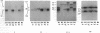Abstract
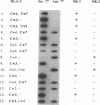
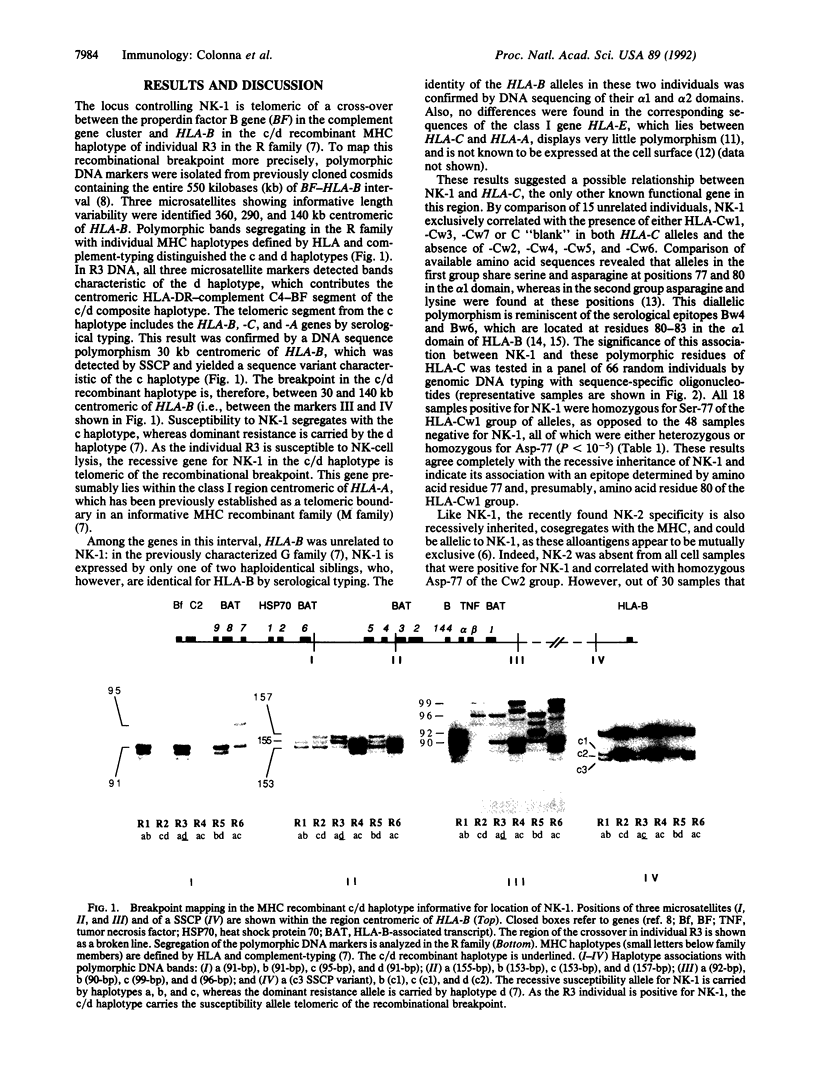
Human natural killer (NK) cells with the CD3- CD16+ phenotype recognize allospecificities on normal T-cell blasts. The NK-defined specificity 1 (NK-1) is recessively inherited and has been mapped to the major histocompatibility complex between the complement gene cluster and HLA-A. A gene for NK-1, however, has not been identified. Here we demonstrate that NK-1 and the recently defined NK specificity 2 (NK-2) are reciprocally associated with homozygosity for a diallelic polymorphism at amino acid positions 77 and 80 in the putative peptide-binding site of HLA-C (P less than 10(-5)). NK-cell recognition of allogeneic cells may, therefore, be controlled by HLA-C itself or by a closely linked gene(s), which dominantly prevents (resistance alleles) or recessively permits (susceptibility alleles) recognition of still-unknown target determinants.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bix M., Liao N. S., Zijlstra M., Loring J., Jaenisch R., Raulet D. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature. 1991 Jan 24;349(6307):329–331. doi: 10.1038/349329a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- Ciccone E., Colonna M., Viale O., Pende D., Di Donato C., Reinharz D., Amoroso A., Jeannet M., Guardiola J., Moretta A. Susceptibility or resistance to lysis by alloreactive natural killer cells is governed by a gene in the human major histocompatibility complex between BF and HLA-B. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9794–9797. doi: 10.1073/pnas.87.24.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone E., Pende D., Viale O., Di Donato C., Tripodi G., Orengo A. M., Guardiola J., Moretta A., Moretta L. Evidence of a natural killer (NK) cell repertoire for (allo) antigen recognition: definition of five distinct NK-determined allospecificities in humans. J Exp Med. 1992 Mar 1;175(3):709–718. doi: 10.1084/jem.175.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone E., Pende D., Viale O., Tambussi G., Ferrini S., Biassoni R., Longo A., Guardiola J., Moretta A., Moretta L. Specific recognition of human CD3-CD16+ natural killer cells requires the expression of an autosomic recessive gene on target cells. J Exp Med. 1990 Jul 1;172(1):47–52. doi: 10.1084/jem.172.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone E., Viale O., Pende D., Malnati M., Biassoni R., Melioli G., Moretta A., Long E. O., Moretta L. Specific lysis of allogeneic cells after activation of CD3- lymphocytes in mixed lymphocyte culture. J Exp Med. 1988 Dec 1;168(6):2403–2408. doi: 10.1084/jem.168.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudkowicz G., Bennett M. Peculiar immunobiology of bone marrow allografts. II. Rejection of parental grafts by resistant F 1 hybrid mice. J Exp Med. 1971 Dec 1;134(6):1513–1528. doi: 10.1084/jem.134.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberman R. B., Ortaldo J. R. Natural killer cells: their roles in defenses against disease. Science. 1981 Oct 2;214(4516):24–30. doi: 10.1126/science.7025208. [DOI] [PubMed] [Google Scholar]
- Höglund P., Ohlén C., Carbone E., Franksson L., Ljunggren H. G., Latour A., Koller B., Kärre K. Recognition of beta 2-microglobulin-negative (beta 2m-) T-cell blasts by natural killer cells from normal but not from beta 2m- mice: nonresponsiveness controlled by beta 2m- bone marrow in chimeric mice. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10332–10336. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren H. G., Kärre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990 Jul;11(7):237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- Ohya K., Kondo K., Mizuno S. Polymorphism in the human class I MHC locus HLA-E in Japanese. Immunogenetics. 1990;32(3):205–209. doi: 10.1007/BF02114975. [DOI] [PubMed] [Google Scholar]
- Rembecki R. M., Bennett M., Kumar V., Potter T. A. Expression of hemopoietic histocompatibility antigens on H-2-loss variants of F1 hybrid lymphoma cells: evidence consistent with trans gene regulation. J Immunol. 1987 Apr 15;138(8):2734–2738. [PubMed] [Google Scholar]
- Santos-Aguado J., Barbosa J. A., Biro P. A., Strominger J. L. Molecular characterization of serologic recognition sites in the human HLA-A2 molecule. J Immunol. 1988 Oct 15;141(8):2811–2818. [PubMed] [Google Scholar]
- Sargent C. A., Dunham I., Campbell R. D. Identification of multiple HTF-island associated genes in the human major histocompatibility complex class III region. EMBO J. 1989 Aug;8(8):2305–2312. doi: 10.1002/j.1460-2075.1989.tb08357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent C. A., Dunham I., Trowsdale J., Campbell R. D. Human major histocompatibility complex contains genes for the major heat shock protein HSP70. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1968–1972. doi: 10.1073/pnas.86.6.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Geraghty D. E., Koller B. H., Orr H. T., DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci U S A. 1988 Jan;85(1):227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies T., Bresnahan M., Strominger J. L. Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8955–8958. doi: 10.1073/pnas.86.22.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storkus W. J., Dawson J. R. Target structures involved in natural killing (NK): characteristics, distribution, and candidate molecules. Crit Rev Immunol. 1991;10(5):393–416. [PubMed] [Google Scholar]
- Storkus W. J., Salter R. D., Alexander J., Ward F. E., Ruiz R. E., Cresswell P., Dawson J. R. Class I-induced resistance to natural killing: identification of nonpermissive residues in HLA-A2. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):5989–5992. doi: 10.1073/pnas.88.14.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Bianchi E., Bass H., Suzuki T., Bender J., Pardi R., Brenner C. A., Larrick J. W., Engleman E. G. Natural killer lines and clones with apparent antigen specificity. J Exp Med. 1990 Aug 1;172(2):457–462. doi: 10.1084/jem.172.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan A. M., Ennis P., Parham P., Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. 1986 Dec 1;137(11):3671–3674. [PubMed] [Google Scholar]