Significance
Madagascar is a conservation priority because of its unique and threatened biodiversity. Lemurs, by acting as seed dispersers, are essential to maintaining healthy and diverse forests on the island. However, in the past few thousand years, at least 17 lemur species, many of which were inferred seed dispersers, have gone extinct. We outline the substantial impact that these extinctions have likely had on Malagasy forests by comparing the gape sizes and diets of living and extinct lemurs to identify large-seeded Malagasy plants that appear to be without extant animal dispersers. Additionally, we identify living lemurs that are endangered yet occupy unique and essential dispersal niches. This information can inform conservation initiatives targeting the protection and restoration of these vulnerable ecosystems.
Keywords: anachronism, extinction, lemurs, Madagascar, seed dispersal
Abstract
Madagascar’s lemurs display a diverse array of feeding strategies with complex relationships to seed dispersal mechanisms in Malagasy plants. Although these relationships have been explored previously on a case-by-case basis, we present here the first comprehensive analysis of lemuriform feeding, to our knowledge, and its hypothesized effects on seed dispersal and the long-term survival of Malagasy plant lineages. We used a molecular phylogenetic framework to examine the mode and tempo of diet evolution, and to quantify the associated morphological space occupied by Madagascar’s lemurs, both extinct and extant. Using statistical models and morphometric analyses, we demonstrate that the extinction of large-bodied lemurs resulted in a significant reduction in functional morphological space associated with seed dispersal ability. These reductions carry potentially far-reaching consequences for Malagasy ecosystems, and we highlight large-seeded Malagasy plants that appear to be without extant animal dispersers. We also identify living lemurs that are endangered yet occupy unique and essential dispersal niches defined by our morphometric analyses.
Madagascar’s vertebrate fauna is characterized by high levels of endemicity and diversity within a small number of taxonomic groups, a pattern attributed to the island’s long isolation (1, 2). Patterns of endemism are particularly noteworthy among native nonflying Malagasy mammals, which comprise four orders: Carnivora, Afrosoricida, Primates, and Rodentia. These mammals occur throughout Madagascar and have radiated to fill a unique variety of life history and dietary niches (1–3). For example, it has been noted that compared with other tropical biomes, Madagascar is depauperate in seed dispersers, with lemurs (the primates of Madagascar) acting as the predominant dispersers (4–8). Despite their importance as seed dispersers of native plants, living Malagasy primates comprise proportionately fewer frugivores than comparable primate assemblages on other continents, with a higher than expected proportion of lemurs eating predominantly leaves (8, 9).
Although the idiosyncratic dietary strategies of Madagascar’s lemurs have been studied in detail (3, 9), relatively less attention has been paid to Madagascar’s recently extinct lemur lineages. However, recent advances in ancient DNA sequencing have now made it possible to incorporate extinct lineages into phylogenetic analyses (10, 11). These advances yield the possibility for a richer understanding of the evolution of lemuriform feeding strategies and their potential relationship to plant dispersal mechanisms. Within the past few thousand years, at least 17 species of Madagascar’s lemurs have gone extinct (12). Based on radiocarbon dating (12), the majority of the giant lemurs went extinct after the intensification of human activity on the island roughly 1,700 y ago (13), with some extinct lineages persisting until at least 500 y ago (12). Many of these extinct lineages likely played important seed-dispersal roles, as determined by tooth morphology, dental wear, and stable isotopes (14–16), and all of them were substantially larger than any surviving lemur species (14, 15). It is widely hypothesized that these extinctions significantly reduced the morphological and ecological diversity of Malagasy seed dispersers (14, 16–18). In turn, these reductions may have had an impact on the structure and function of Madagascar’s flora (14, 16, 19), especially large-seeded plant species that would have relied on correspondingly large-bodied animals for dispersal (14, 16–18). Here, we investigate the evolutionary history of diet in Madagascar’s lemurs, both extinct and extant, and show multiple evolutionary origins of folivory. We demonstrate that a significant proportion of the dietary niche occupied by lemurs was lost with the extinctions of these large-bodied lineages. This reduction, in the context of seed dispersers, has created multiple “orphaned” Malagasy plant lineages/large-seeded taxa, bereft of their dispersers.
Results and Discussion
Evolution and Morphological Range of Dietary Strategies.
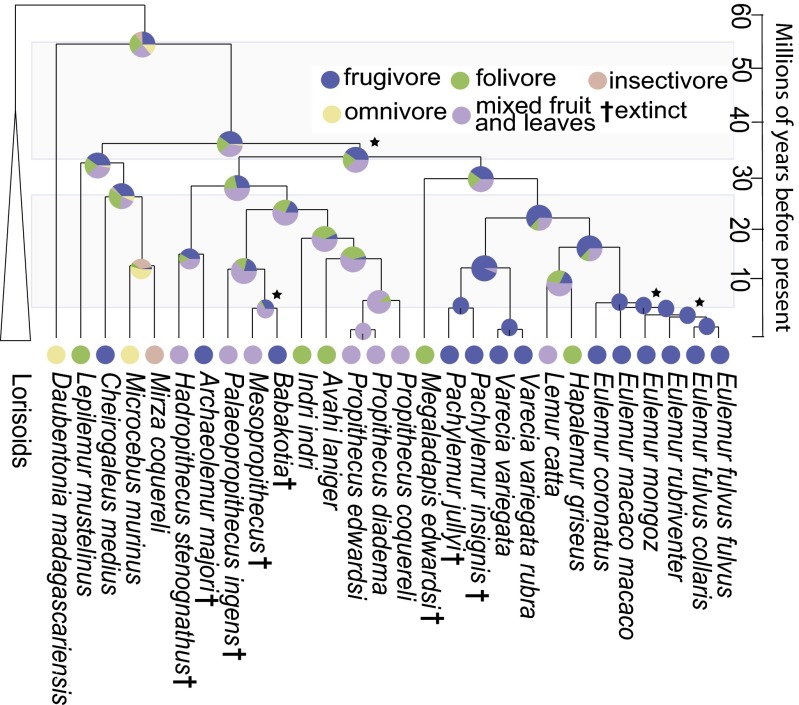
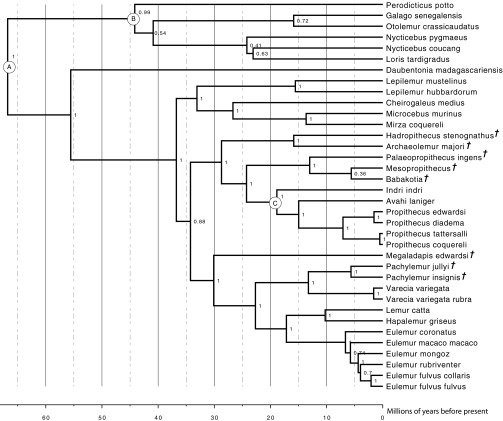
To investigate the evolution of lemur feeding strategies, we estimated a time-calibrated molecular phylogeny that included all major extinct and extant lineages (Fig. 1). We assigned diet according to behavioral observations of extant taxa (assignment information is provided in Dataset S1). For extinct taxa, diet assignments were based on inferences made in the literature using tooth morphology, dental wear, and stable isotope ratios (Dataset S1). Extinct taxa that were probable seed dispersers, and also inferred to have had a significant dietary component of leaves, were assigned to a mixed fruit-and-leaves dietary category (Dataset S1). If an organism’s diet contains low levels of C4 vegetation, it can be difficult to distinguish between folivory and frugivory using stable isotope analyses because C3 plants often exhibit high isotopic variability (20); a mixed fruit-and-leaves diet category provides a conservative assignment for potentially ambiguous diets. We simulated the evolutionary history of diet using stochastic character mapping, a method for inferring discrete character changes that accommodates phylogenetic uncertainty (21, 22) (Fig. 1). The lemuriform ancestral dietary state was inferred to have had equal probabilities of being frugivorous, being folivorous, or exhibiting a mixed diet of fruit and leaves (Fig. 1). Folivory was inferred to have evolved independently between one and five times, with phylogenetic analyses of ancient DNA demonstrating that Megaladapis, an extinct folivorous lineage, was not a sister to the strictly folivorous Lepilemur clade. Before molecular phylogenetic analyses, the two lineages were considered to form a monophyletic group on the basis of morphology (10). Thus, whereas folivory was initially assumed to have evolved only once in the putative Lepilemur-Megaladapis clade, ancient DNA analysis has revealed that it must have evolved independently at least twice. Correspondingly, frugivory was inferred to have evolved independently between one and four times. Although contemporary frugivores are concentrated within the Varecia and Eulemur clades, stem lineages leading to the Indri and Avahi clades also are inferred to have been frugivorous, indicating that frugivory was once present in an evolutionarily diverse array of lemur lineages (Fig. 1).
Fig. 1.
Evolution of lemuriform dietary strategies. Time-calibrated lemur phylogeny with stochastic character mapping of feeding strategy (color legend at the base of the figure). Unless starred, all nodes represent a Bayesian posterior probability (BPP) of 0.95 and higher, whereas starred nodes have a BPP of 0.94 and lower. Extinct taxa are indicated by a dagger (†).
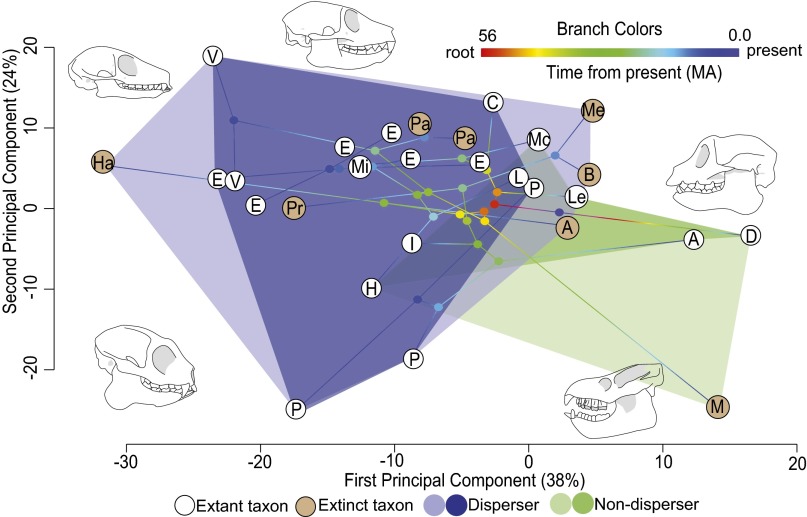
To understand the morphological breadth lost with the extinction of Madagascar’s large-bodied seed dispersers, we compiled a dataset of cranial, dental, and mandibular measurements for the species sampled in our phylogeny (specimens measured are given in Dataset S2, and illustrations of the measurements taken are shown in Fig. S1). Using our phylogenetic framework in conjunction with a phylogenetic principal components analysis (PCA), we projected the dataset of morphological measurements into multidimensional morphospace (22, 23). Visualizing the major axes of morphological variation within a phylogenetic context revealed species to be partitioned by dietary strategy (i.e., seed dispersers vs. nondispersers) rather than phylogenetic relatedness (Fig. 2). In other words, dietary strategy is a better predictor of morphology, and phylogenetic relatedness does not imply morphological conservatism. Several extinct lineages occupied the margins of the morphological space belonging to their respective dietary categories, and when extinct seed-dispersing lineages were excluded from the analysis, a substantial area (32%) of the space occupied by seed dispersers was lost (Fig. 2). This reduction of morphological space implies a proportionate loss of seed dispersal ability with potentially far-reaching consequences for the Malagasy flora (Fig. 2). Similarly, the extinction of Megaladapis resulted in a significant reduction of the morphological space associated with folivory, which, although not the focus of this study, could have an impact on the flora by changing patterns of herbivory, as has been detailed in other systems where large herbivores have been extirpated (19). Those few extant species approaching areas of morphospace once occupied by extinct lineages, such as Varecia (Fig. 2), should be considered conservation priorities because they likely perform irreplaceable ecosystem functions, including the dispersal of large seeds.
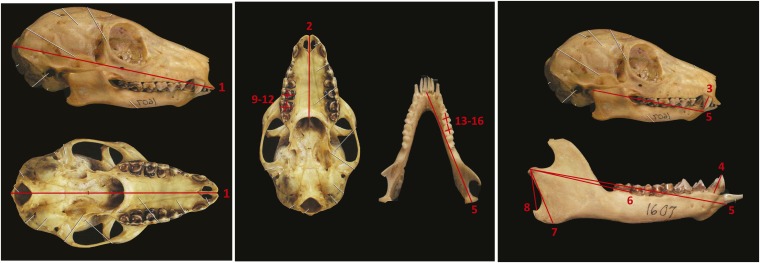
Fig. S1.
Schematic of cranial and mandibular measurements. (1) Cranium length: prosthion to inion (i.e., distance from the most anterior point on the maxillary alveolar process, between the central incisors, to the most prominent projection on the occipital bone at the posteroinferior part of the skull). (2) Palate length: prosthion to alveolare (i.e., distance from the most anterior point on the maxillary alveolar process, between the central incisors, to the posterior palatine). (3) Upper canine length: length of the upper canine from gumline to tip. (4) Lower p2 length: length of the lower second (caniniform) premolar from gumline to tip. (5) Mandible length: infradentale to the posterior edge of the mandibular condyle. (6) Superficial masseter origin: measured from the posterior edge of the mandibular condyle to the posterior margin of m2. (7) Mandibular height: measured from the superior edge of the mandibular condyle to the base of the mandibular corpus. (8) Superficial masseter insertion: measured from the superior edge of the mandibular condyle to the superior edge of the angular process. (9) Upper M1 length: length at the longest point on the upper M1. (10) Upper M1 width: width at the broadest point on the upper M1. (11) Upper M2 length: length at the longest point on the upper M2. (12) Upper M2 width: width at the broadest point on the upper M2. (13) Lower m1 length: length at the longest point on the lower m1. (14) Lower m1 width: width at the broadest point on the lower m1. (15) Lower m2 length: length at the longest point on the lower m2. (16) Lower m2 width: width at the broadest point on the lower m2.
Fig. 2.
Phylogenetic PCA of phylogenetic residuals of lemur morphological measurements projected into morphological space. Colored phylogenetic branches indicate time since the root. Tips of the phylogeny are abbreviated: A, Avahi; B, Babakotia; C, Cheirogaleus; D, Daubentonia; E, Eulemur; H, Hapalemur; Ha, Hadropithecus; I, Indri; L, Lemur; Le, Lepilemur; M, Megaladapis; Me, Mesopropithecus; Mc, Microcebus; Mz, Mirza; P, Propithecus; Pa, Pachylemur; Pr, Paleopropithecus; V, Varecia. Tips with colored white circles indicate extant taxa, and tips with orange circles indicate extinct taxa. Colored polygons represent seed dispersers vs. nondispersers, with darker shaded areas indicating the morphological space occupied solely by extant lemurs. Lemur skull illustrations show morphologies occupying particular areas of morphological space. MA, million years before present.
Seed Size, Orphaned Plants, and Maximum Ingestible Food Size.
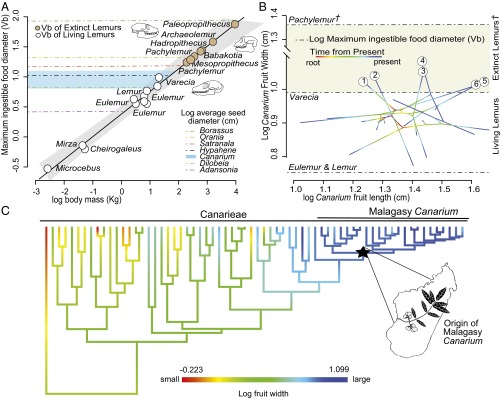
The Malagasy flora has multiple examples of large-seeded plants that, based on their morphology, appear to require animal-mediated dispersal yet have no observed relationship with frugivores (14). These plants are hypothesized to be “orphans,” bereft of their now-extinct animal dispersers (14). We compiled a database, using species descriptions, of seed size measurements for those Malagasy lineages that had previously been identified in the literature as orphans (14); these lineages included representative species of Adansonia (Malvaceae), Borassus (Arecaceae), Dilobeia (Proteaceae), Hyphaene (Arecaceae), Orania (Arecaceae), and Satranala (Arecaceae) (seed size averages are given in Dataset S3). To infer whether extinct lemurs would have been capable of ingesting and dispersing seeds from these putatively orphaned Malagasy plant lineages, we used a linear regression to estimate the maximum ingestible food size (Vb) (24) among extinct and extant frugivorous and mixed fruit-and-leaf–eating lemurs using the allometric relationship between body mass and Vb detailed by Perry and Hartstone-Rose (24) (Fig. 3A and Dataset S3). We found that the majority of these plant lineages are too large-seeded for consumption by extant lemurs, but could easily have been consumed by extinct species (Fig. 3A and Datasets S3 and S4). That extant lemurs cannot consume these seeds supports the hypothesis that extinct lemurs played a major role as seed dispersers in a niche that has narrowed significantly since their extinction, and could easily narrow further (14). For instance, in contemporary forests, the critically endangered Varecia variegata and Varecia rubra (25) occupy an otherwise empty area of dietary morphospace (Fig. 2), and are the only extant frugivores with Vbs able to accommodate many of the large-seeded plants analyzed in this study (Figs. 2 and 3). Their extinction could jeopardize the future of these plant lineages.
Fig. 3.
Seed size and evolution and the limits of Vb for extinct and extant frugivorous lemurs. (A) Linear model of relationships between frugivorous lemur body weight and Vb (n = 8; R2 = 0.96, P = 3.27 e−7). The light gray surrounding the regression line represents the 95% confidence range of Vb predictions. Orange circles represent Vb predictions for extinct lemurs, and white circles represent Vb predictions for extant lemurs. The horizontal blue area shows the 95% confidence range of Malagasy Canarium fruit diameters. Horizontal colored dashed lines indicate the seed diameter of other putatively orphaned angiosperm lineages. (B) Log-transformed Malagasy Canarium fruit length and width data projected into a 2D phylogenetic morphospace. Numbered lineages represent those Canarium species inferred to be outside of the Vb of extant lemur lineages (1, C. betamponae; 2, C. galokense; 3, C. longistipulatum; 4, C. elegans; 5, C. ampasindavae; 6, C. planifolium). Dashed lines indicate Vb of lemur lineages. (C) Maximum likelihood reconstruction of fruit width on the time-calibrated Canarieae phylogeny; the black star represents the crown of the Malagasy Canarium.
With a time-calibrated phylogeny (26) and data collected on fruit size for 335 fruits from 23 species (information on specimens measured is provided in Dataset S4), we examined the tempo and mode of fruit size evolution of one such lineage: the large-seeded and ecologically dominant Malagasy rainforest trees, Canarium (ca. 33 species in Madagascar), which fall within the broader, pantropically distributed Canarieae of the Burseraceae (ca. 250 species) (26, 27). For Canarium species, we measured dry fruit size rather than seed size because the flesh is quite hard and thin, surrounding a bony endocarp (or stone) that contains the seed(s) (27). When lemurs ingest Canarium, they tend to hold the fruit in their cheeks, using the buccal cusps of their teeth to scrape the flesh from the endocarp before swallowing it whole. To account for possible biases due to measuring entire fruit rather than solely the endocarp, we measured all Canarium fruit dry in the herbarium, where there is minimal difference between fruit size and endocarp size. The Southeast Asian clade containing the Malagasy Canarium displays a persistent tendency toward increased fruit size relative to the rest of the clade, with Malagasy fruit being, on average, the largest in the entire Canarieae (Fig. 3C). In the case of Malagasy Canarium, this fruit gigantism may be to the lineage’s detriment, given that Varecia are the only remaining Malagasy mammals capable of ingesting most Canarium species fruit (5, 26, 27; also ref. 28, which documents some Eulemur dispersal in the absence of Varecia) (Fig. 3 A and B). Further, it appears that the range of variation for some Canarium species falls entirely outside of Varecia’s Vb (Fig. 3 A and B). Extinct lemurs, Pachylemur especially, have been hypothesized to play a major role in Canarium dispersal (14), and their Vbs would have easily accommodated the consumption of fruit from all Canarium species (Fig. 3).
Seed germination rates may increase after passage through a disperser’s gut due to abrasion and chemical factors (29). Additionally, extinct large-bodied lemurs would likely have promoted effective seed dispersal away from the parent tree by swallowing and subsequently passing seeds during the course of their daily movements. Dispersal to growing sites removed from parent trees enhances gene flow, reduces competition among related individuals, and promotes escape from specialized herbivores (30, 31). Dispersal alternatives for large-seeded orphans include the strong winds of cyclones and secondary dispersal through the movement of fallen seeds by rodents (18, 32, 33). Although effective in some circumstances (34), secondary dispersal has generally been shown to be inefficient and unreliable (18, 32, 33), and cyclone-mediated seed dispersal is, at best, an extreme and unreliable dispersal mechanism (35).
Conclusion
The inferred evolutionary trend toward larger seeds that might once have contributed to the success of these Malagasy trees may now facilitate their decline. Although large-seeded orphans persist today, this persistence is likely due to their long generation times and uncertain and inefficient secondary dispersal agents, such as Rattus rattus or strong winds during cyclones (18, 32, 33, 35). Our study suggests that their long-term survival may be tenuous. Additional studies are needed to understand more fully the impact of these extinctions on the Malagasy flora, especially focusing on seed germination and seedling survival. It will also be important to consider other recently extinct and potentially important large-bodied seed dispersers, such as elephant birds (genus Aepyornis) (36). It is possible that with the extirpation of large-bodied dispersers, smaller bodied dispersers will select for reductions in seed size (37). However, in order for such selection to occur, the natural range of variation in seed size must permit dispersal by at least one extant dispersal agent (37). Although this natural range of variation may still be the case for some of the large-seeded species of Canarium, the seed sizes of some orphaned lineages appear to be too large to be handled by any other extant disperser (Fig. 3). Without intervention, orphans that have not yet adapted to Madagascar’s altered seed dispersal landscape may be in jeopardy. This conclusion has practical, philosophical, and political ramifications for the conservation and management of Madagascar’s orphaned plants. Our analyses identify certain extant lemurs occupying otherwise empty areas of morphological space, such as Varecia (Fig. 2), as performing unique and irreplaceable ecosystem functions. Given the dominant role of primates in providing seed dispersal for many of Madagascar’s trees, the identification of orphans and the limits of extant Vb can inform conservation strategies to ameliorate the potential cascading effects of extinction on the structure and function of Malagasy forests.
Methods
Estimating Lemur Divergence Times and Inferring the Evolutionary History of Their Feeding Strategies.
To infer a time-calibrated phylogeny of lemurs, we used publicly available full mitochondrial genome data from the GenBank (www.ncbi.nlm.nih.gov/genbank/), when possible (Genbank information is provided in Dataset S5). When full mitochondrial genome data were unavailable, we used all publicly available data for mitochondrial coding regions (Dataset S5). Our sampling included representatives of all major lineages and was based on a recently published phylogeny (11), as well as those species studied by Perry and Hartstone-Rose (24). For those extinct taxa lacking any genetic data, we conditioned the placement of the taxa following the topology of Karanth et al. (10), which was based on both ancient DNA and morphology.
For all DNA sequence data, we aligned the mitochondrial coding regions to a complete mtDNA reference sequence (V. variegata, GI:671760213), using MUSCLE v3.7 (38) implemented in Geneious v7.1.2 (39) with alignments checked by eye. We used PartitionFinder (40) to infer both the best-fitting nucleotide substitution models and partitioning scheme simultaneously. The candidate pool of potential partitions ranged from a single partition per locus to partitions that divided each protein-coding locus by codon position.
Phylogenetic time trees were estimated using a Bayesian statistical framework in BEAST v1.7.5 (41). All analyses were performed using an uncorrelated lognormal relaxed clock to avoid the assumption of rate correlation among lineages (41). To ensure the conditional placement of fossil lineages, we generated a starting tree for the Bayesian analyses using maximum likelihood in RAxML (42). BEAST analyses were each run for 800 million generations (sampled every 500–5,000 generations). Substitution and clock models were unlinked among partitions, yielding more precise molecular rate estimates (43), and a birth–death speciation process on branching times was specified as the tree prior for each analysis (41). The alignment was partitioned based on PartitionFinder analyses (40) (PartitionFinder results are provided in Dataset S6). Convergence between runs and adequacy of the burn-in period were both assessed using Tracer v1.5 (41). Adequate sampling of the posterior distribution was diagnosed by quantification of effective sample size (ESS) values in Tracer from combined runs, with ESS values above 200 indicating effective sampling (41). We used Tree Annotator to summarize the posterior probability distribution of trees using a maximum clade credibility tree with median node heights.
Because we used fossil-tip dating techniques and not every individual with genetic data had a fully sequenced mitochondrial genome, we constrained the topology to reflect the relationships inferred by Karanth et al. (10). To time-calibrate the phylogeny, we used two fossil-based prior age calibrations following the methodology of Kistler et al. (11). Briefly, we dated the split between the Lorisiformes and Lemuriformes as being between 52 and 66 Ma (Fig. S2, node A) and the split between Nycticebus and Perodicticus as being 36.9 Ma (Fig. S2, node B), and we also placed a secondary calibration at the crown of the indriids as being between 14 and 20 Ma (Fig. S2, node C).
Fig. S2.
Lemuriform molecular phylogeny. Maximum clade credibility tree summarizing the results of Bayesian dating analyses (fossil calibration nodes marked with letters A–C) with normal probability priors on nodes B and C and a lognormal probability prior on node A. Numbers at the nodes show Bayesian posterior probabilities (BPPs). Daggers (†) next to species names indicate extinct taxa.
To infer the evolutionary history of feeding strategy, we assigned species to five broad categories: (i) frugivory, (ii) folivory, (iii) insectivory, (iv) omnivory, and (v) mixed fruit-and-leaves. For extant taxa, feeding strategy was assigned based on behavioral studies surveyed from the literature (Dataset S1). Frugivory, folivory, and insectivory were assigned to those taxa eating 50% or more of fruit, leaves, or insects, respectively. Those taxa with preponderantly folivorous diets, but still considered important seed dispersers, were assigned to a mixed fruit-and-leaves category (Dataset S1). Extinct taxa were assigned feeding strategies based on morphological and stable isotope analyses, as determined in the literature (Dataset S1). As the basis for the ancestral state assignments, we inferred the phylogenetic history of feeding strategy using stochastic character mapping. This method allows for the inference of discrete character changes while accommodating uncertainty in both topology and branch lengths (44). All analyses were conducted using the “phytools” package (22) in the statistical programming platform R (45) (version 3.2.3), and all simulations were run 1,000 times under a model of Brownian motion.
Morphological Data Collection and Vb Modeling.
To quantify the morphological variation of extant and extinct lemurs, we examined 169 adult specimens from collections at the Duke Primate Center (DPC), American Museum of Natural History, and University of Antananarivo (Dataset S2), and measured 16 morphometric traits that are detailed and illustrated in Fig. S1. Because lemurs are not sexually dimorphic in terms of size (14, 46), we did not separate our measurements by sex. Perry and Hartstone-Rose (24) calculated the Vb of lemur species at the DPC, and demonstrated an allometric relationship between Vb and body mass in frugivorous lemurs. This relationship allows the construction of predictive models for Vb based on body mass. Skeletal measurements, such as mandible length and articulated mouth size, are insufficient for estimating Vb because they do not take into account craniofacial musculature, which can restrict the maximum range of motion observed in a disarticulated skull. Because the frugivorous lemur species studied by Perry and Hartstone-Rose were included in our phylogenetic and morphometric analyses, we were able to use their observed Vb measurements (24) regressed against body mass estimates gathered from the literature (Dataset S7) to predict the Vb of the remaining seed dispersers in our analyses. Vb estimates and their 95% confidence intervals were predicted using a linear regression with the “nlme” package in the statistical programming platform R (45, 47) (Fig. 3A).
We collected fruit length and width data to quantify phenotypic variation in the fruit size of Canarieae species, using both species descriptions (48) and measurements from 325 individuals representing 23 species of Malagasy Canarium, and seed size measurements gathered from the literature for other putatively orphaned lineages (Datasets S3 and S4). The evolutionary history of log-transformed Canarieae fruit width was estimated on the time-calibrated phylogeny of Federman et al. (26) with maximum likelihood ancestral character state estimation for continuous trait data using the package “ape” in R (45, 49) (Fig. 3C).
Morphological Analyses.
Considering morphological variation within a phylogenetic context is useful because it can reflect patterns of convergence and independent evolutionary origins of a trait or traits. To visualize general overlap and range of variation along the axes of fruit size variation within a phylogenetic context, we log-transformed fruit length and width and projected these data into a 2D phylomorphospace (50) using both the time-calibrated Canarieae and Malagasy Canarium phylogenies (26) (Fig. 3). All phylomorphospaces were generated using the “phytools” package (22) in R (45), which estimates ancestral character states using a maximum likelihood framework. To examine the partitioning of morphological diversity and diet type among lemur subclades, we conducted a PCA using phylogenetic residual values of individual traits that were regressed against body mass (22, 23, 51) (Dataset S8). PCA reduces the redundancy and dimensionality of multivariate data such that individual principal components can be fit to univariate models. This transformation allows for the investigation of evolutionary patterns and processes across multiple traits (52). However, standard PCA does not take evolutionary processes into account, which, due to the nonindependence of shared ancestry, can be misleading (52, 53). A phylogenetic PCA incorporates this nonindependence among lineages by assuming a multivariate Brownian motion process of trait evolution, and using the expected covariance among traits to calculate principal component axes and scores (53). Regressing against body mass is preferable to conducting a PCA and removing the axis that correlates with body size to control for body size in linear measurements (54) because both simulated and empirical evidence has found regression-based approaches that allow covariance with body size to be more robust to statistical artifacts (55). We then used the first two principal components in conjunction with the time-calibrated lemur phylogeny to project patterns of morphological evolution into a phylomorphospace (21). The phylomorphospace provides a visual representation of the morphological space occupied by seed-dispersing and non–seed-dispersing lemurs. To quantify the dispersal ability lost with the megafaunal extinction, we calculated the area of morphospace lost with the exclusion of extinct taxa using convex hulls with the “siar” package in R (45, 56).
Supplementary Material
Acknowledgments
We thank the University of Antananarivo, Gregg Gunnell (DPC), Amanda Mancini (Hunter College), and Eileen Westwig and Aja Marcato [American Museum of Natural History (AMNH)] for facilitating morphometric analyses; April Lamb and Willa Brooks for aiding with the figures; Aaron Reuben for editorial assistance; the M.J.D. and Near laboratories for support; and the AMNH and DPC for access to their collections. This work was supported by National Science Foundation Graduate Research Fellowship DGE-1122492 (to S.F.). This report is Duke Lemur Center publication 1314.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Phylogenetic and morphological datasets used in this study, along with any associated R scripts, are available in the Zenodo repository, https://zenodo.org/ (DOI 10.5281/zenodo.45471).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523825113/-/DCSupplemental.
References
- 1.Dewar RE, Richard AF. Evolution in the hypervariable environment of Madagascar. Proc Natl Acad Sci USA. 2007;104(34):13723–13727. doi: 10.1073/pnas.0704346104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu Rev Ecol Evol Syst. 2006;37:405–431. [Google Scholar]
- 3.Wright PC. Lemur traits and Madagascar ecology: Coping with an island environment. Am J Phys Anthropol. 1999;110(29) Suppl 29:31–72. doi: 10.1002/(sici)1096-8644(1999)110:29+<31::aid-ajpa3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Razafindratsima OH, Mehtani S, Dunham AE. Extinctions, traits and phylogenetic community structure: Insights from primate assemblages in Madagascar. Ecography. 2013;36(1):47–56. [Google Scholar]
- 5.Wright PC, et al. Frugivory in four sympatric lemurs: Implications for the future of Madagascar’s forests. Am J Primatol. 2011;73(6):585–602. doi: 10.1002/ajp.20936. [DOI] [PubMed] [Google Scholar]
- 6.Valenta K, et al. Colour and odour drive fruit selection and seed dispersal by mouse lemurs. Sci Rep. 2013;3:2424. doi: 10.1038/srep02424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman S, Ganzhorn J. Rarity of figs (Ficus) on Madagascar and its relationship to a depauperate frugivore community. Revue D'Ecologie. 1997;52(4):321–329. [Google Scholar]
- 8.Fleagle JG, Reed KE. Comparing primate communities: A multivariate approach. J Hum Evol. 1996;30(6):489–510. [Google Scholar]
- 9.Richard AF, Dewar RE. Lemur ecology. Annu Rev Ecol Evol Syst. 1991;22:145–175. [Google Scholar]
- 10.Karanth KP, Delefosse T, Rakotosamimanana B, Parsons TJ, Yoder AD. Ancient DNA from giant extinct lemurs confirms single origin of Malagasy primates. Proc Natl Acad Sci USA. 2005;102(14):5090–5095. doi: 10.1073/pnas.0408354102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kistler L, et al. Comparative and population mitogenomic analyses of Madagascar’s extinct, giant ‘subfossil’ lemurs. J Hum Evol. 2015;79:45–54. doi: 10.1016/j.jhevol.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Crowley BE. A refined chronology of prehistoric Madagascar and the demise of the megafauna. Quat Sci Rev. 2010;29(19):2591–2603. [Google Scholar]
- 13.Burney DA, et al. A chronology for late prehistoric Madagascar. J Hum Evol. 2004;47(1-2):25–63. doi: 10.1016/j.jhevol.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Godfrey LR, Jungers WL, Schwartz GT, Irwin MT. Ghosts and orphans. In: Fleagle JG, Gilbert CC, editors. Elwyn Simons: A Search for Origins. Springer; New York: 2008. pp. 361–395. [Google Scholar]
- 15.Jungers WL, et al. Ecomorphology and behavior of giant extinct lemurs from Madagascar. In: Plavcan MJ, Kay RF, Jungers WL, van Schaik CP, editors. Reconstructing Behavior in the Primate Fossil Record. Springer; New York: 2002. pp. 371–411. [Google Scholar]
- 16.Crowley BE, Godfrey LR, Irwin MT. A glance to the past: Subfossils, stable isotopes, seed dispersal, and lemur species loss in Southern Madagascar. Am J Primatol. 2011;73(1):25–37. doi: 10.1002/ajp.20817. [DOI] [PubMed] [Google Scholar]
- 17.Terborgh J. The big things that run the world—A sequel to E.O. Wilson. Conserv Biol. 1988;2(4):402–403. [Google Scholar]
- 18.Janzen DH, Martin PS. Neotropical anachronisms: The fruits the gomphotheres ate. Science. 1982;215(4528):19–27. doi: 10.1126/science.215.4528.19. [DOI] [PubMed] [Google Scholar]
- 19.Johnson CN. Ecological consequences of Late Quaternary extinctions of megafauna. Proc Biol Sci. 2009;276(1667):2509–2519. doi: 10.1098/rspb.2008.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blumenthal SA, Chritz KL, Rothman JM, Cerling TE. Detecting intraannual dietary variability in wild mountain gorillas by stable isotope analysis of feces. Proc Natl Acad Sci USA. 2012;109(52):21277–21282. doi: 10.1073/pnas.1215782109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52(2):131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 22.Revell LJ. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3(2):217–223. [Google Scholar]
- 23.Revell LJ. Graphical methods for visualizing comparative data on phylogenies. In: Garamszegi LZ, editor. Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology. Springer; New York: 2014. pp. 77–103. [Google Scholar]
- 24.Perry JM, Hartstone-Rose A. Maximum ingested food size in captive strepsirrhine primates: Scaling and the effects of diet. Am J Phys Anthropol. 2010;142(4):625–635. doi: 10.1002/ajpa.21285. [DOI] [PubMed] [Google Scholar]
- 25.Schwitzer C, et al. Lemurs of Madagascar: A Strategy for Their Conservation 2013–2016. IUCN SSC Primate Specialist Group, Bristol Conservation and Science Foundation, and Conservation International; Bristol, UK: 2013. [Google Scholar]
- 26.Federman S, et al. The biogeographic origin of a radiation of trees in Madagascar: Implications for the assembly of a tropical forest biome. BMC Evol Biol. 2015;15(1):216. doi: 10.1186/s12862-015-0483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daly DC, Raharimampionona J, Federman S. A revision of Canarium L.(Burseraceae) in Madagascar. Adansonia. 2015;37(2):277–345. [Google Scholar]
- 28.Bollen A, Elsacker LV, Ganzhorn JU. Relations between fruits and disperser assemblages in a Malagasy littoral forest: A community-level approach. J Trop Ecol. 2004;20(6):599–612. [Google Scholar]
- 29.Schaefer HM, Ruxton GD. Plant-Animal Communication. Oxford Univ Press; Oxford: 2011. [Google Scholar]
- 30.Connell JH. On the role of natural enemies in preventing competitive exclusion in some marine animals and in rain forest trees. In: den Boer PJ, Gradwell GR, editors. Dynamics of Populations. PUDOC; Wageningen, The Netherlands: 1971. pp. 298–312. [Google Scholar]
- 31.Janzen DH. Herbivores and the number of tree species in tropical forests. Am Nat. 1970;104(940):501–528. [Google Scholar]
- 32.Chang G, Zhang Z. Functional traits determine formation of mutualism and predation interactions in seed-rodent dispersal system of a subtropical forest. Acta Oecol (Montruge) 2014;55:43–50. [Google Scholar]
- 33.Loayza AP, Carvajal DE, García-Guzmán P, Gutierrez JR, Squeo FA. Seed predation by rodents results in directed dispersal of viable seed fragments of an endangered desert shrub. Ecosphere. 2014;5(4):art43. [Google Scholar]
- 34.Jansen PA, et al. Thieving rodents as substitute dispersers of megafaunal seeds. Proc Natl Acad Sci USA. 2012;109(31):12610–12615. doi: 10.1073/pnas.1205184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nathan R. Long-distance dispersal of plants. Science. 2006;313(5788):786–788. doi: 10.1126/science.1124975. [DOI] [PubMed] [Google Scholar]
- 36.Hansen DM, Galetti M. Ecology. The forgotten megafauna. Science. 2009;324(5923):42–43. doi: 10.1126/science.1172393. [DOI] [PubMed] [Google Scholar]
- 37.Galetti M, et al. Functional extinction of birds drives rapid evolutionary changes in seed size. Science. 2013;340(6136):1086–1090. doi: 10.1126/science.1233774. [DOI] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 41.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 43.Ho SY, Lanfear R. Improved characterisation of among-lineage rate variation in cetacean mitogenomes using codon-partitioned relaxed clocks. Mitochondrial DNA. 2010;21(3-4):138–146. doi: 10.3109/19401736.2010.494727. [DOI] [PubMed] [Google Scholar]
- 44.Bollback JP. SIMMAP:Stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics. 2006;7(1):88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. R Development Core Team (2015) R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
- 46.Godfrey LR, Lyon SK, Sutherland MR. Sexual dimorphism in large-bodied primates: The case of the subfossil lemurs. Am J Phys Anthropol. 1993;90(3):315–334. doi: 10.1002/ajpa.1330900306. [DOI] [PubMed] [Google Scholar]
- 47.Pinheiro J, Bates D, DebRoy S, Sarkar D. R Development Core Team . nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-103. R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]
- 48.Leenhouts P. Revision of the Burseraceae of the Malaysian area in the wider sense. X a. Canarium. Blumea. 1959;9(2):275–475. [Google Scholar]
- 49.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 50.Sidlauskas B. Continuous and arrested morphological diversification in sister clades of characiform fishes: A phylomorphospace approach. Evolution. 2008;62(12):3135–3156. doi: 10.1111/j.1558-5646.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 51.Revell LJ. Phylogenetic signal and linear regression on species data. Methods Ecol Evol. 2010;1(4):319–329. [Google Scholar]
- 52.Uyeda JC, Caetano DS, Pennell MW. Comparative analysis of principal components can be misleading. Syst Biol. 2015;64(4):677–689. doi: 10.1093/sysbio/syv019. [DOI] [PubMed] [Google Scholar]
- 53.Revell LJ. Size-correction and principal components for interspecific comparative studies. Evolution. 2009;63(12):3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- 54.McCoy MW, Bolker BM, Osenberg CW, Miner BG, Vonesh JR. Size correction: Comparing morphological traits among populations and environments. Oecologia. 2006;148(4):547–554. doi: 10.1007/s00442-006-0403-6. [DOI] [PubMed] [Google Scholar]
- 55.Berner D. Size correction in biology: How reliable are approaches based on (common) principal component analysis? Oecologia. 2011;166(4):961–971. doi: 10.1007/s00442-011-1934-z. [DOI] [PubMed] [Google Scholar]
- 56.Parnell A, Inger R, Bearhop S, Jackson A. SIAR: Stable Isotope Analysis in R. R Package Version 3. R Foundation for Statistical Computing; Vienna: 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.