the A site could unfold a partially misfolded protein outside the peptide exit tunnel, giving a second chance for the nascent peptide to refold. It is noteworthy that EF-4 was reported to increase both the rate of protein synthesis (16) and the fraction of active synthesized proteins (9).
Despite that EF-4 is a universally conserved translation factor in bacteria and organelles, all recent efforts have failed to uncover its cellular role. We have shown here that EF-4 remodels the A-site tRNA by recognizing its L-shaped conformation, causing a displacement of the acceptor-stem of the tRNA away from the PTC. Additional studies are required to understand the functional significance of the A/L distortion of the A-site tRNA.
Materials and Methods
mRNA, tRNAs, Ribosomes, and the L9–EF-4 Protein Fusion.
The mRNA with a Shine–Dalgarno sequence and an initiation codon in the P site was synthesized by Integrated DNA Technologies with the sequence 5′-GGC AAG GAG GUA AAA AUG UUC UAA-3′. The start codon in the P site is bold, and the A-site codon is underlined. The fMet-tRNAfMet and the Phe-tRNAPhe were prepared as previously described (39). We used the same L9–EF-4 protein fusion, expression, and purification procedures as described previously (14). The 70S ribosomes (70S:L91–58) were prepared from the Thermus thermophilus HB8 strain that carries a truncated ribosomal protein uL9 in its genome (6, 14).
Complex Formation and Crystallization.
Ribosome complexes were prepared and crystallized essentially as described previously with some minor modifications (14). Briefly, 4 µM 70S:L91–58 ribosomes were incubated with 8 µM mRNA in 5 mM Hepes-KOH (pH 7.5), 10 mM Mg(CH3COO)2, 50 mM KCl, 10 mM NH4Cl, and 6 mM β-mercaptoethanol at 55 °C for 5 min. The P-site fMet-tRNAfMet was added to a final concentration of 8 µM, and the complex was further incubated at 55 °C for 5 min. Following the addition of 20 µM A-site Phe-tRNAPhe and incubation at 55 °C for 10 min, which resulted in the formation of a PRE ribosome complex, the L9–EF-4 protein fusion and the nucleotide were added directly to the mixture. The crystals grew to full size at 20 °C within 7–10 d in sitting drop trays in which 3 µL of ribosome complex was mixed with 3–4.5 µL of reservoir solution containing 100 mM Tris⋅HCl (pH 7.6), 2.9% (wt/vol) PEG 20000, 9% (vol/vol) 2-methyl-2,4-pentanediol, 150 mM l-arginine, and 0.5 mM β-mercaptoethanol. The crystals were transferred stepwise to a cryoprotectant solution containing 100 mM Tris⋅HCl, pH 7.6, 10 mM Mg acetate, 50 mM KCl, 10 mM NH4Cl, 6 mM β-mercaptoethanol, 2.9% (wt/vol) PEG 20000, 40% (vol/vol) 2-methyl-2,4-pentanediol, and 1 mM GDPCP, and left to equilibrate overnight at 20 °C before being frozen in a liquid nitrogen stream at 80 K as described (14).
Data Collection and Structure Determination.
X-ray diffraction data were collected at beamline 24ID-C at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) using 0.3° oscillations. Data were integrated and scaled with the XDS software package (40). Molecular replacement was performed using PHASER (41). As with EF-4–GDP (14), one 70S ribosome was present in the asymmetric unit of the crystal. The L9–EF-4 protein fusion bound to GDPCP, the mRNA and tRNAs in the A, P, or E sites were built into the unbiased Fobs – Fcalc difference Fourier electron density map using COOT (42), and the structure was refined in PHENIX (43). Because we used two different tRNAs to assemble the PRE ribosome complex, the tRNA bound in the E site of the ribosome likely represents a mixture of deacylated tRNAfMet and tRNAPhe. Based on the electron density, the E-site tRNA in our structure is modeled as tRNAPhe that is present in a 2.5-fold molar excess than tRNAfMet. The site occupancies for EF-4, the A-, P-, and E-site tRNAs are 97%, 90%, 96%, and 94%, respectively. The final refinement statistics are provided in Table S1.
Table S1.
Data collection and refinement statistics of the EF-4–GDPCP–70S ribosome complex
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c, Å | 213.1, 271.7, 436.8 |
| α, β, γ, ° | 90.0, 90.0, 90.0 |
| Resolution, Å | 50–2.59 (2.75–2.59)* |
| Rmerge, % | 25.7 (151.7) |
| I/σI | 8.23 (1.13)† |
| Completeness, % | 98.7 (95.1) |
| No. of reflections | |
| Observed | 3,199,533 (497,190) |
| Unique | 766,326 (118,578) |
| Redundancy | 4.18 (4.19) |
| Refinement | |
| Resolution, Å | 50–2.6 |
| Rwork/Rfree, % | 19.4/26.0 |
| No. of atoms | |
| RNA | 101,413 |
| Protein | 51,764 |
| Ions | 933 |
| Water | 966 |
| Average B factors | |
| RNA | 49.1 |
| Protein | 55.0 |
| Ions | 47.0 |
| Water | 37.1 |
| rms deviations | |
| Bond lengths, Å | 0.007 |
| Bond angles, ° | 1.246 |
Parentheses indicate statistics from the outer shell.
I/σI = 2 at ∼2.8 Å.
Figures.
All figures were generated using PyMOL (www.pymol.org), and structure alignments were carried out by superposing the 23S rRNA, unless indicated otherwise.
Note.
While this paper was under revision, a cryo-EM reconstruction at 3.2-Å resolution of EF-4–GDPNP bound to a PRE ribosome was published (44). Although we did not have access to these structural data, the conformation of the A-site tRNA described in our structure appears to be very similar to that seen in this cryo-EM reconstruction. However, the main conclusion made by the authors is different than the functional implications proposed here. The central premise of their paper is based on the observation that the CTD of EF-4 disrupts the base pairing between the P-loop of the 23S rRNA and the CCA-end of the P-site tRNA in the POST ribosome complex reconstructed at a resolution of 3.7 Å. This unusual conformation of the CCA-end of the P-site tRNA is proposed to be at the origin of the back-translocation of tRNAs mediated by EF-4 on the ribosome. However, in the previous POST ribosome complex bound to EF-4, determined at a resolution of 2.9 Å, the CCA-end of the P-site tRNA is involved in canonical base pairing with the P-loop in the ribosome without any apparent distortion (14).
Acknowledgments
We thank Yury Polikanov (University of Illinois at Chicago) for valuable discussions, suggestions, and critical reading of the manuscript. We thank Robert Grodzicki for preparing fMet-tRNAfMet and Phe-tRNAPhe. We also thank the staffs of the Advanced Photon Source beamline 24-ID for help with data collection and the Richards Center facility at Yale University for computational support. This work was supported by National Institutes of Health Grant GM022778 (to T.A.S.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (Grant P41 GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by NIH–ORIP HEI Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5J8B).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522932113/-/DCSupplemental.
References
- 1.Clementi N, Polacek N. Ribosome-associated GTPases: The role of RNA for GTPase activation. RNA Biol. 2010;7(5):521–527. doi: 10.4161/rna.7.5.12467. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina MV, et al. GTPases mechanisms and functions of translation factors on the ribosome. Biol Chem. 2000;381(5-6):377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- 3.Allen GS, Frank J. Structural insights on the translation initiation complex: Ghosts of a universal initiation complex. Mol Microbiol. 2007;63(4):941–950. doi: 10.1111/j.1365-2958.2006.05574.x. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson J, Nissen P. Elongation factors on the ribosome. Curr Opin Struct Biol. 2005;15(3):349–354. doi: 10.1016/j.sbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 6.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160(1-2):219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisselev LL, Buckingham RH. Translational termination comes of age. Trends Biochem Sci. 2000;25(11):561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 8.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci USA. 2008;105(12):4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin Y, et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127(4):721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Pan D, Pech M, Cooperman BS. Interrupted catalysis: The EF4 (LepA) effect on back-translocation. J Mol Biol. 2010;396(4):1043–1052. doi: 10.1016/j.jmb.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell SR, et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15(9):910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 12.De Laurentiis EI, Wieden HJ. Identification of two structural elements important for ribosome-dependent GTPase activity of elongation factor 4 (EF4/LepA) Sci Rep. 2015;5:8573. doi: 10.1038/srep08573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42(21):13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagnon MG, Lin J, Bulkley D, Steitz TA. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science. 2014;345(6197):684–687. doi: 10.1126/science.1253525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Qin Y. The paradox of elongation factor 4: Highly conserved, yet of no physiological significance? Biochem J. 2013;452(2):173–181. doi: 10.1042/BJ20121792. [DOI] [PubMed] [Google Scholar]
- 16.Pech M, et al. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci USA. 2011;108(8):3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, et al. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc Natl Acad Sci USA. 2011;108(39):16223–16228. doi: 10.1073/pnas.1103820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie. 2010;92(2):157–163. doi: 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, et al. Ribosomal elongation factor 4 promotes cell death associated with lethal stress. MBio. 2014;5(6):e01708. doi: 10.1128/mBio.01708-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Li Z, Hao J, Qin Y. EF4 knockout E. coli cells exhibit lower levels of cellular biosynthesis under acidic stress. Protein Cell. 2014;5(7):563–567. doi: 10.1007/s13238-014-0050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selmer M, Gao YG, Weixlbaumer A, Ramakrishnan V. Ribosome engineering to promote new crystal forms. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 5):578–583. doi: 10.1107/S0907444912006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111(5):721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 23.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292(5518):897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340(6140):1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340(6140):1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340(6140):1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18(2):230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci USA. 2011;108(38):15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res. 2012;40(1):360–370. doi: 10.1093/nar/gkr623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrne RT, Konevega AL, Rodnina MV, Antson AA. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010;38(12):4154–4162. doi: 10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326(5953):688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 34.Rojiani MV, Jakubowski H, Goldman E. Effect of variation of charged and uncharged tRNA(Trp) levels on ppGpp synthesis in Escherichia coli. J Bacteriol. 1989;171(12):6493–6502. doi: 10.1128/jb.171.12.6493-6502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70(5):1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agirrezabala X, et al. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14(9):811–816. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haseltine WA, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238(5364):381–384. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 38.Goldman DH, et al. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348(6233):457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jünemann R, et al. In vivo deuteration of transfer RNAs: Overexpression and large-scale purification of deuterated specific tRNAs. Nucleic Acids Res. 1996;24(5):907–913. doi: 10.1093/nar/24.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26(6):795–800. [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Zhang D, et al. EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome. Nat Struct Mol Biol. 2016;23(2):125–131. doi: 10.1038/nsmb.3160. [DOI] [PubMed] [Google Scholar]
- 45.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 46.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21(9):787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]