Significance
The physical basis for the long timescale of memory has been mysterious. The formation of a functional prion-like fiber in the neuronal system may resolve the question. This work shows that the energy landscapes of a candidate prion, cytoplasmic polyadenylation element binding (CPEB) protein, allow external mechanical forces to facilitate the structural transition to a fiber form. The mechanical coupling thus allows a positive feedback loop between CPEB prion transitions and cytoskeletal actions to mark synapses.
Keywords: long-term memory, mechanical prion, protein aggregation, Q-rich protein
Abstract
Aplysia cytoplasmic polyadenylation element binding (CPEB) protein, a translational regulator that recruits mRNAs and facilitates translation, has been shown to be a key component in the formation of long-term memory. Experimental data show that CPEB exists in at least a low-molecular weight coiled-coil oligomeric form and an amyloid fiber form involving the Q-rich domain (CPEB-Q). Using a coarse-grained energy landscape model, we predict the structures of the low-molecular weight oligomeric form and the dynamics of their transitions to the β-form. Up to the decamer, the oligomeric structures are predicted to be coiled coils. Free energy profiles confirm that the coiled coil is the most stable form for dimers and trimers. The structural transition from α to β is shown to be concentration dependent, with the transition barrier decreasing with increased concentration. We observe that a mechanical pulling force can facilitate the α-helix to β-sheet (α-to-β) transition by lowering the free energy barrier between the two forms. Interactome analysis of the CPEB protein suggests that its interactions with the cytoskeleton could provide the necessary mechanical force. We propose that, by exerting mechanical forces on CPEB oligomers, an active cytoskeleton can facilitate fiber formation. This mechanical catalysis makes possible a positive feedback loop that would help localize the formation of CPEB fibers to active synapse areas and mark those synapses for forming a long-term memory after the prion form is established. The functional role of the CPEB helical oligomers in this mechanism carries with it implications for targeting such species in neurodegenerative diseases.
It is widely believed that learning involves the modification in number, strength, and structure of specific localized synaptic connections. Although short-term memory formation occurs without protein synthesis, establishing long-term memory (LTM) involves protein synthesis localized at the synapse area, thus requiring a stable translational regulatory system that activates mRNA and protein synthesis in the synapse (1–3). It has long been recognized that the relatively short half-life of most proteins in eukaryotic cells (4) poses questions about the long timescales over which memories can be retained. It has been suggested that forming a very stable prion could provide a mechanism for achieving memory longevity (5, 6). An excellent candidate species has emerged from the works of Kandel and coworkers (1–3) and Lindquist and coworker (6) on cytoplasmic polyadenylation element binding (CPEB). It has been established, in Aplysia, that Aplysia cytoplasmic polyadenylation element binding (ApCPEB) activates mRNA and mediates synaptic protein synthesis for at least 72 h and that it does so by taking on a functional prion-like form (3, 7). Forming the prion is, thus, a key element in LTM formation and maintenance.
Prions were first proposed as protein-only infectious particles causing Creutzfeldt–Jakob and related neurological diseases (8–10). In these cases, the prion fails to fold to its native conformation but instead, leads to self-propagating aggregation into fibers rich in β-sheet secondary structure. Many Q/N-rich proteins in yeast have also been shown to display this prion-like property. Most Q/N-rich proteins are rather insoluble and thus, difficult to study by conventional structural techniques in the laboratory. In this paper, we carry out computational studies to explore the structure of oligomers of the Q-rich domain of cytoplasmic polyadenylation element binding (CPEB-Q) and characterize the energy landscapes for their transition into the functional prion form. These studies suggest the possibility of an interesting role for mechanical forces exerted by the cytoskeleton in facilitating the transition to the functional prion form.
ApCPEB has a C-terminal RNA binding domain and an N-terminal prion domain that is rich in glutamine (11). Like other Q-rich proteins, the Q-rich region confers a self-perpetuating prion-like property on ApCPEB (3, 6, 12, 13). In the neuron, ApCPEB exists both in a soluble oligomeric form and as an insoluble fiber (13, 14). The soluble oligomer form has been confirmed to be an α-helical–rich coiled-coil structure (15). Both solid-state NMR and trypsin proteolysis show that, in the insoluble fibrous form, the glutamine-rich domain constitutes the core of a fiber with β-rich structure (14). As well as increasing the excitatory postsynaptic potential of the neuron (a measurement of LTM), challenging the neuron with 5-HT (serotonin) increases the concentration of CPEB and enhances formation of CPEB aggregates (3, 12). Thus, we see that a transition between two forms of CPEB must be at the heart of LTM formation.
Polyglutamine expansions cause nine human neurodegenerative diseases in which β-strand amyloid fibers and intracellular deposits form pathologically. However, the functional role of CPEB-Q suggests that its structural transition may be controlled, because making functional fibers must be tightly and locally regulated. We, therefore, explore the details of the structures of CPEB-Q oligomers and the energy landscapes of their structural transitions.
Our calculations first show that different forms of oligomeric structures are locally stable. We show that a coiled-coil structure is generally the thermodynamically dominant oligomer form. Under an external driving force, such as a very high effective concentration or mechanical tension, a transition from the α-helical structure to an extended β-sheet structure is thermodynamically and kinetically possible. The controllable relative stability of the two forms arises from the simultaneous presence of an amyloid-forming region and a coiled-coil heptad repeat feature in the CPEB-Q sequence. Both structural forms seem to have been under selection pressure. A systematic survey of the CPEB interactome shows that CPEB’s interaction partners are very often cytoskeleton-related proteins. We, therefore, propose that, if the cytoskeleton is intact and mechanically active, elements of the cytoskeleton can bind to CPEB and exert mechanical forces to facilitate the structural transition of CPEB oligomers. The positive feedback between forming CPEB fibers that activate actin mRNA and forming cytoskeletal filaments that can stretch the CPEB oligomers into their prion functional form helps localize CPEB fibers and actins to the synapse area, thus marking a synapse for the formation of LTM.
Coiled-Coil Oligomeric Structures of CPEB-Q
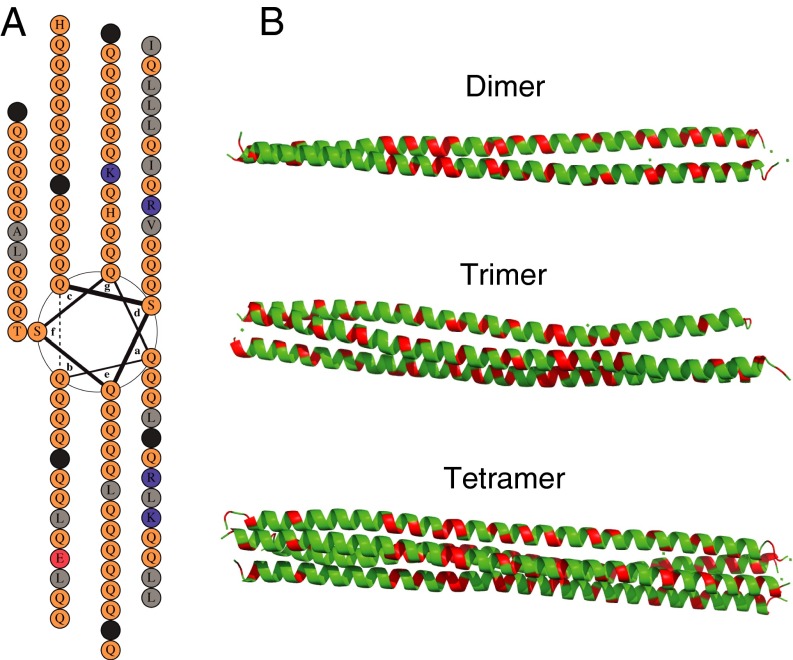
ApCPEB is composed of an N-terminal prion-like domain and a C-terminal domain bearing two mRNA recognition motifs. Our focus is on the energy landscapes of the Q-rich region (90-aa residues, with 72 being glutamines). Secondary structure predictions for CPEB-Q suggest that it has a strong α-helix coiled-coil propensity. The heptad repeat pattern of hydrophobic residues (I/L/V) on the predicted coiled-coil interface also agrees with this prediction (Fig. 1A).
Fig. 1.
Coiled-coil structure of the CPEB-Q oligomers, which corresponds to residues 53–142 in the prion domain. In the full protein, the prion domain constitutes residues 1–246, whereas the remainder residues 246–689 constitute the RNA binding domain. These additional regions of CPEB are not explicitly considered in our simulations. (A) Heptad repeat pattern in a helical wheel diagram. Modified from ref. 15. (B) Simulated coiled-coil structures for the CPEB-Q dimer, trimer, and tetramer with side view. The glutamine residues are colored green, whereas the nonglutamine residues are red.
To study the structures and dynamics of the oligomer, we use molecular dynamics simulation of the oligomers using the Associative Memory, Water-Mediated, Structure, and Energy Model (AWSEM) force field. The AWSEM is a coarse-grained predictive force field, where the parameters are learned based on the energy landscape formulation of protein folding theory (16). The force field explicitly includes transferable physically motivated interactions, such as hydrogen bonding and solvent-mediated interactions, to model the formation of tertiary structure and incorporates system-specific “memory” terms by bioinformatic matches of locally similar sequences that provide the forces guiding the formation of local sequence structural elements. Both kinds of interactions effectively sculpt the landscapes of naturally occurring proteins so as to be highly funneled toward the native state. Simulations with the AWSEM have proved successful in predicting structures of both protein monomers (16) and dimers (17). They also have provided a quantitative treatment of the initial stages of misfolding and aggregation (18).
Simulated annealing using the AWSEM shows that dimers, trimers, and tetramers of CPEB-Q, indeed, are coiled-coil structures. Structures with more monomer units are predicted to be more loosely organized clusters composed of dimer, trimer, and tetramer coiled-coil subunits (SI Appendix, Fig. S5). These predictions for CPEB-Q agree with the experimental evidence that the soluble forms of CPEB are helix-rich (14). The subunit organization of the higher oligomers explains how such oligomers can remain soluble without much additional aggregation. To determine the most likely oligomer states under physiological conditions, we evaluated the thermal stability of the different oligomeric coiled-coil structures (SI Appendix, Fig. S4A). The trimer and the tetramer have a higher melting temperature than the larger or smaller constructs do, and therefore, we consider them to be the dominant stabilized forms. By computing a 2D free energy profile using umbrella sampling, we see that the free energy of a trimer is relatively lower than that of a tetramer, even at high concentration (SI Appendix, Fig. S4B). Our prediction results compare well with experimental evidence that, in solution, the dominant oligomer forms are a trimer or a larger assembly having roughly 10 monomer subunits.
α-Helix to β-Sheet Transition in Coiled-Coil Oligomers
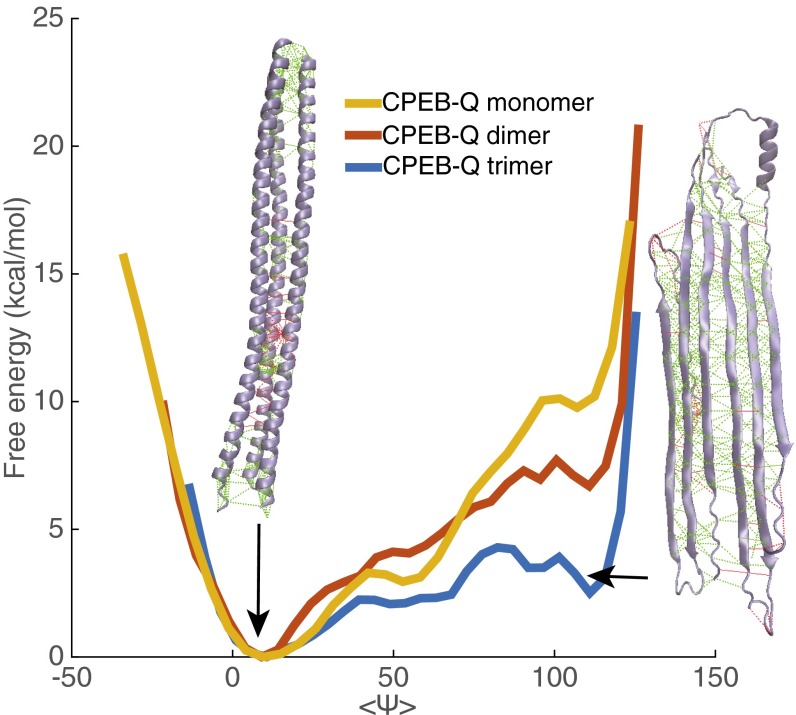
Although the α-helical oligomeric form is the most favorable form in solution, the aggregated form is a β-rich structure. To understand how fibers form, we then probe the transition from α to β in specific oligomers using umbrella sampling with an order parameter measuring β-sheet content.
For CPEB-Q monomers, dimers, and trimers using the AWSEM force field, we compute 1D free energy profiles with respect to an order parameter measuring β-sheet content: the average backbone angle . In all of these low-molecularity clusters, we see that helical structure (average around 0) dominates thermodynamically over the β-rich structure (average around 110). The β-rich structure does become easier to form in the trimer, needing only about ∼3 kcal/mol of excitation, whereas in a dimer, the necessary free energy input is ∼7 kcal/mol, and in the monomer, the necessary free energy input is ∼10 kcal/mol (Fig. 2). The predicted high free energy barrier between the two states in these oligomers suggests that aggregation can be well-controlled, because the barrier can only be overcome by a sufficiently strong external stimulus.
Fig. 2.
Free energy profiles as a function of , the average backbone angle, in CPEB-Q monomer, dimer, and trimer quantifying the transition to β-rich structures are shown. Representative structures of coiled-coil state and coiled coil-rich state of the trimer and their frustratograms are illustrated. Minimally frustrated contacts are shown in green, and highly frustrated contacts are shown in red. Although both structures are overall only weakly frustrated, we see that localized frustration exists in both forms.
In other systems, increasing the number of preassociating domains increases the local effective concentration (19). Likewise, here, the formation of a trimer coiled-coil intermediate facilitates formation of interstrand backbone hydrogen bonds by bringing up the local effective concentration in comparison with the situations for both the monomer and the dimer. Following the law of mass action, our simulations, thus, are in harmony with experimental evidence that increasing the concentration of CPEB facilitates aggregate formation both in vivo and in vitro (12).
Initiation of the α-Helix to β-Sheet Transition in a Coiled Coil Is Sequence Dependent
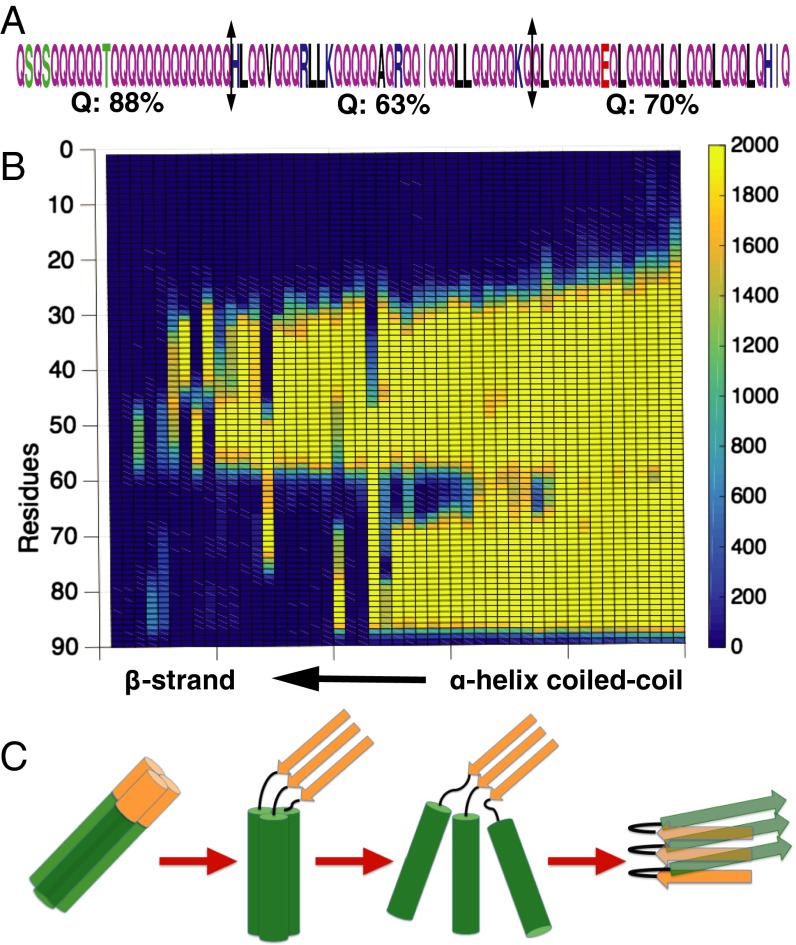
By following the structural transition as the β-sheet order parameter is increased, we uncover the dominant pathway for the transition. The CPEB-Q sequence is divided into three fragments having somewhat different glutamine fractions [88% for fragment 1 (residues 1–25), 68% for fragment 2 (residues 26–65), and 72% for fragment 3 (residues 66–90)] (Fig. 3A). Fragment 1 with the highest Q fraction undergoes the β-sheet transition first followed by the other high Q segment, fragment 3, whereas the low Q fragment 2 in the middle remains the longest in its α-helical conformation (Fig. 3B). We see that a higher fraction of glutamine leads to a greater tendency to form amyloid. This observation agrees well with laboratory studies on polyglutamine proteins, such as those leading to amyloid formation in Huntington’s disease.
Fig. 3.
Transition dynamics from a coiled-coil structure to the β-strand form in each subunit correlate with sequence. (A) The Q-rich region can be divided into three fragments having different glutamine fractions. (B) The frequency of each residue being found in α-helix conformation is shown during the transition trajectory. (C) A proposed mechanism for the coiled-coil to β-strand transition. The different fragments are color-coded, with brown representing the amyloid-initiating region.
After a β-strand forms in the amyloidogenic fragment 1, these initial β-strands nucleate further β-strand formation by binding the helices closer together, after which interchain backbone hydrogen bonds and β-strands form (Fig. 3C). We see then that the heptad repeat coiled-coil feature of the sequence and the amyloid tendency of CPEB-Q compete with each other (i.e., there is a frustrated energy landscape) (20). We propose that both of the two stable forms (coiled coil vs. fiber) are evolutionarily encoded in the protein sequence. This sequence-encoded structural dualism gives rise to a controllable structural transition that only can occur under certain conditions, such as very high concentration or when mechanical forces are applied. In the low-concentration and low-mechanical force regime, backbone hydrogen bonds have little chance to form, and the coiled coil is the dominant structure in the soluble state. Under mechanical force pulling, however, hydrogen bonds that stabilize the helices can be broken. After the helix is lengthened, interchain backbone hydrogen bonds can be made, trading their stability in the β-sheet for the stability of the α-helical hydrogen bonds in the low-molecular weight oligomer ground state.
α-Helix to β-Sheet Transition in a Coiled Coil Is Promoted by Mechanical Force
Although the dual sequence features encode a bistable energy landscape, the structural transition even in the trimer is still not favored thermodynamically. This observation suggests that an external driving force will be needed in vivo. The α-helix to β-sheet (α-to-β) transition in other coiled coils has been shown to be enhanced by mechanical force (21, 22), high temperature (23), pH change (24), and high protein concentration (25). A mechanical force-induced α-to-β transition has been observed in fibrin (22), collagen, and vimentin (26). The simulations in the works by Qin et al. (26) and Zhmurov et al. (22) on the α-to-β transition in the coiled coils of fibrin and vimentin indicate that mechanical force directly drives the transition. Their simulations also show that a β-seeded region should transform first in harmony with our results for CPEB-Q.
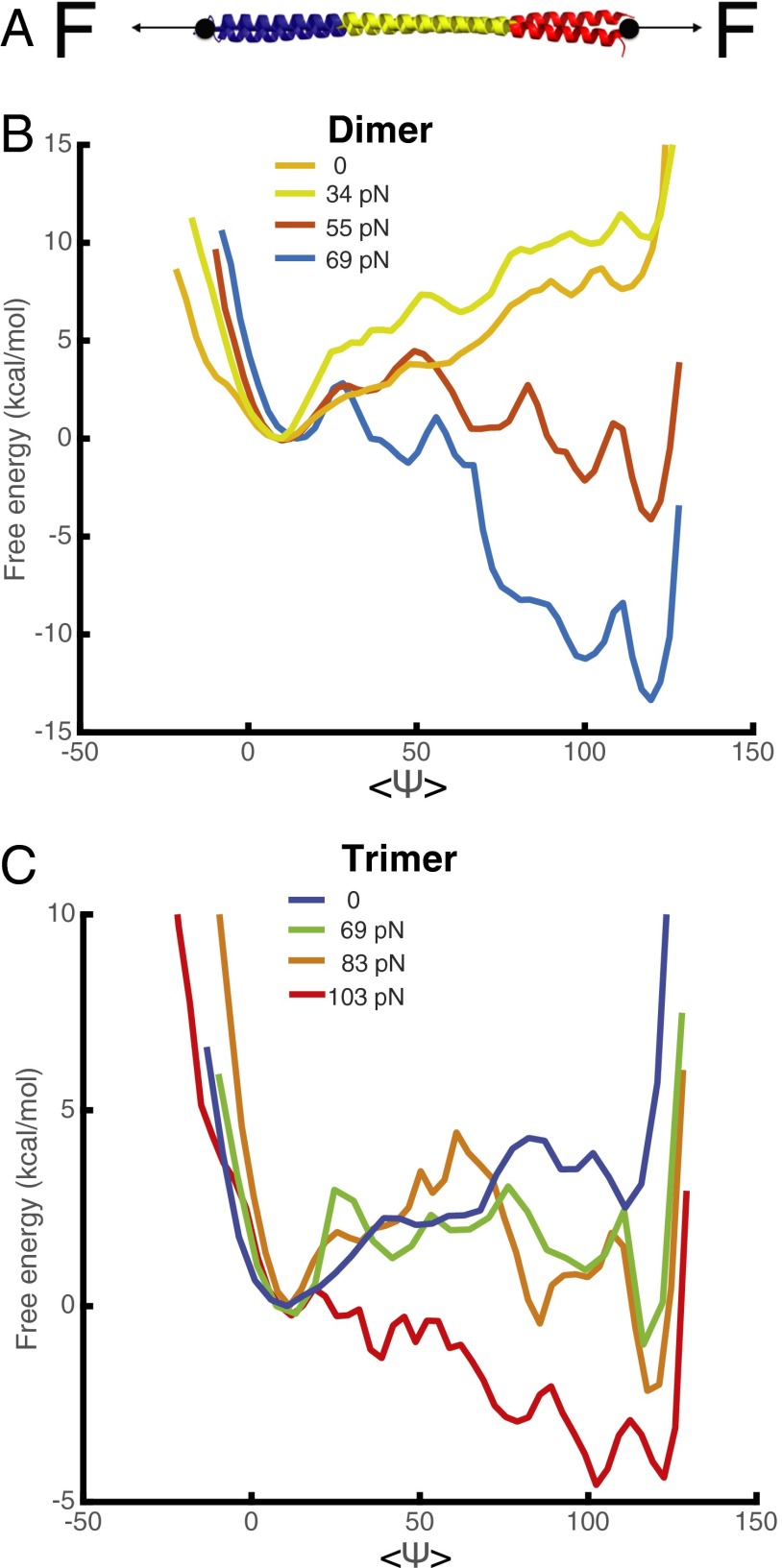
To see the effect of different mechanical forces on the structural transition, we computed free energy profiles under mechanical force for several of the oligomer species (Fig. 4A). As shown in Fig. 5B for a dimer, when the mechanical force is around 55 pN in laboratory units, the coiled-coil structures and β-strand structures become equally favorable in a thermodynamic sense. At higher tension, the β-sheet aggregated form clearly becomes favored (Fig. 4B). A similar trend is also observed in the trimer, and when the force is greater than about 70 pN, the aggregated form is still more favored (Fig. 4C). The sequence of events for the transition under tension follows that seen in umbrella sampling using the β-sheet order parameter (SI Appendix, Fig. S6).
Fig. 4.
Free energy profiles as a function of , the average backbone angle, in CPEB-Q dimer and trimer to β-strand transition are shown for different values of an external force. (A) Diagram of pulling simulation of the structural transition. Force is applied to both ends of the chains. (B) Free energy curves for the coiled-coil to β-strand transition in a dimer under different mechanical forces. (C) Free energy curves for the coiled-coil to β-strand transition in a trimer under different mechanical forces.
Fig. 5.
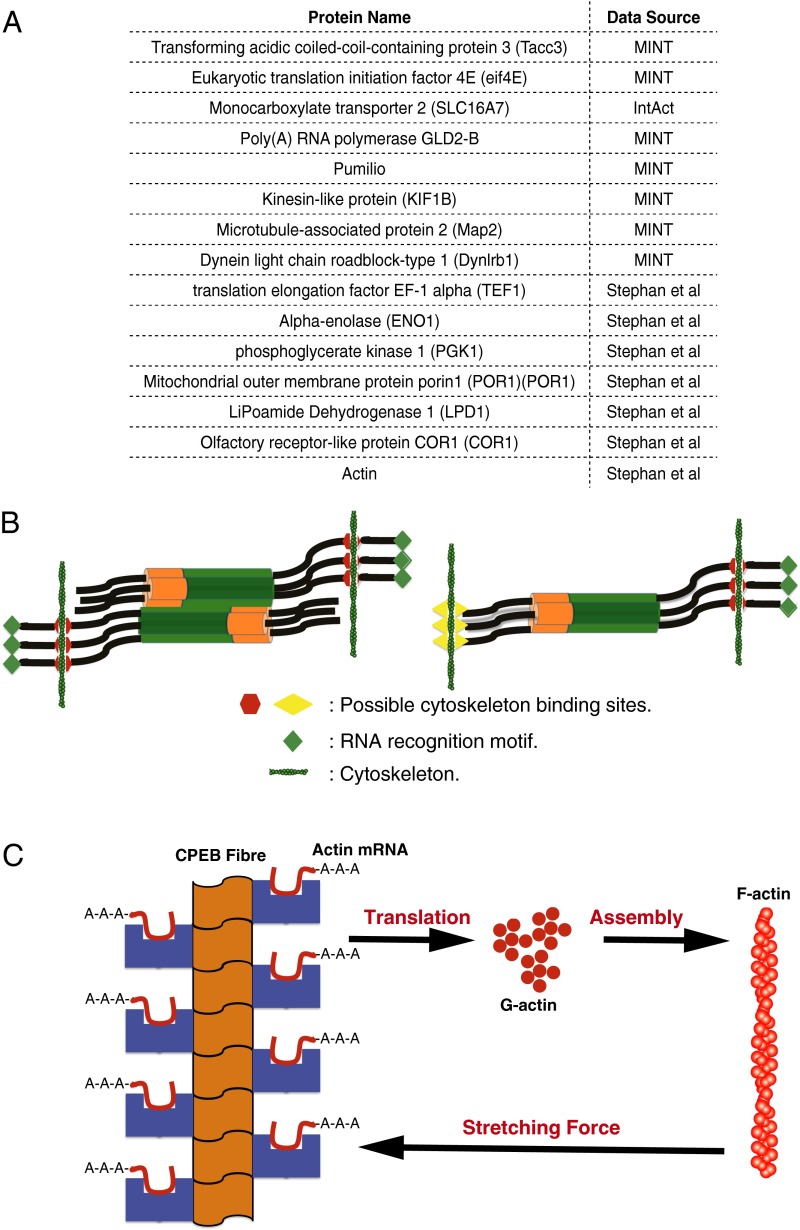
The cytoskeleton as the origin of mechanical force on the CPEB oligomers. (A) A table of known interacting partners for CPEB. (B) Possible scenarios for exerting mechanical forces on the CPEB oligomers by cytoskeleton. (C) Forming CPEB fibers and actin filaments compose a positive feedback loop. MINT, molecular interaction database.
Interactome Analysis Shows That CPEB Interacts with the Cytoskeleton
What could the origin of the mechanical force be in vivo? The cytoskeleton seems a strong possibility. Stephan et al. (27) have shown that CPEB3, a homolog of ApCPEB in mouse, interacts and colocalizes with actin filaments in coimmunoprecipitation and fluorescence labeling experiments. Furthermore, a systematic survey of the CPEB interactome reveals 15 hits in the molecular interaction database (MINT) database, the IntAct database, and the work by Stephan et al. (27). Nine of the hits are cytoskeleton or actin-related proteins (Kinesin-like protein KIF1B, Microtubule-associated protein 2, Dynein light-chain roadblock-type 1, translation elongation factor EF-1α, α-enolase, phosphoglycerate kinase 1, Mitochondrial outer membrane protein porin 1, LiPoamide Dehydrogenase 1, Olfactory receptor-like protein COR1, and actin) (Fig. 5A). The variety of cytoskeleton binding partners suggests the existence of multiple cytoskeleton binding sites on CPEB.
Discussion
Coiled Coil-Mediated Structural Transition in a Functional Prion.
Disease-causing prions also switch to a self-perpetuating conformation having β-strand structure. Many of these disease-related prions, however, do not have alternate stable intermediate oligomer structures that would allow aggregation to be regulated. In Huntington’s disease, the aggregates for Huntington form in vitro without an obvious intermediate oligomer state. Aggregation proceeds through a monomeric nucleus when the repeat length is greater than 25 (28). In Aβ-aggregation in Alzheimer’s disease, Aβ40 seems to form prefibrillar oligomers, but these oligomers already possess the β-strand features that then go on to form fibers (29).
The existence of a coiled-coil intermediate state distinct from the β-sheet structure allows aggregation to be brought under control by stabilizing the coiled-coil state with hydrophobic interfaces. Prions cannot propagate rapidly without the additional stimulation needed to cross the predicted free energy barrier. If CPEB molecules could directly self-associate to form β-strands, runaway aggregation kinetics would be expected. By having a stable coiled-coil intermediate state in the oligomer, at cellular concentration, the CPEB oligomers must await stimulation to aggregate: the coiled coil-mediated structural transition allows CPEB to be a controllable, functional, mechanical prion.
Forming Actin Filaments and Forming CPEB Fibers Compose a Positive Feedback Loop.
Actin mRNA is a target of CPEB (3). Actin filaments can form when sufficient G actin is available, and the resulting filaments, in turn, can also exert mechanical force on CPEB oligomers, thus facilitating the structural transition of CPEB into its functional fiber form. Stephan et al. (27) have already proposed that G-actin synthesis and CPEB prion formation could form a positive feedback loop. We see from the energy landscape analysis how such a loop could be completed by CPEB oligomers being stretched by actin filaments or other cytoskeletal motors so as to undergo their structural transition. CPEB fibers, after they are formed, enhance the translation of actin mRNA, thus eventually producing more F actins, closing the feedback loop (Fig. 5C).
Earlier, Francis Crick proposed that a synapse could be marked by modifying proteins posttranslationally or forming localized aggregates in the synapse, leading to LTM formation and maintenance (4). In this model, the positive feedback loop between a mechanical prion transition and the dynamics of cytoskeletal assemblies would result in the continuous formation of actin filaments and CPEB fibers in the synapse area. It is possible and indeed likely in our view, that specific structures of CPEB in interaction with the cytoskeleton are involved. After a disordered or ordered assembly is formed, the colocalized actin filaments block the transportation of CPEB fibers away from the synapse. The presence of such an actin/CPEB assembly would thereby mark the synapse for LTM formation.
How Does the Cytoskeleton Exert Force in the Right Place?
We propose two possible scenarios by which the cytoskeleton could exert the necessary tension to stretch the coiled-coil domain to facilitate the structural transition. If CPEB possesses only a single binding site for elements of the cytoskeleton, it will be necessary for larger coiled-coil oligomers to associate together in an antiparallel pattern so that mechanical force on different oligomer subunits could stretch the coiled-coil domains (Fig. 5B). Indeed, the AWSEM simulations show that higher oligomers can have this antiparallel orientation (SI Appendix, Fig. S5). If CPEB itself possesses two binding sites for cytoskeleton to either actin or other cytoskeleton-related proteins, the cytoskeleton could exert tension on even a single oligomer (Fig. 5B). The magnitude of the total necessary mechanical force predicted from our simulations is 69 pN (around 23 pN on each chain). This magnitude for each chain is in the range of the stall force (10–30 pN) found in single-molecule experiments on actin filaments.
Implications of the Mechanical Prion for Therapeutic Strategies.
Unlike the large critical nucleus needed for forming sickle cell fibers (30), polyglutamine repeat aggregation requires only a fairly small nucleus size (four for shorter repeats and one for repeat length greater than 25) (28). Aggregation kinetics, thus, are not nearly as strongly affected by protein concentration level in the neuronal system as in the sickle cell example (30). The pathophysiology of Q-rich neurodegenerative disease-related proteins must, therefore, be quite different from that for sickle cell disease. Interactions with the cytoskeleton are well-known for several other Q-rich proteins (Huntington, Sup35, and Ure2). Our model suggests that the mechanical force exerted by the cytoskeleton might also facilitate the transition to a fiber form for Q-rich disease-causing proteins. If so, then the Q-region itself may not be the best drug target, but rather, the domains that bind with the cytoskeletal components should be targeted.
Recent work on neurodegenerative disease has identified oligomer species in some cases as being cytotoxic. This work has led to the suggestion that targeting “toxic” oligomers should be an effective therapeutic strategy (29, 31). The coiled-coil oligomer species, far from being pathogenic, are also key intermediates for controlling the formation of the functional fibers; thus, targeting such α-helical oligomers could interfere with neuronal activity.
Materials and Methods
A detailed description of the materials and methods is given in SI Appendix, SI Materials and Methods. Briefly, the simulations were carried out using the AWSEM force field for proteins in the LAMMPS open source software package. Free energy profiles were computed by carrying out umbrella sampling using the β-sheet order parameter as the reaction coordinate. Constant force was applied to the system in pulling simulations. Interactome analysis was carried out using the IntAct database.
Supplementary Material
Acknowledgments
We thank the Data Analysis and Visualization Cyberinfrastructure funded by National Science Foundation Grant OCI-0959097. This work was supported by National Institute of General Medical Sciences Grant R01 GM44557. Additional support was also provided by D. R. Bullard-Welch Chair at Rice University Grant C-0016.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602702113/-/DCSupplemental.
References
- 1.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5(1):14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell. 2014;157(1):163–186. doi: 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Si K, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115(7):893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 4.Crick F. Memory and molecular turnover. Nature. 1984;312(5990):101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- 5.Tompa P, Friedrich P. Prion proteins as memory molecules: An hypothesis. Neuroscience. 1998;86(4):1037–1043. doi: 10.1016/s0306-4522(98)00148-1. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich SU, Lindquist S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB) Proc Natl Acad Sci USA. 2011;108(7):2999–3004. doi: 10.1073/pnas.1019368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miniaci MC, et al. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59(6):1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith JS. Self-replication and scrapie. Nature. 1967;215(5105):1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 9.Alper T, Cramp WA, Haig DA, Clarke MC. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214(5090):764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 10.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Schwartz JH. The cytoplasmic polyadenylation element binding protein and polyadenylation of messenger RNA in Aplysia neurons. Brain Res. 2003;959(1):68–76. doi: 10.1016/s0006-8993(02)03729-0. [DOI] [PubMed] [Google Scholar]
- 12.Si K, Choi Y-B, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140(3):421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115(7):879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 14.Raveendra BL, et al. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat Struct Mol Biol. 2013;20(4):495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143(7):1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davtyan A, et al. AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J Phys Chem B. 2012;116(29):8494–8503. doi: 10.1021/jp212541y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng W, Schafer NP, Davtyan A, Papoian GA, Wolynes PG. Predictive energy landscapes for protein-protein association. Proc Natl Acad Sci USA. 2012;109(47):19244–19249. doi: 10.1073/pnas.1216215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W, Schafer NP, Wolynes PG. Frustration in the energy landscapes of multidomain protein misfolding. Proc Natl Acad Sci USA. 2013;110(5):1680–1685. doi: 10.1073/pnas.1222130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng W, Schafer NP, Wolynes PG. Free energy landscapes for initiation and branching of protein aggregation. Proc Natl Acad Sci USA. 2013;110(51):20515–20520. doi: 10.1073/pnas.1320483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreiro DU, Komives EA, Wolynes PG. Frustration in biomolecules. Q Rev Biophys. 2014;47(4):285–363. doi: 10.1017/S0033583514000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin Z, Buehler MJ. Molecular dynamics simulation of the α-helix to β-sheet transition in coiled protein filaments: Evidence for a critical filament length scale. Phys Rev Lett. 2010;104(19):198304. doi: 10.1103/PhysRevLett.104.198304. [DOI] [PubMed] [Google Scholar]
- 22.Zhmurov A, et al. Mechanical transition from α-helical coiled coils to β-sheets in fibrin(ogen) J Am Chem Soc. 2012;134(50):20396–20402. doi: 10.1021/ja3076428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricchiuto P, Brukhno AV, Paci E, Auer S. Communication: Conformation state diagram of polypeptides: A chain length induced α-β transition. J Chem Phys. 2011;135(6):061101. doi: 10.1063/1.3624928. [DOI] [PubMed] [Google Scholar]
- 24.Pagel K, et al. Intramolecular charge interactions as a tool to control the coiled-coil-to-amyloid transformation. Chemistry. 2008;14(36):11442–11451. doi: 10.1002/chem.200801206. [DOI] [PubMed] [Google Scholar]
- 25.Pagel K, et al. Random coils, β-sheet ribbons, and α-helical fibers: One peptide adopting three different secondary structures at will. J Am Chem Soc. 2006;128(7):2196–2197. doi: 10.1021/ja057450h. [DOI] [PubMed] [Google Scholar]
- 26.Qin Z, Kreplak L, Buehler MJ. Hierarchical structure controls nanomechanical properties of vimentin intermediate filaments. PLoS One. 2009;4(10):e7294. doi: 10.1371/journal.pone.0007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephan JS, et al. The CPEB3 protein is a functional prion that interacts with the actin cytoskeleton. Cell Reports. 2015;11(11):1772–1785. doi: 10.1016/j.celrep.2015.04.060. [DOI] [PubMed] [Google Scholar]
- 28.Kar K, Jayaraman M, Sahoo B, Kodali R, Wetzel R. Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat Struct Mol Biol. 2011;18(3):328–336. doi: 10.1038/nsmb.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaton WA, Hofrichter J. Sickle cell hemoglobin polymerization. In: Anfinsen CB, Edsall JT, Richards FM, Eisenberg DS, editors. Advances in Protein Chemistry. Vol 40. Elsevier; Amsterdam: 1990. pp. 63–279. [DOI] [PubMed] [Google Scholar]
- 31.Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.